W5 L12: Translation in Eukaryotes
L1:
The central Dogma
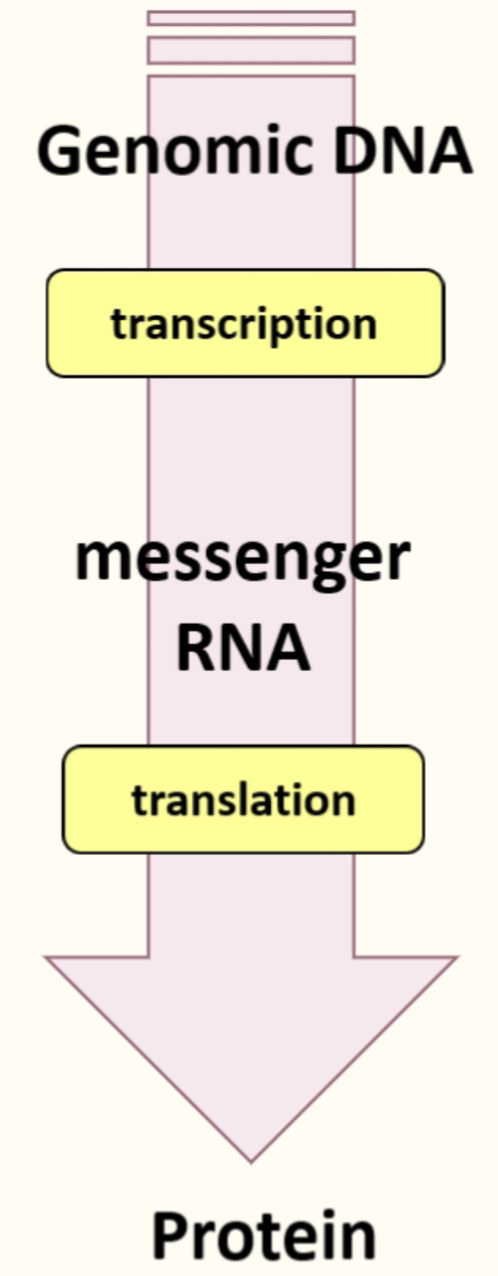
Coding region of mRNA specifies what protein is going to be produced
Ribosomes translate mRNA into proteins, translation factors promote this pathways & tRNA deliver the relevant AA in the right order
Amino acids are the raw materials used in translation
Uses ATP & GTP hydrolysis to provide E for process
Multiple surveillance pathways to make sure protein production & reading the code is done accurately - makes sure process only occurs when needed and when E is available
Ribosome composition
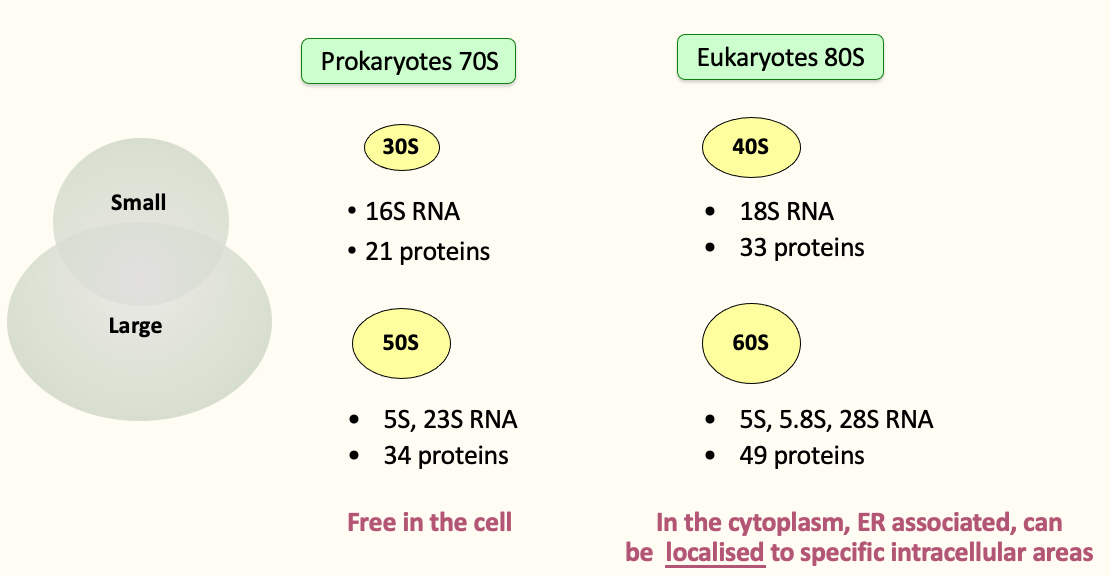
An RNA scaffold coated by proteins
Ribosome - History & background
1970s → biochemical investigation of translation
2000 → 1st high resolution structure of ribosome (x-ray) by Venki Ramakrishnan
2000 onwards → dozens of structures of various functional state of the ribosome - rationalisation of more than 30 yrs of biochemical findings
2009 → Nobel Prize (chem.) to Yonath, Steitz & Ramakrishnan for their structural investigations of the ribosome
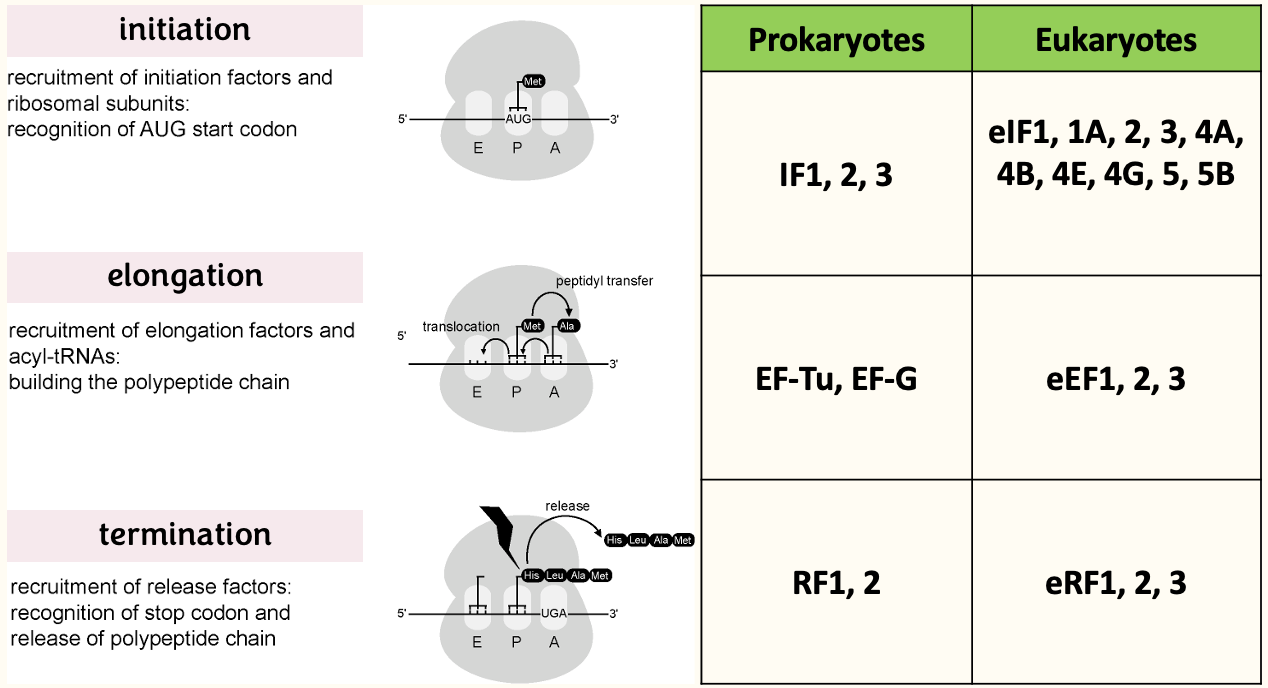
→ translation pathways deciphered (initiation-elongation termination)
→ antibiotics mode of action unravelled
Ribosomes
Many RP paralogs produced in higher eukaryotes
RPs can be post-translationally modified
Variable rRNA modification
Variability in composition → specialised ribosomes
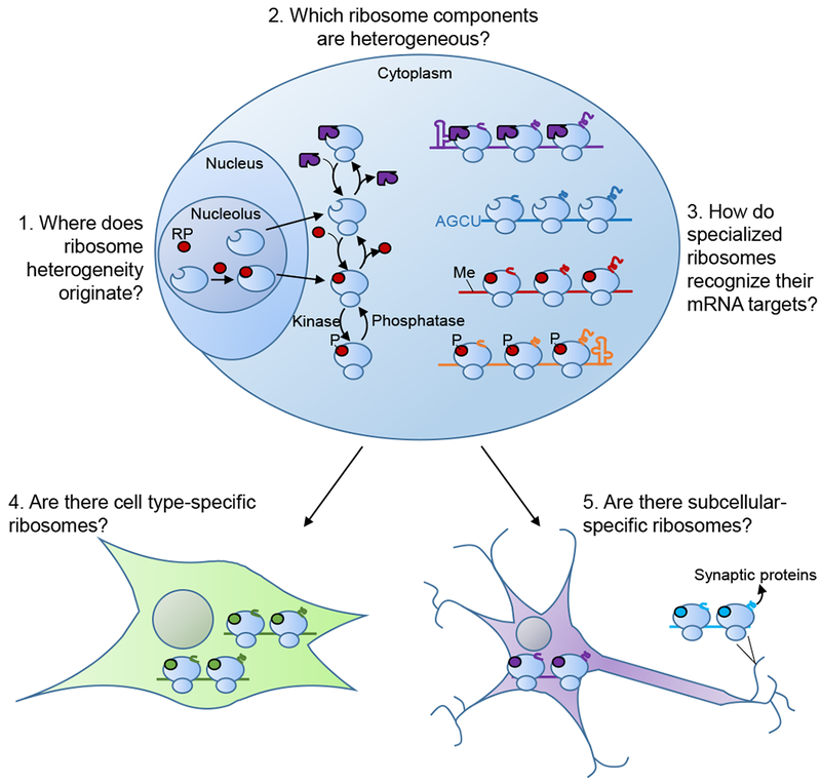
Ribosomes - protein synthesis in 3 steps
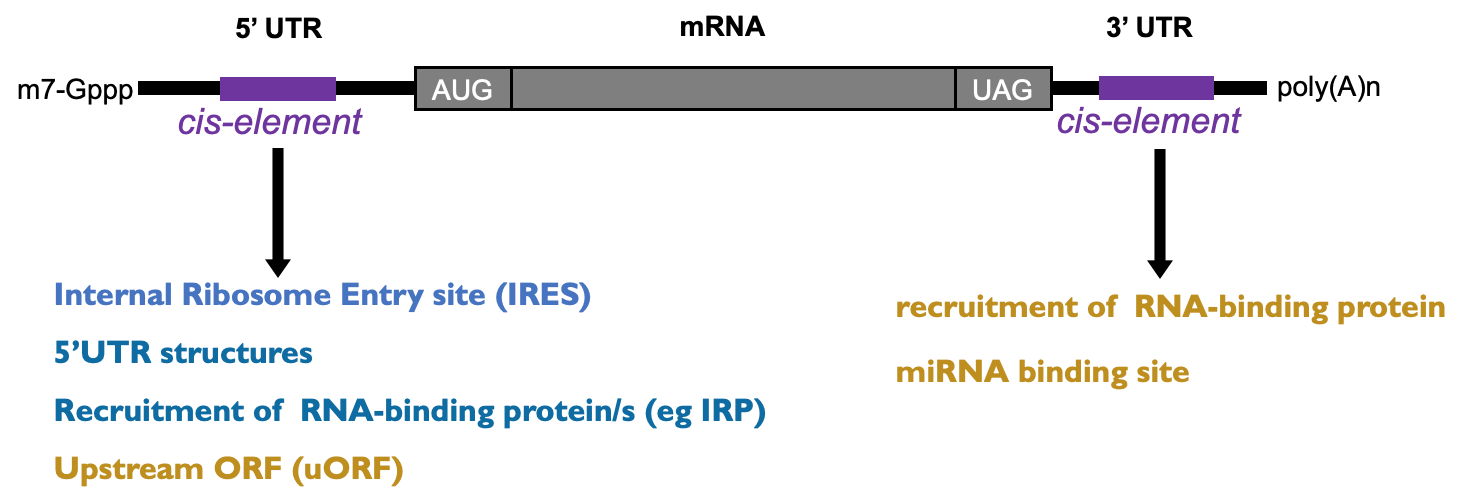
mRNA features that influence eukaryotic translation
Eukaryotic DNA capped at 5’ end w/ m7G cap → protective for degradation & crucial for translation
ORF contributes to protein code being produced - section from start codon to end codon
3’ poly(A) tail contributes to translational control
Most eukaryotic translation is cap dependant
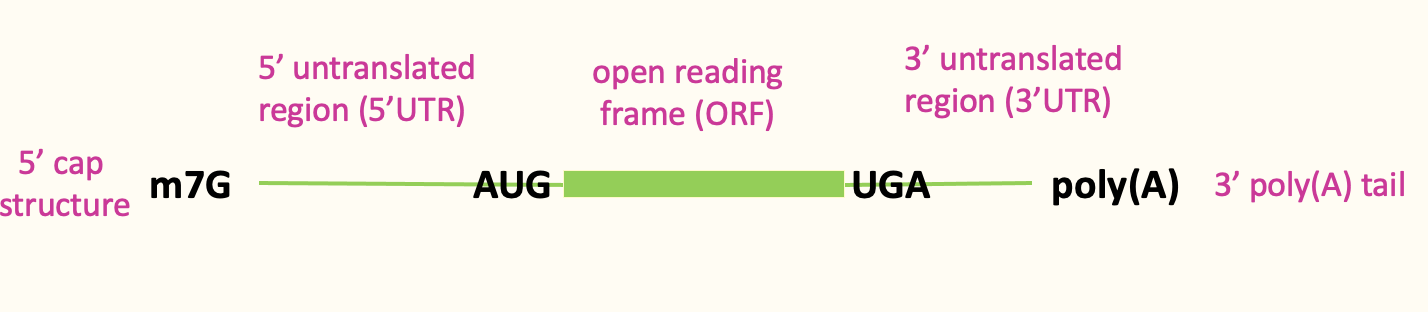
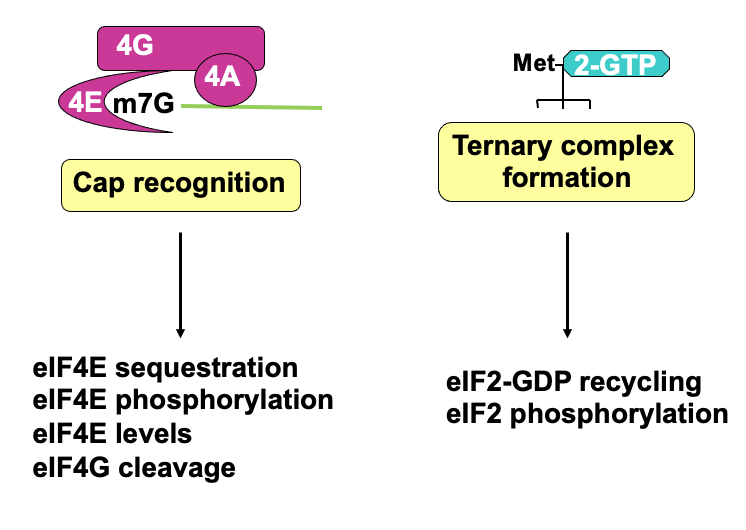
Translation initiation: cap binding
Cap binding complex (eIF4F complex) which helps signify the 5’ end
eIF4F complex → 3 diff. proteins that contribute to cap recognition
eIF4E → high affinity for the cap & binds it directly
eIF4G → bigger protein which binds 4E
eIF4A → associated w/ 4G & its an RNA helicase
Poly(A) tail on 3’ end - bound by poly(A)-binding protein which interacts w/ eIF4G & helps stabilise 4F complex & binding at the 5’ end
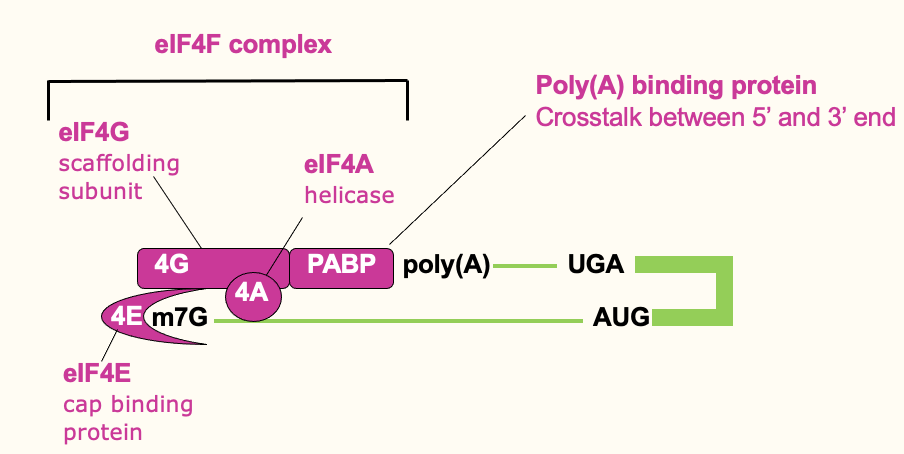
Translation initiation: ribosome recruitment
Small ribosome (40s) forms a pre-initiation complex w/ other initiation factors
eIF3 directly binds 4F & the ribosome → help bridge & bring that ribosome in
eIF1 & 1A have proofreading roles
eIF2 part of a Ternary complex which comes in & delivers initiator tRNA (tRNA loaded w/ initiator methionine) & comes in w/ eIF5 → provides GTPase activity (release E)
43S initiation complex brought in to 5’ prime end of mRNA through bridging interactions of eIF3
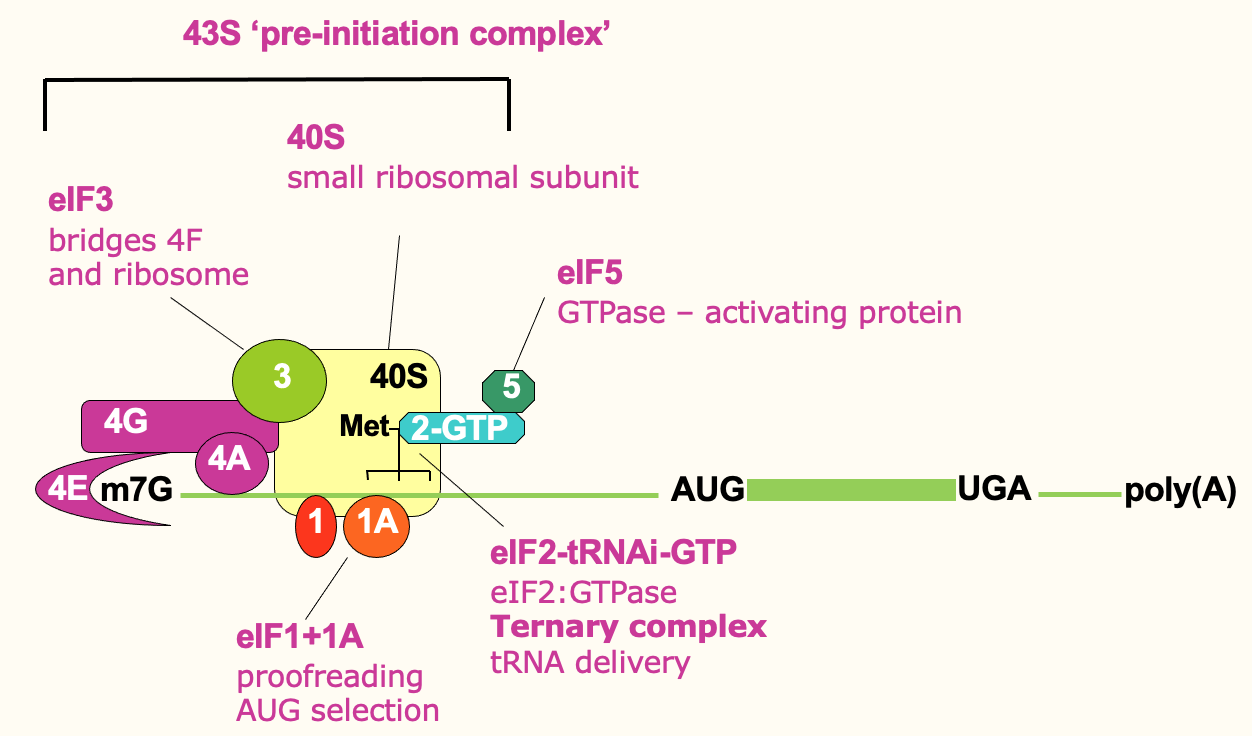
Translation initiation: scanning
To find the correct start codon, it scans along the mRNA, 5’ to 3’ until it finds AUG
eIF4A is helicase used to unwinds RNA structure to be scanned properly
Use E through ATP hydrolysis
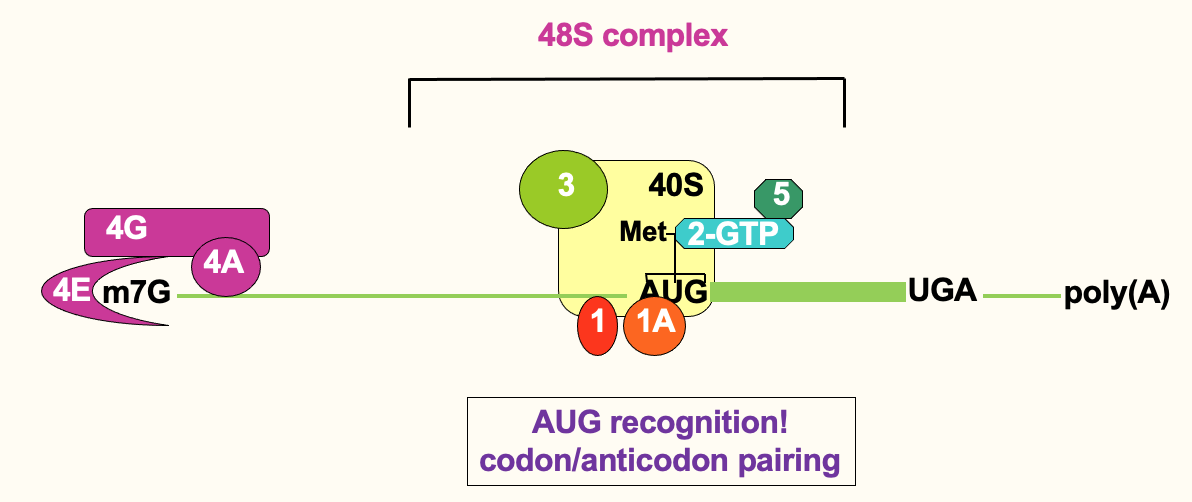
Recognition of AUG is evident through base pairing
anticodon of initiator tRNA base pairing w/ AUG codon
Once scanning ribosome has located AUG → 48S complex
Translation initiation: AUG recognition
This triggers release of eIF5 & eIF2 - using ATP hydrolysis
eIF1 & 1A - important for locating correct AUG → to avoid frame shifts
by sitting at correct interface b/w tRNA anticodon & mRNA
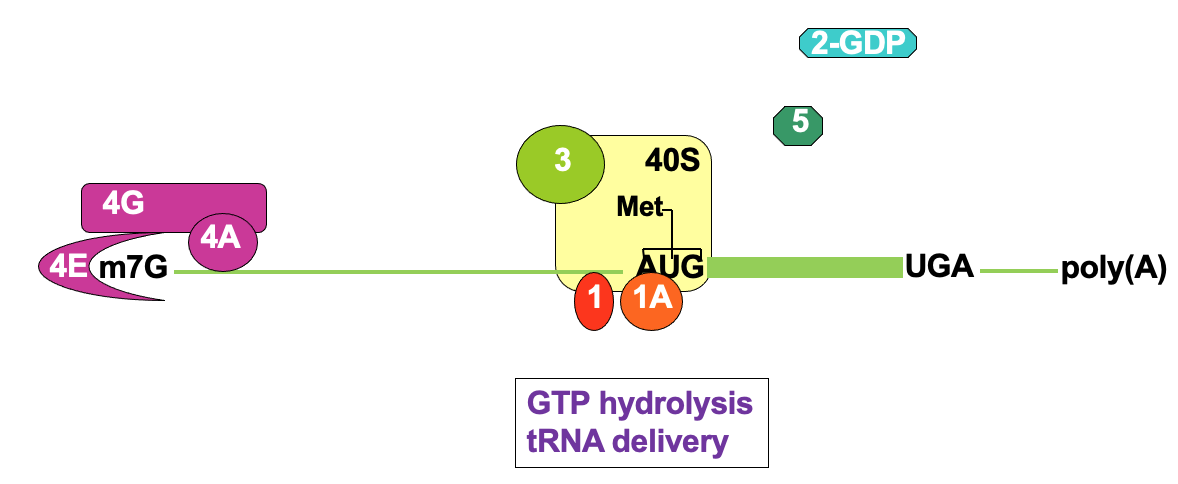
Translation initiation: 80S assembly
Joining of 60s large subunit w/ 40s→ facilitated by eIF5B
using GTP hydrolysis
Ejection of remaining initiation factors - not needed anymore
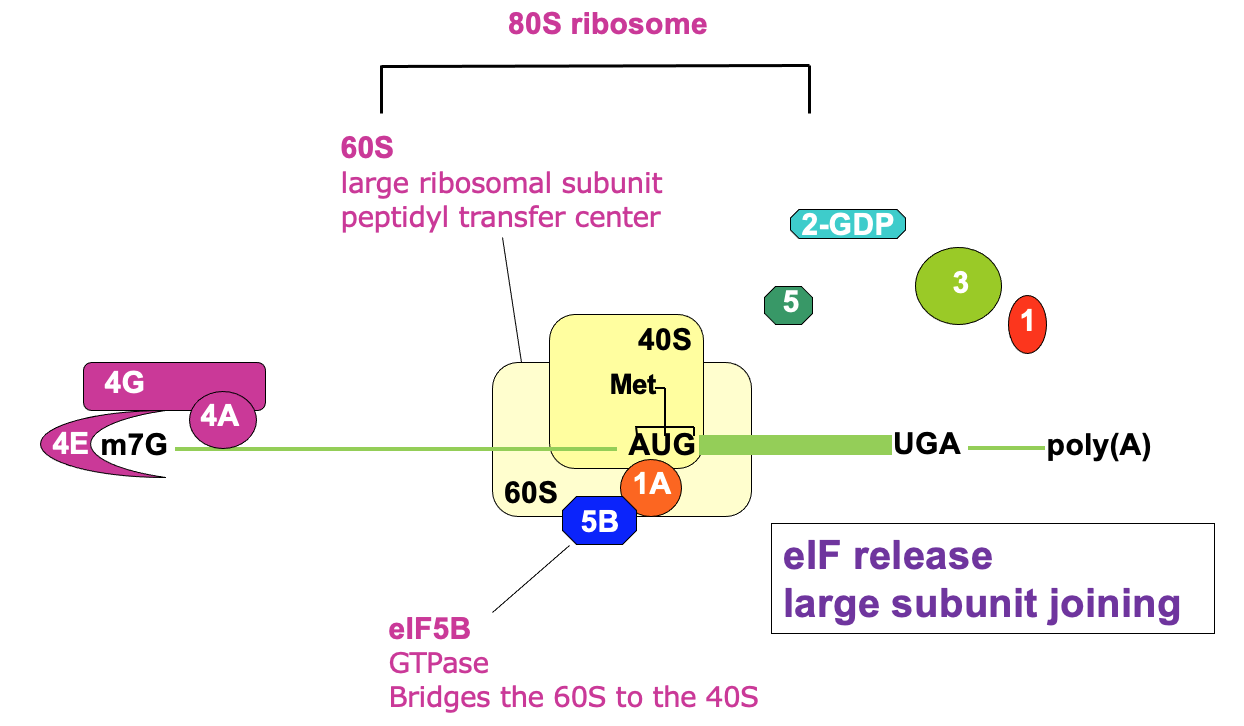
Translation initiation complete
80s ribosome also known as monosome → ready to elongate
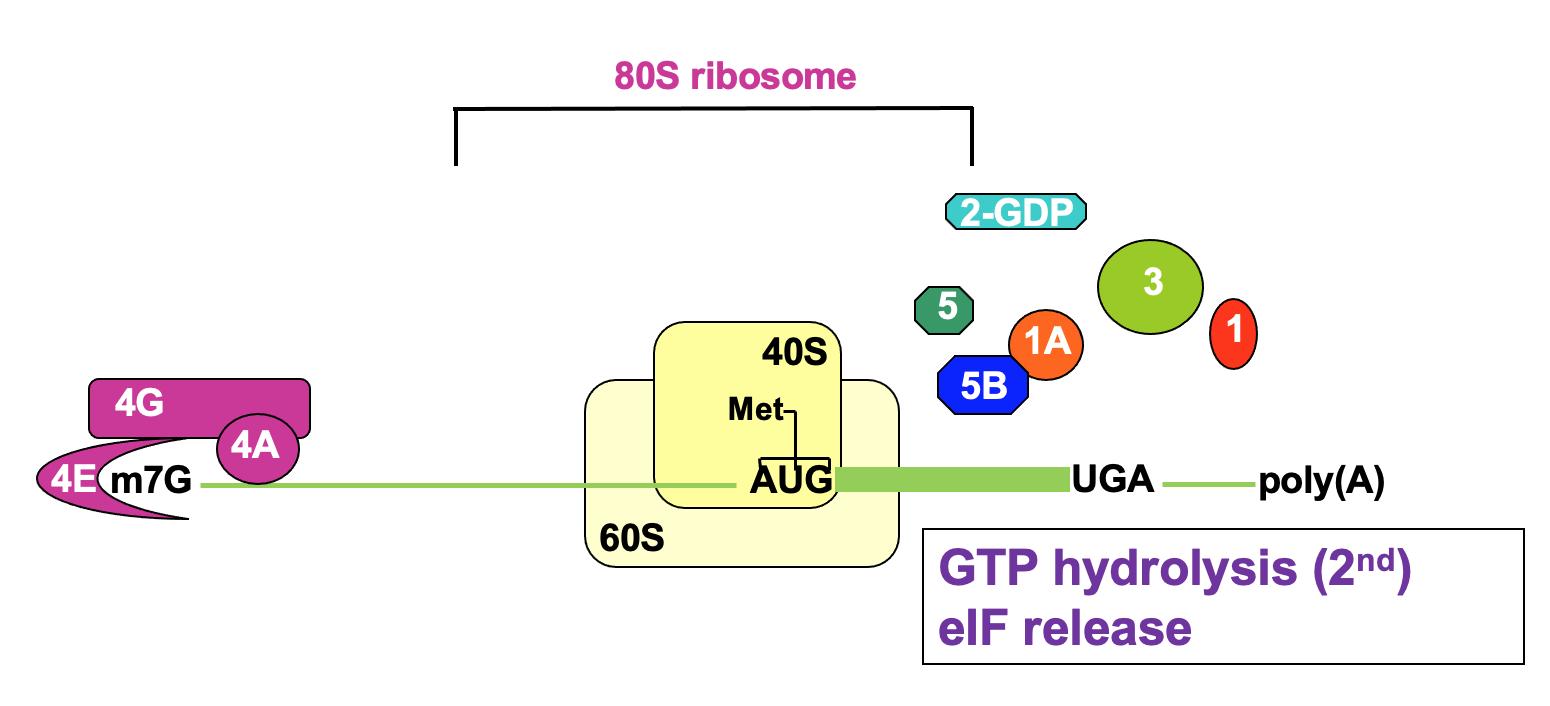
Comparison of eukaryotic & prokaryotic translation
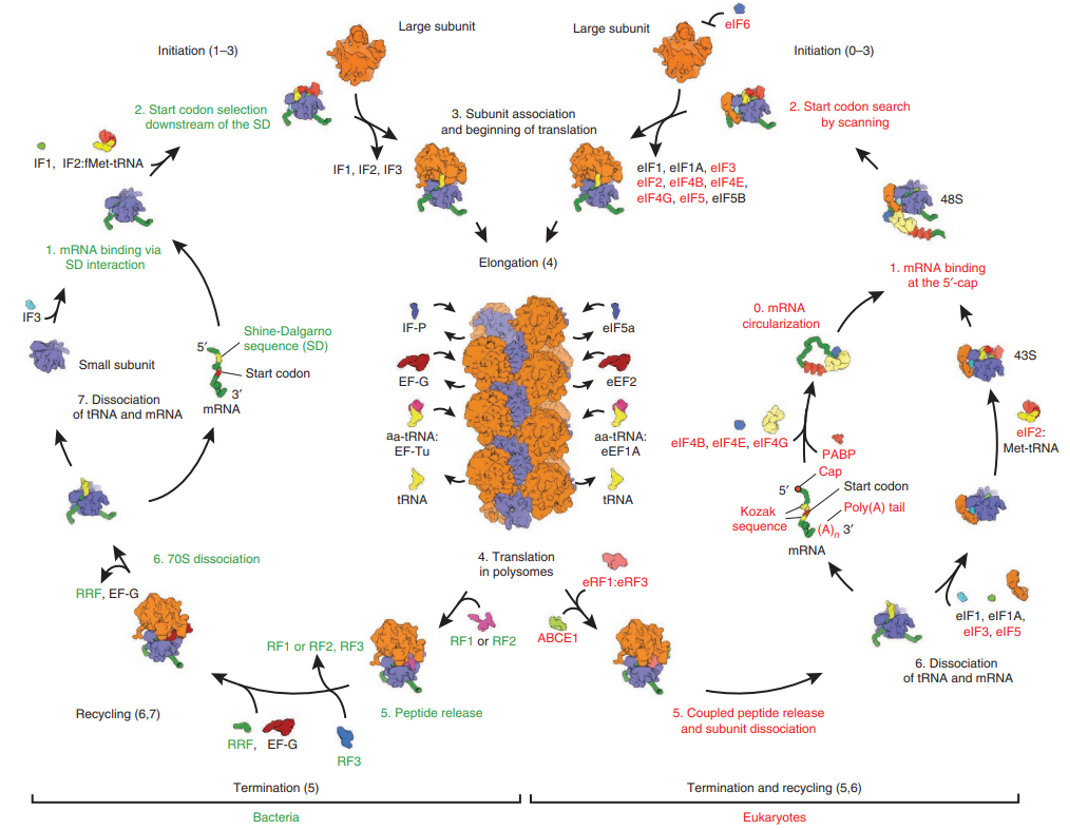
L2:
Translation control & disease
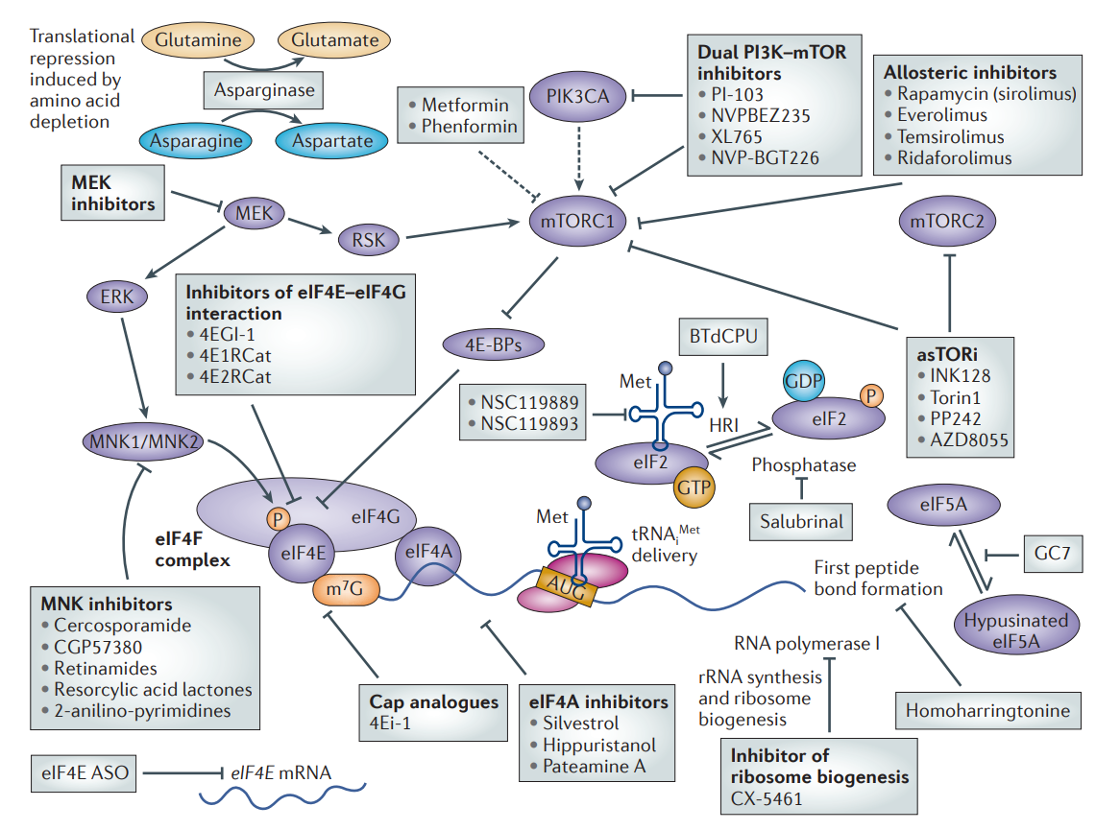
Rate limiting steps are key targets for regulation & dysregulation
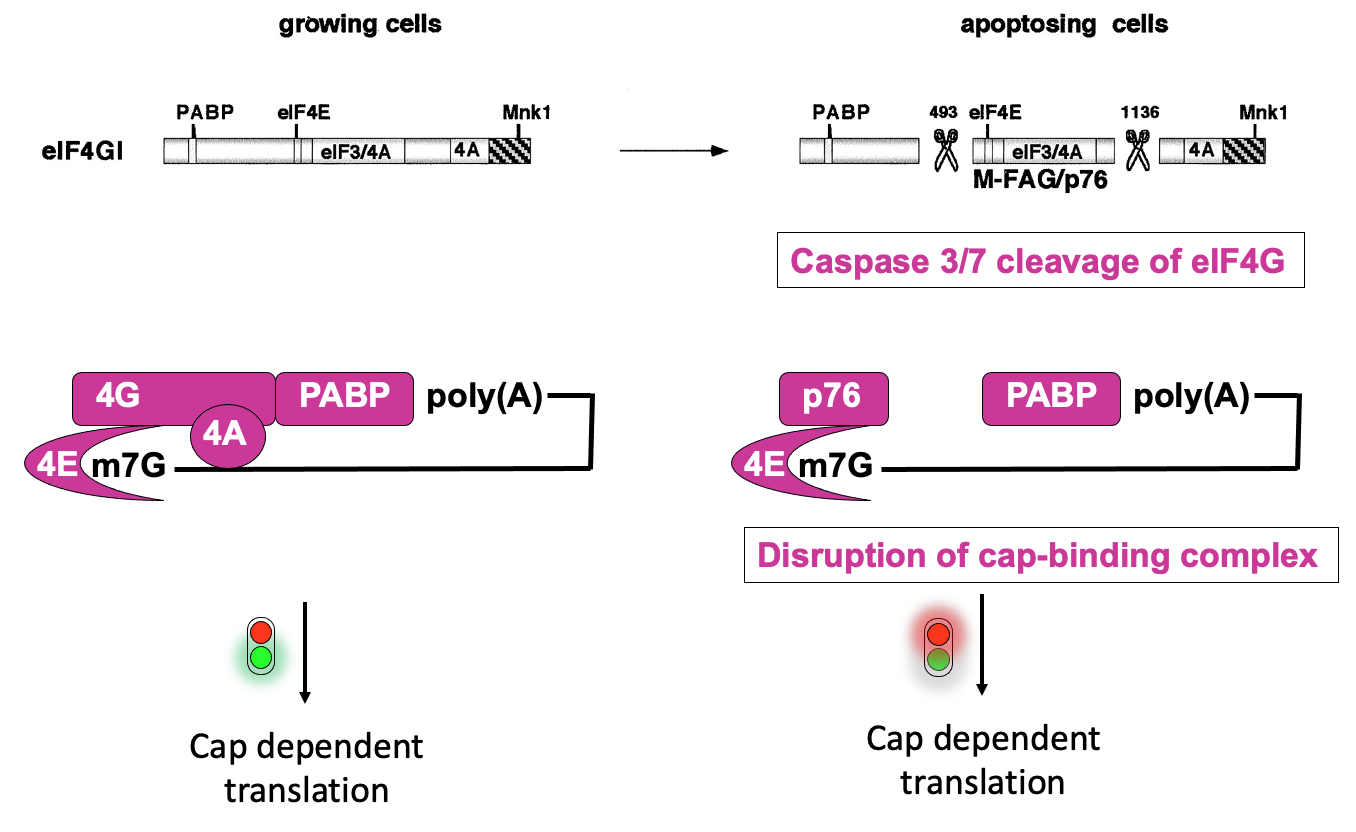
eIF4G structure & regulation during apoptosis
In normal cells, eIF4F complex helps to recruit 43s ribosome pre-initiation complex - through interactions that eIF4G has w/eIF3
eIF4F complex also interacts w/ poly(A)-binding protein which is bound at the 3’ end
communication b/w 5’ & 3’ helps stabilise the 4F complex association w/ mRNA & promote translation
When all interactions proceed, cap-dependant translation occurs
Apoptosis→ important in development - to get rid of cells that are no longer needed & in normal tissue homeostasis to eliminate infected cells
During apoptosis - there is activation of caspases (enzymes that can cut other proteins)
they proteolytic cleave eIF4G in 2 positions → cuts separate the parts of the proteins that interact w/ protein interactors that promote translation initiation
left w/ middle fragment (p76) that interacts w/ eIF4E, but not w/ poly(A)-binding protein & impaired in ability to promote translation initiation
∴ disruption of cap-binding complex activity & overall deregulation of translation in apoptosising cells
Some specific messages that are required to proceed w/ apoptosis, need to be produced
through alternate mechanism → cap-independent translation by internal ribosome entry site (IRES) - e.g. XIAP (inhibitor of apoptosis)
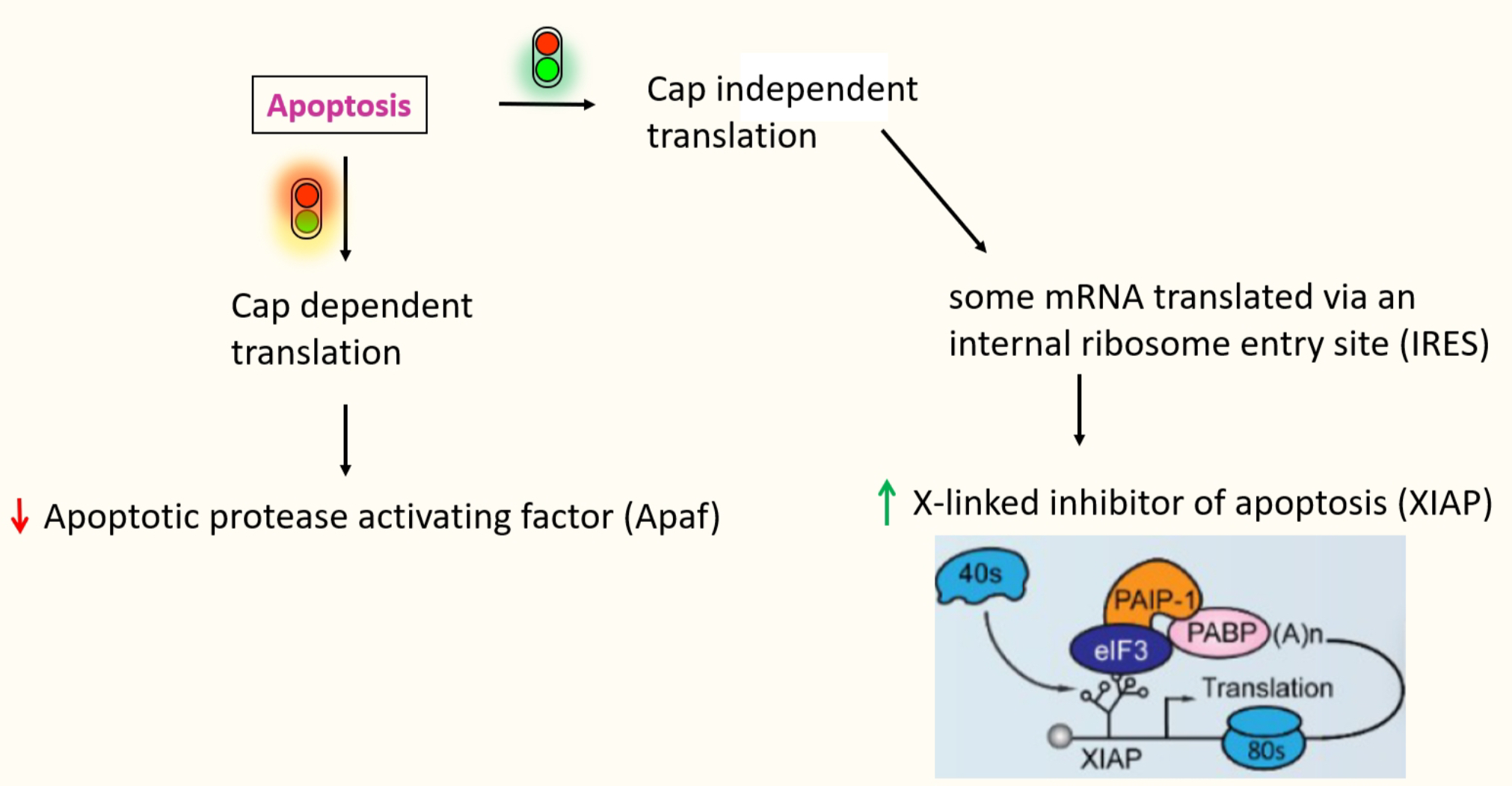
IRES dependant translation
IRES → structure RNA domain that recruits the ribosome
facilitates recruitment of a ribosome internally to the mRNA - rather than recruiting a ribosome directly to the 5’ cap
No need for the cap recognition & scanning - don’t need eIF4E binding for the cap
Require only few eIFs, specific subset varies w/ each IRES
Contains stem loops → highly stable & helps recruit in these proteins
e.g.XIAP → IRES recruits in eIF3 directly
to allow positioning of 40s ribosome directly onto the AUG
mediates 5’ -3’ communication the same way 4G does
Present in both cellular mRNAs & viral RNAs
evolved in viruses to produce their proteins even when the cell is apoptosing
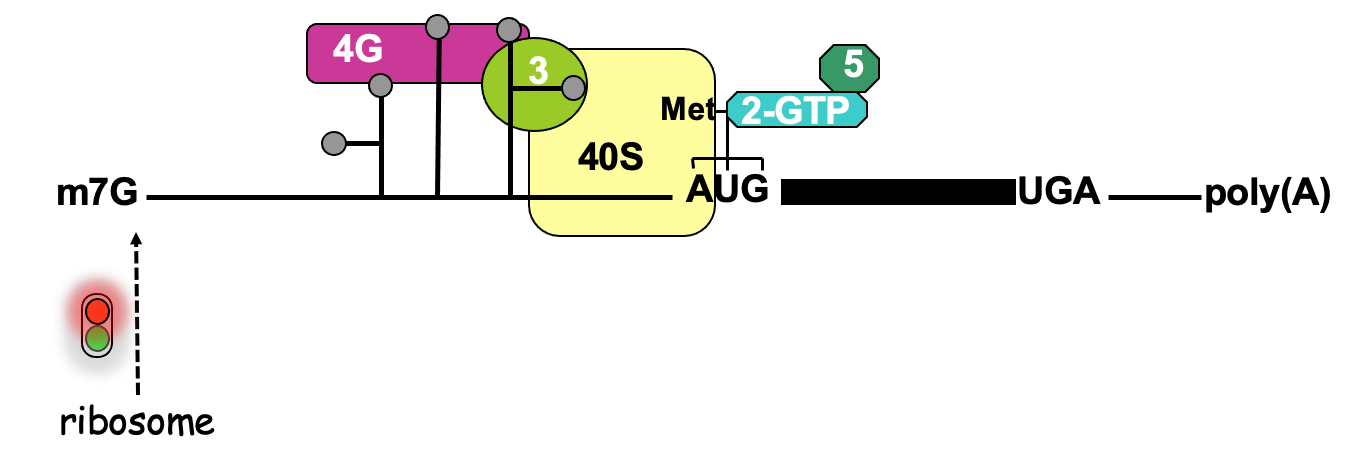
IRES dependant translation
An assay used to define the function of an IRES - in place of cistrons - fluorescent proteins of diff . colours
2 cistrons (open reading frames that code for reporter proteins) are added to an mRNA
Positioning 2 open reading frames next to each other in mRNA that’s capped
In eukaryotic cell via cap-dependant translation
1st cistron will be translated to produce the product & at end of translation, ribosome will terminate & ejected from mRNA protein being made
no translation of downstream 2nd cistron
To test whether a seq. identified in RNA is functioning as an IRES - the sequence is put b/w 2 cistrons
1st cistron - translated in cap-dependant manner → after cap binding complex has recruited the ribosome at cap & allowed 1st cistron to be translated
direct recruitment of a ribosome to IRES seq. - if functioning will allow production of 2nd protein → finds out whether seq. can function as IRES & what the requirements are e.g. what eIFs are important for functioning of IRES
Cap-independent translation → no translation of either cistrons
Highly stable stem loop added to 5’ UTR → prevents unwinding activity of eIF4A → ∴ no translation from 1st cistron but effective translation from 2nd cistron using IRES
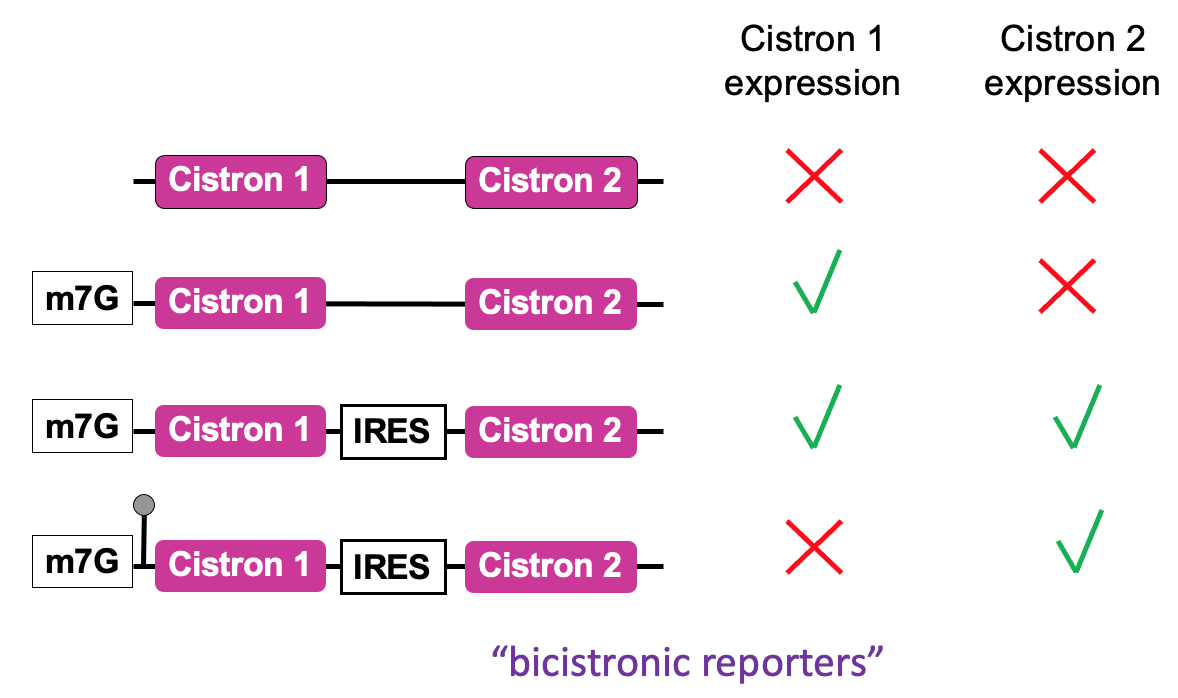
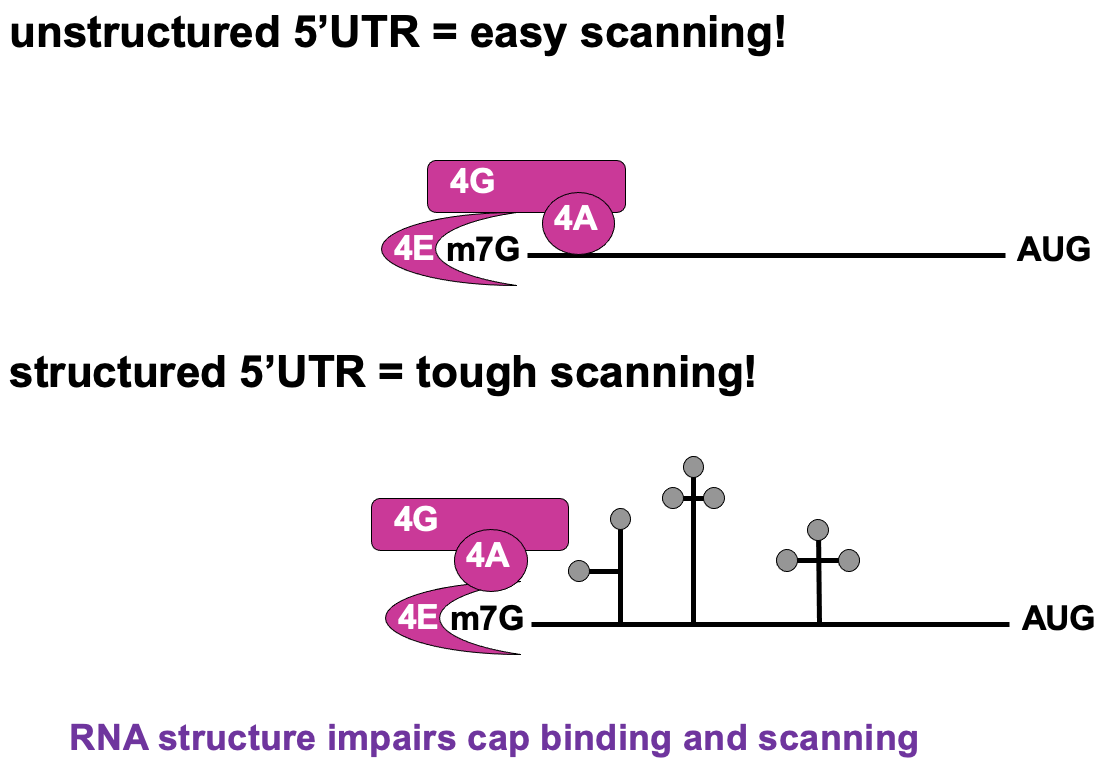
Regulation of cap recognition
Unstructured 5’UTR → less dependant on 4A activity ∴ not using as much E
Structured 5’UTR → e.g. stem loops present - require lots of E from 4F complex to unwind & allow translation to occur → poorly translated in cap-dependant translation
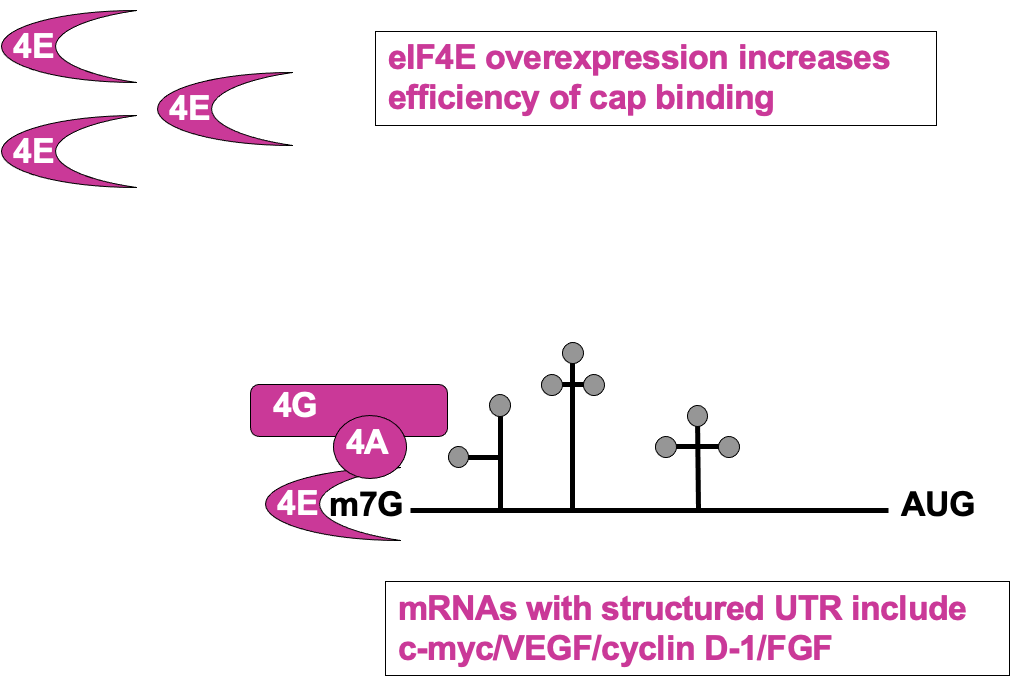
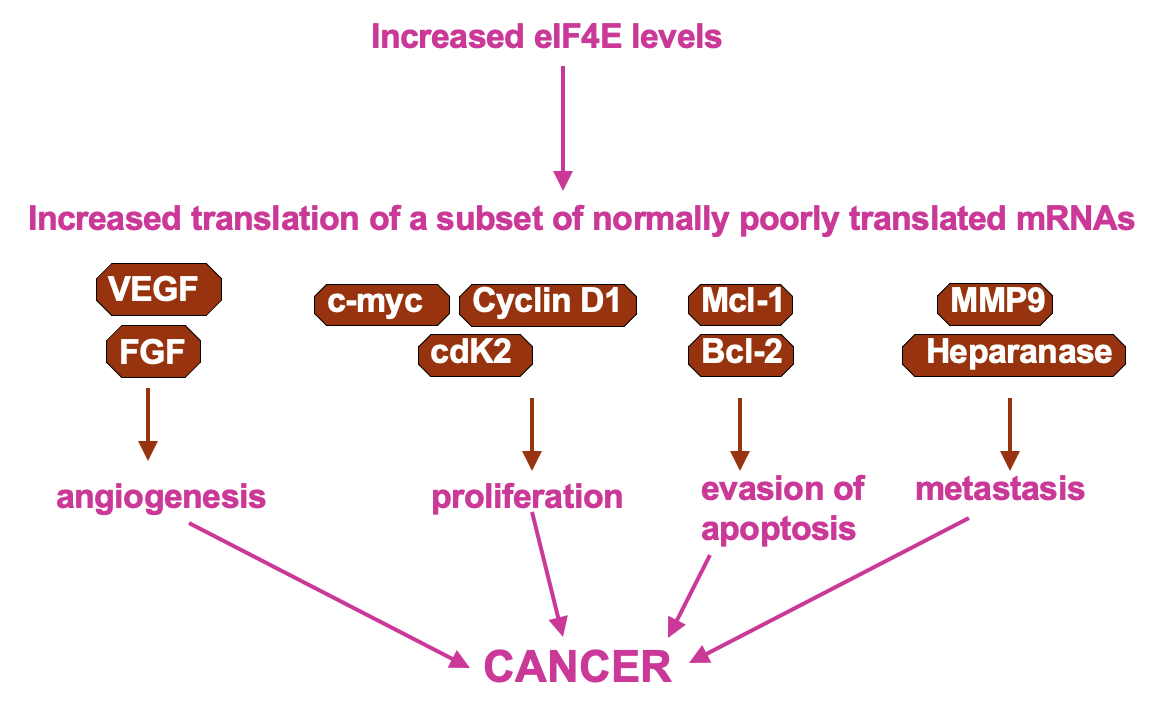
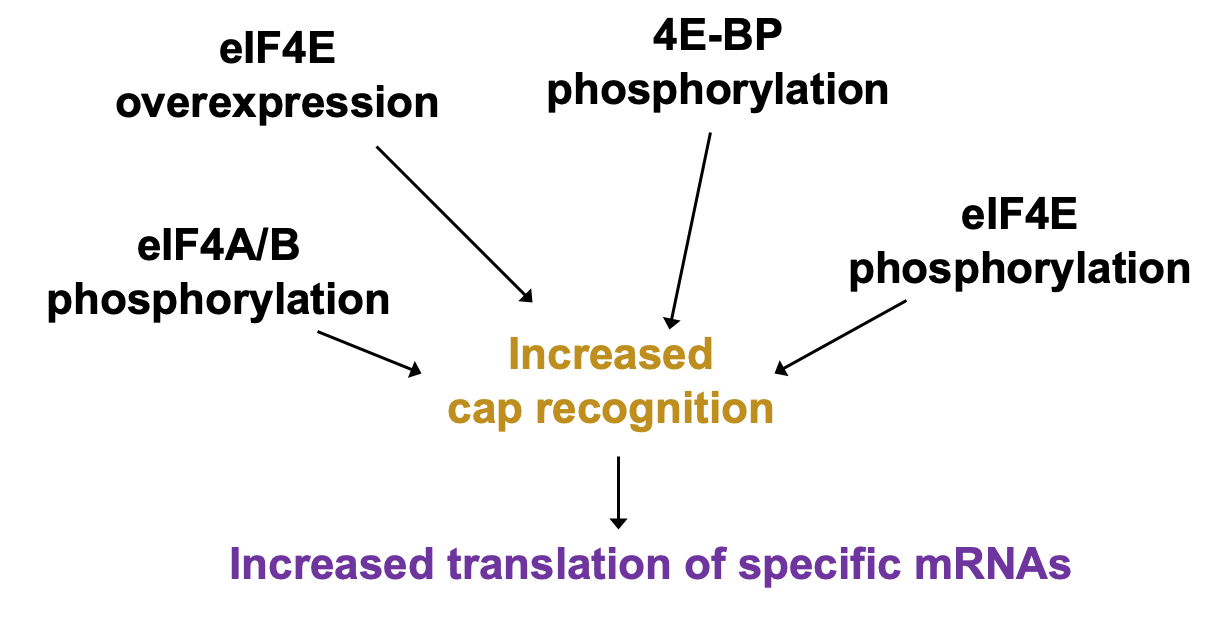
Regulation of cap recognition: eIF4E levels
UTR containing mRNAs disproportionately benefit when 4E levels are higher
higher 4E levels → help translate poorly translated mRNAs
pro-proliferative factors e.g Cyclin D1 & C-myc contain these - upregulation of their expression may cause uncontrolled growth of the cells
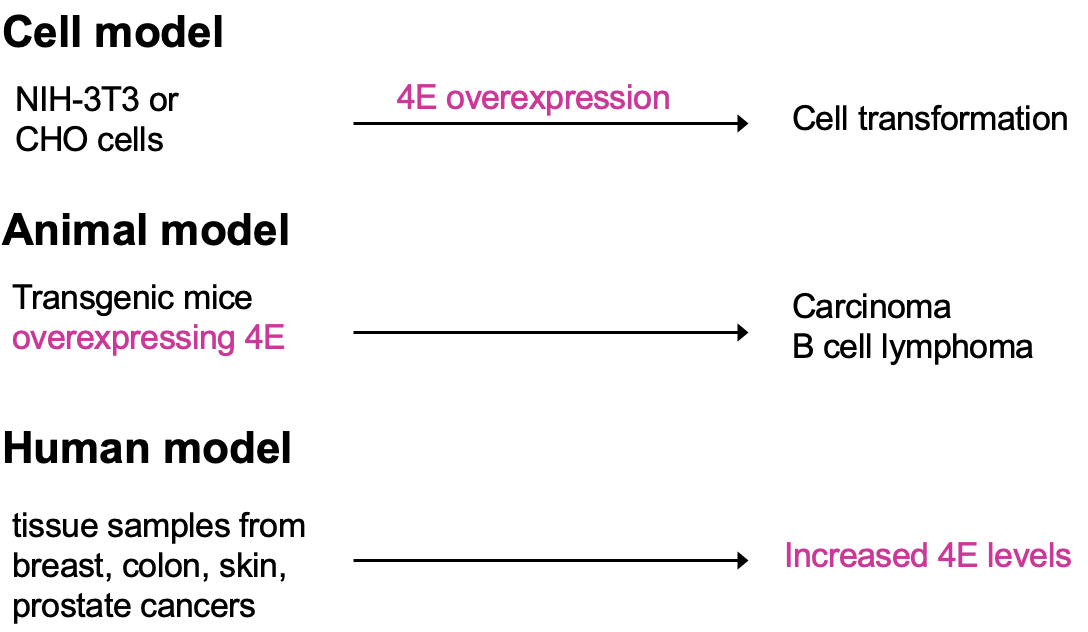
Cultured cells that have overexpressed eIF4E observe cell transformation (transformation to phenotype resembling cancer)
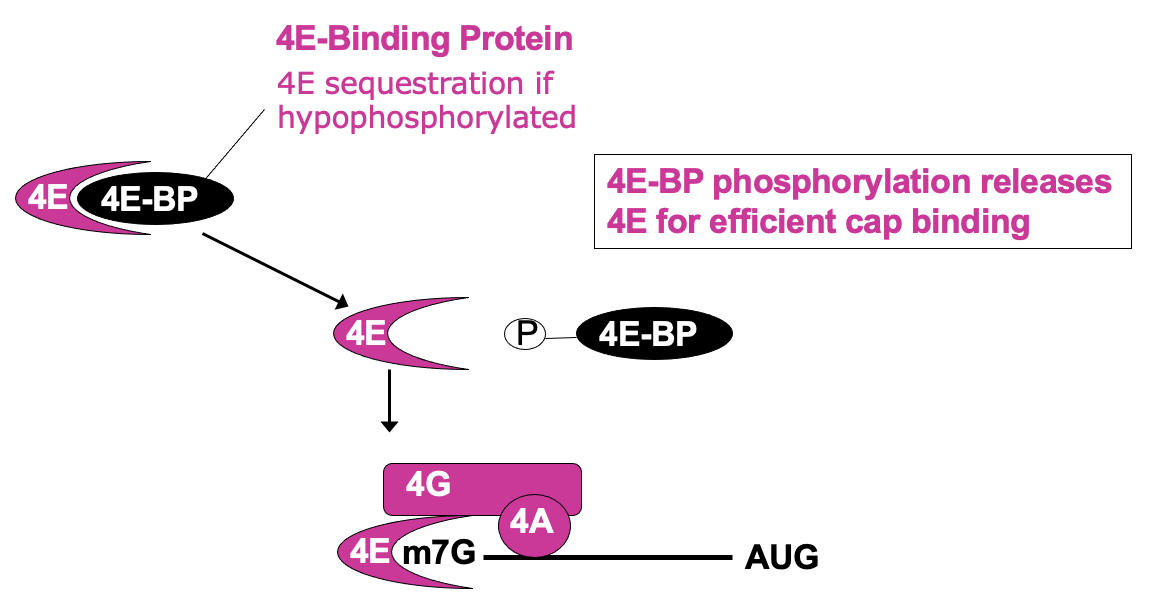
Regulation of cap recognition: eIF4E sequestration
Δ availability of eIF4E in cell can be regulated in its activity
4E bound by eIF4E binding protein (4E-BP) - 4e no longer able to bind cap & participate in translation initiation
4e-BP phosphorylation releases 4E for efficient cap binding
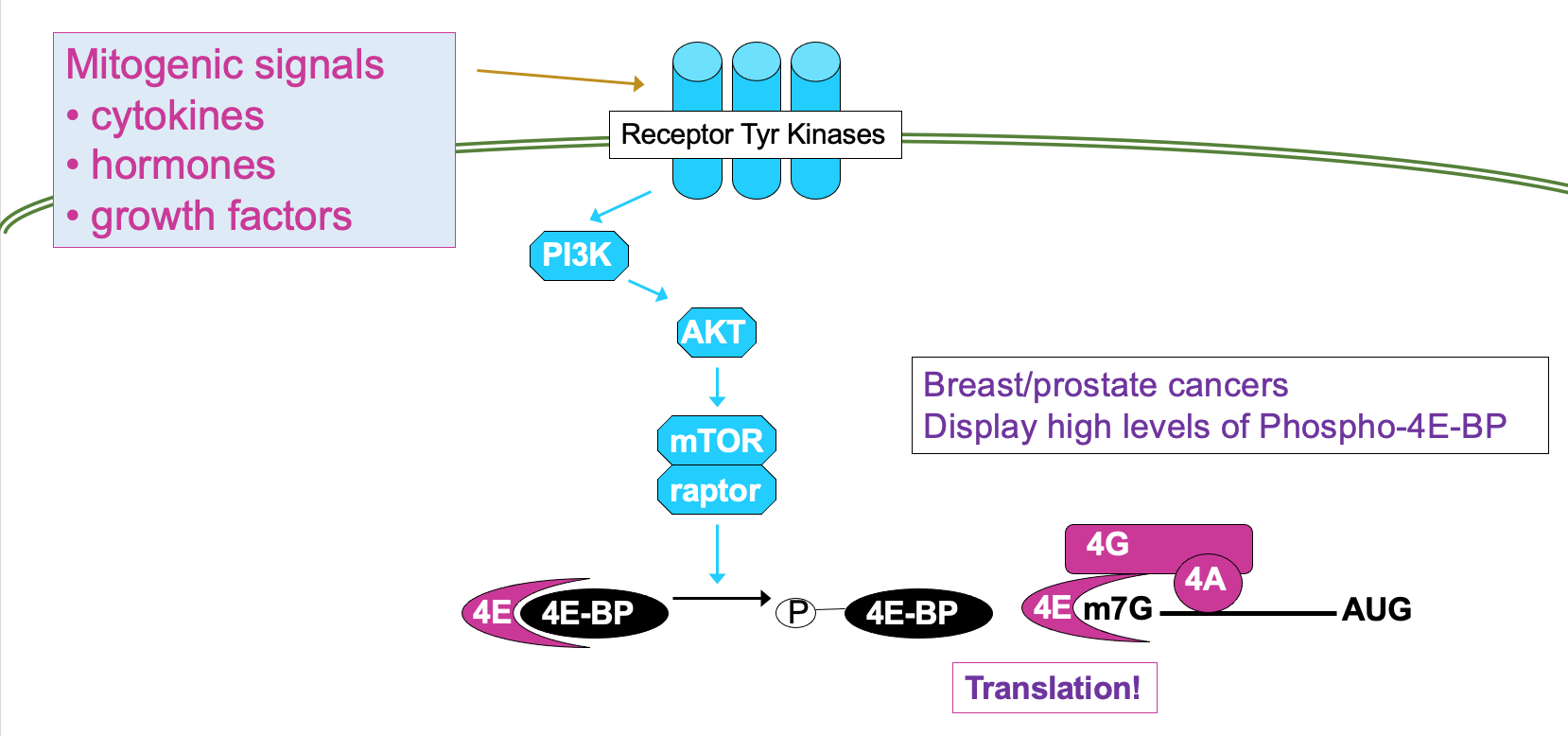
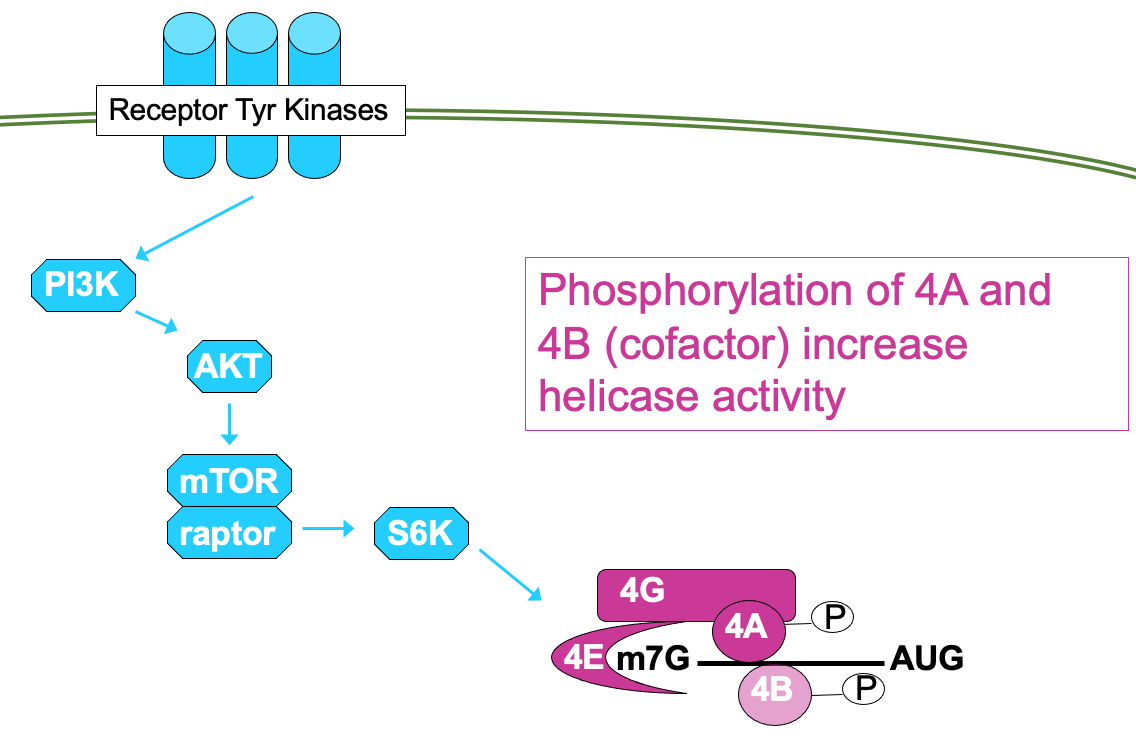
This is regulated through signalling cascade - mTOR signalling pathway →
processes mitogenic signals from cytokines, hormones etc. that are interpreted by receptor Tyr kinases
through cascade of phosphorylation, culminating in activation of mTOR → results in phosphorylation of 4E-BP activating translation
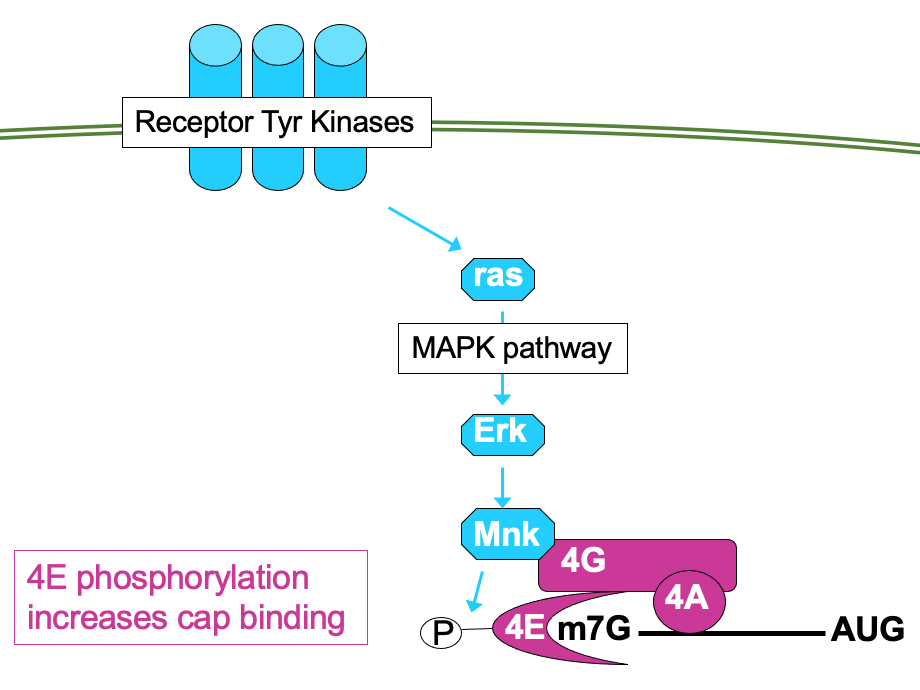
4E can be directly phosphorylated by kinase Mnk → ↑ affinity for cap so functions better
Mnk activated through MAP kinase pathway - also activated by mitogenic signals
Signalling pathways can impact the phosphorylation activity of other eIFs e.g.
mTOR pathway can also lead to phosphorylation of eIF4A - helicase in eIF4F translation initiation complex & phosphorylation of cofactor 4B
phosphorylation of both factors is associated w/ ↑ activity
Targeting translation as cancer therapy
Translational control & disease
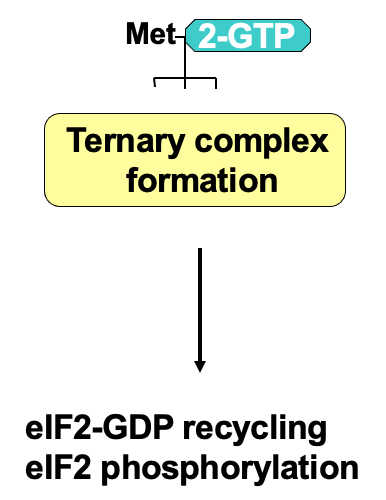
Regulation of ternary complex formation
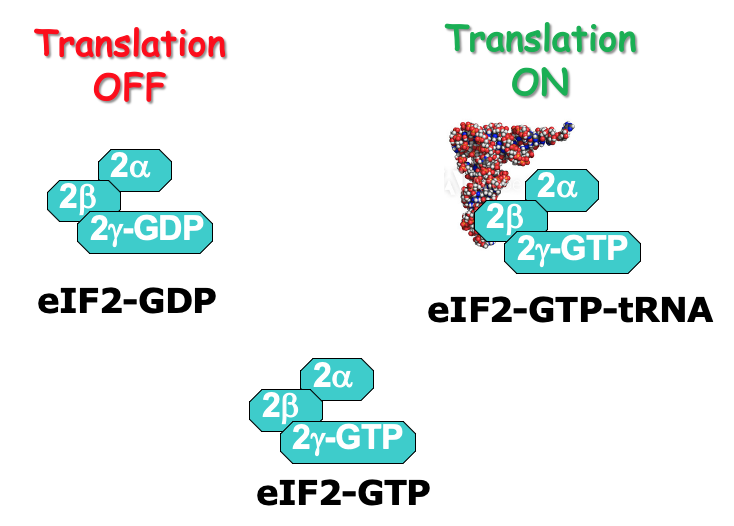
At the end of translation, eIF2 associates w/ GDP due to GTP hydrolysis during initiation
eIF2 has 3 subunits: α, β, γ
γ is bound to GTP or GDP
GTP required for translation initiation - associates w/ initiator tRNA & the ternary complex can participate in translation
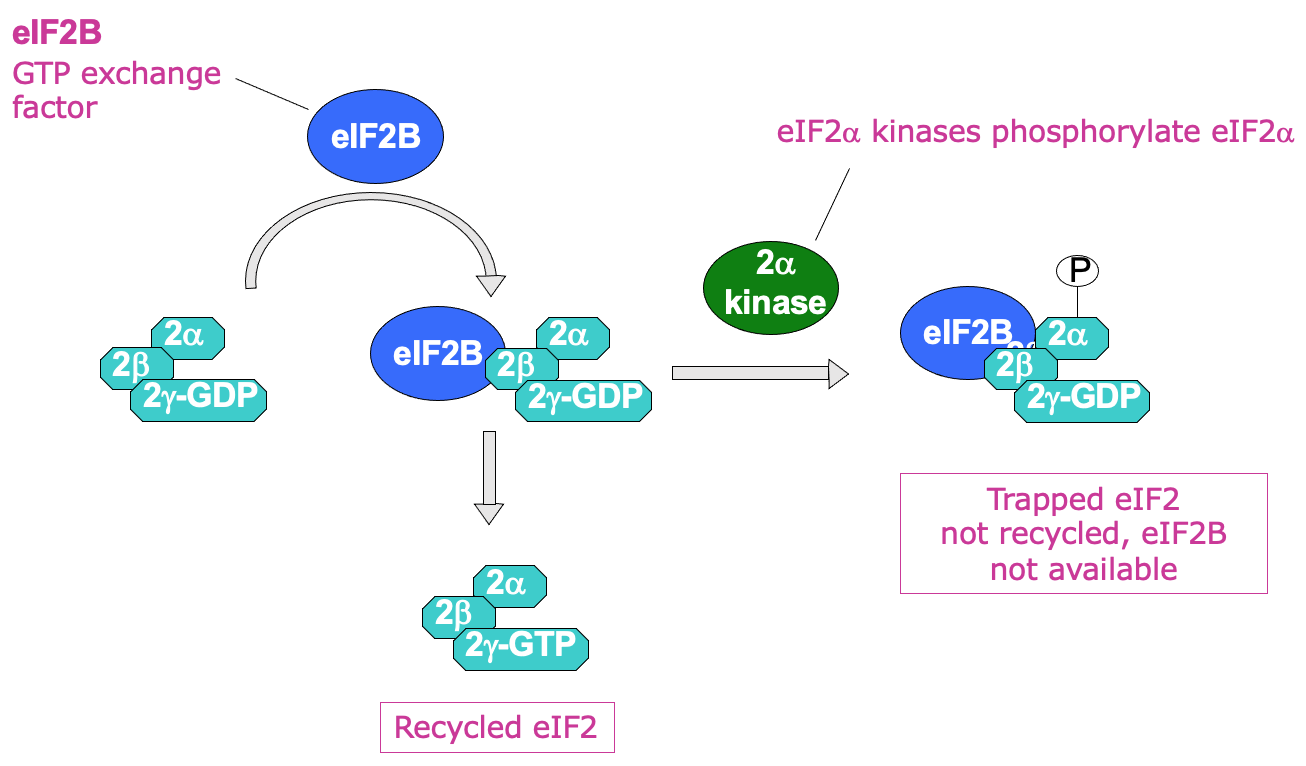
eIF2 recycling → eIF2B (GTP exchange factor) required to help promote transfer of GDP to become GTP
by directly binding eIF2
Process of recycling can be regulated by phosphorylation of eIF2 α subunit using eIF2 α kinases
when phosphorylated GDP bound eIF2 can’t be recycled into GTP
Phosphorylated eIF2 binds eIF2B v/ tightly → issue due to less eIF2B in the cell than eIF2
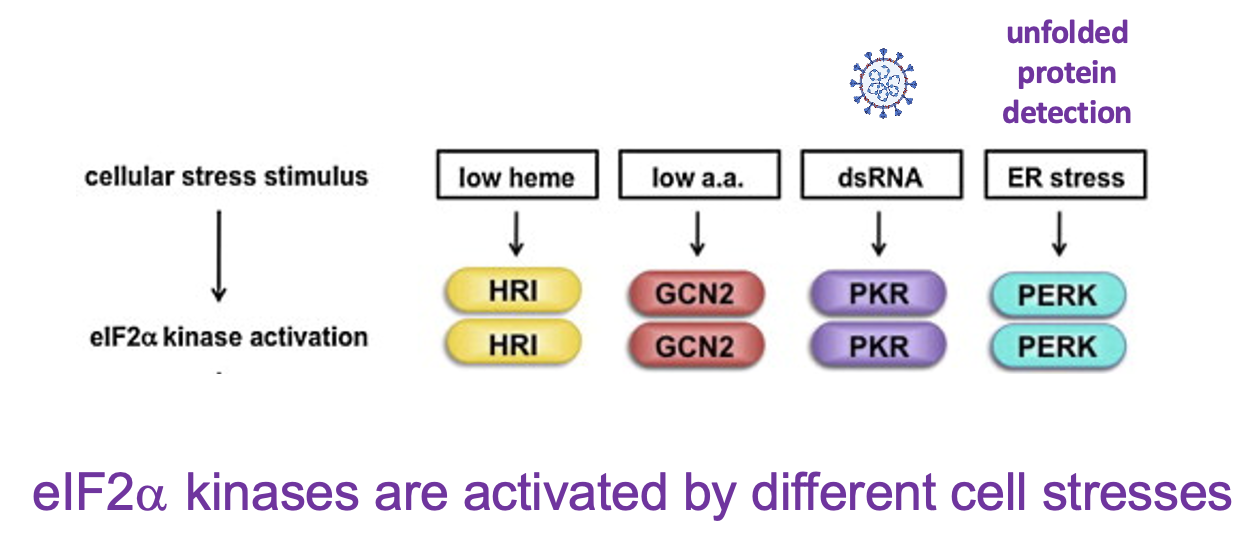
4 diff. types of eIF2 alpha kinases in mammalian cells → each processes diff. stress responses:
HRI → sense low haem levels & inhibits translation of proteins to reduce production of globins
GCN2 → sense low amino acid levels
PKR → activated by dsRNA → hallmark of virus infection
PERK → senses ER stress - response to presence of unfolded proteins in ER
Q: why shut off translation when cells are stressed?
turn off translation when AA are low
prevents the wrong AA from being translated
prevents E from being wasted on wrong resources
turn off translation if cells infected w/ virus
viruses that infect cells contain own RNA/DNA or enzymes → so stops virus infection by preventing viral cells from using host ribosomes & stopping protein synthesis of viral proteins
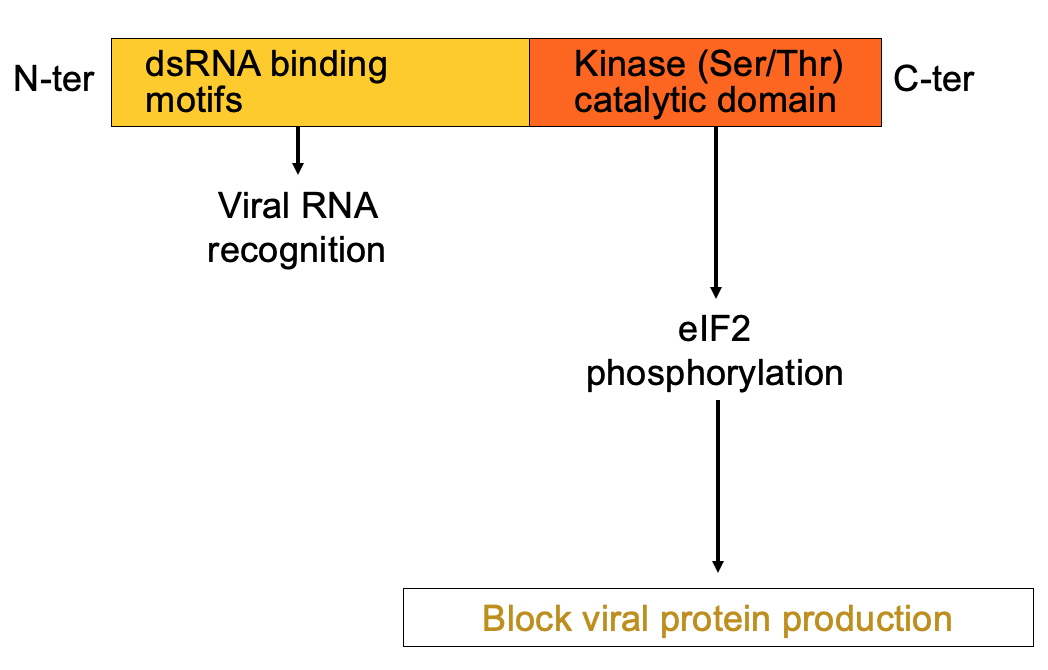
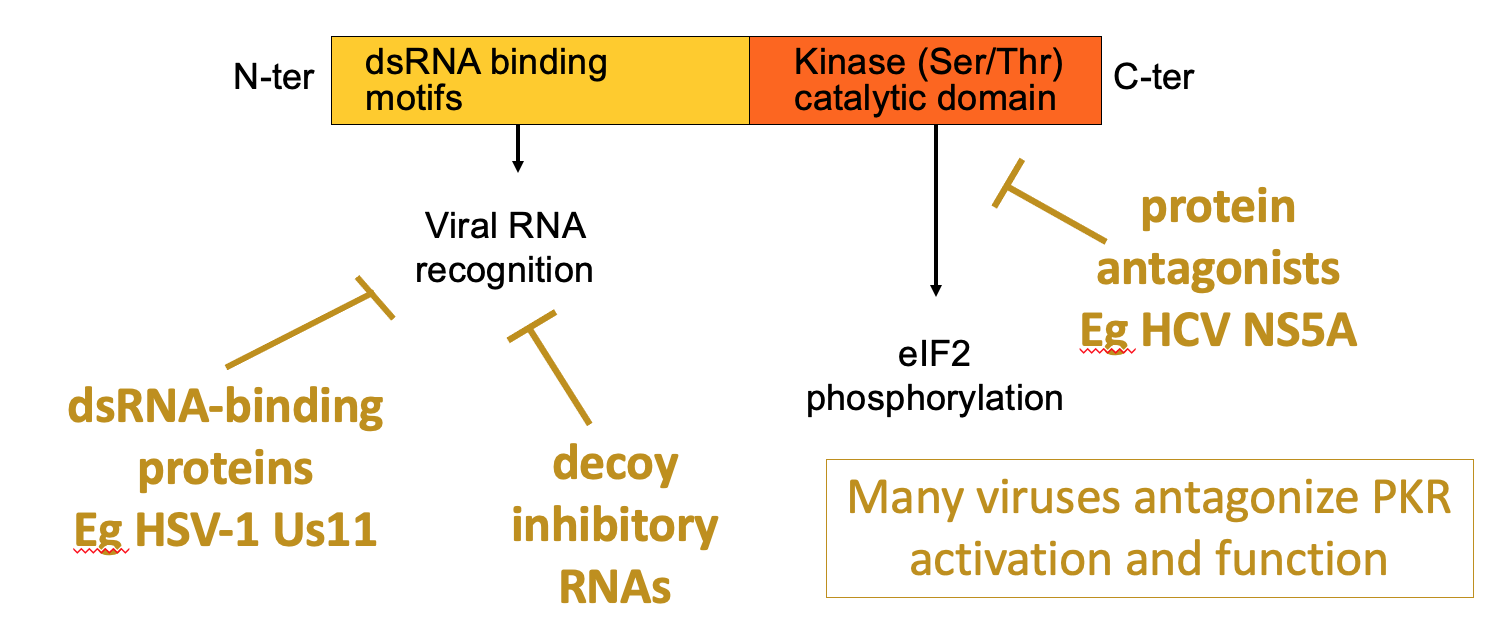
PKR structure, function & regulation
dsRNA is hallmarker for viral infection
PKR has 1 domain that helps bind dsRNA & kinase domain at C terminus that’s activated when PKR binds the dsRNA → leads to eIF2 alpha phosphorylation → blocks viral protein production
Viruses have evolved ways to antagonise PKR
e.g. viruses e.g. HSV1 Us11 - produce dsRNA binding proteins - shield recognition of dsRNA
some viruses may decoy RNAs which are bound by PKR but don’t activate it
HCV NS5A - make antagonists of kinase activity of PKR - so they directly stop activity or kinase domain
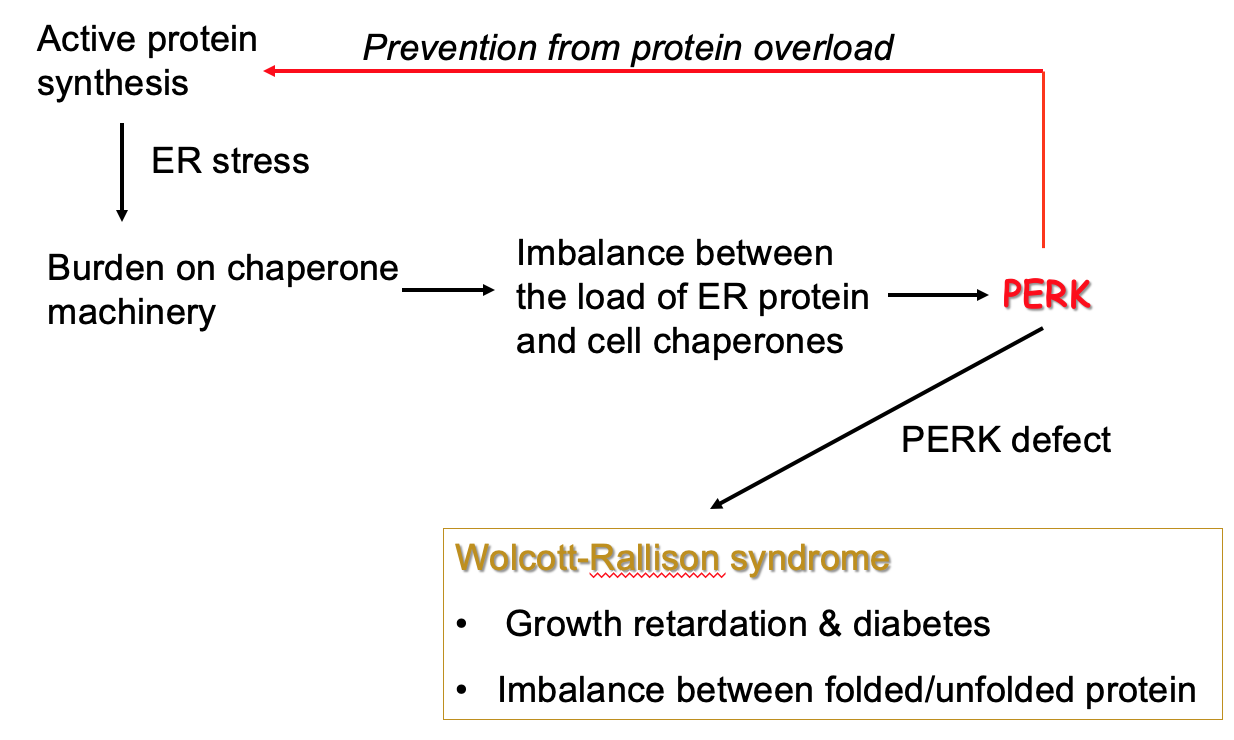
PKR-like ER localised kinase (PERK)
Sense unfolded protein buildup on ER → burden on machinery that folds proteins into active form & causes imbalance
Unfolded proteins may be toxic - need to stop build up
PERK inhibits protein synthesis so that backlog is reduced
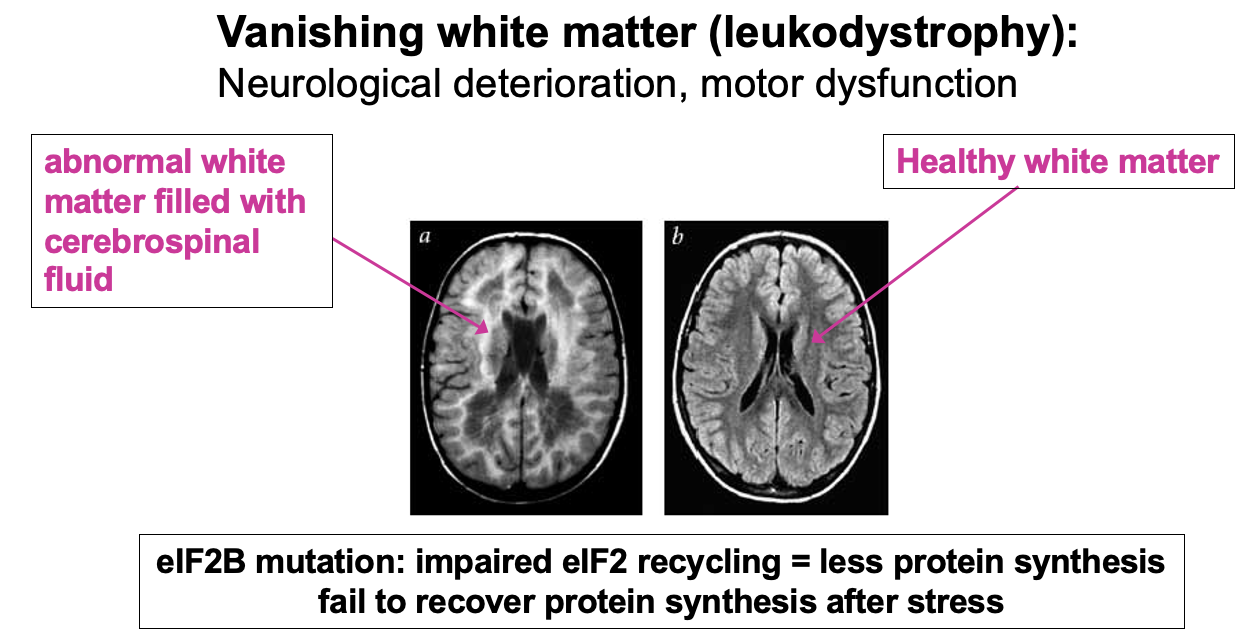
eIF2B mutations cause disease
Example of failure to fully recycle eIF2 & maintain normal levels of ternary complex is in the disease Vanishing white matter (leukodystrophy)re
specifically affects myelinated cells (white matter)
L3:
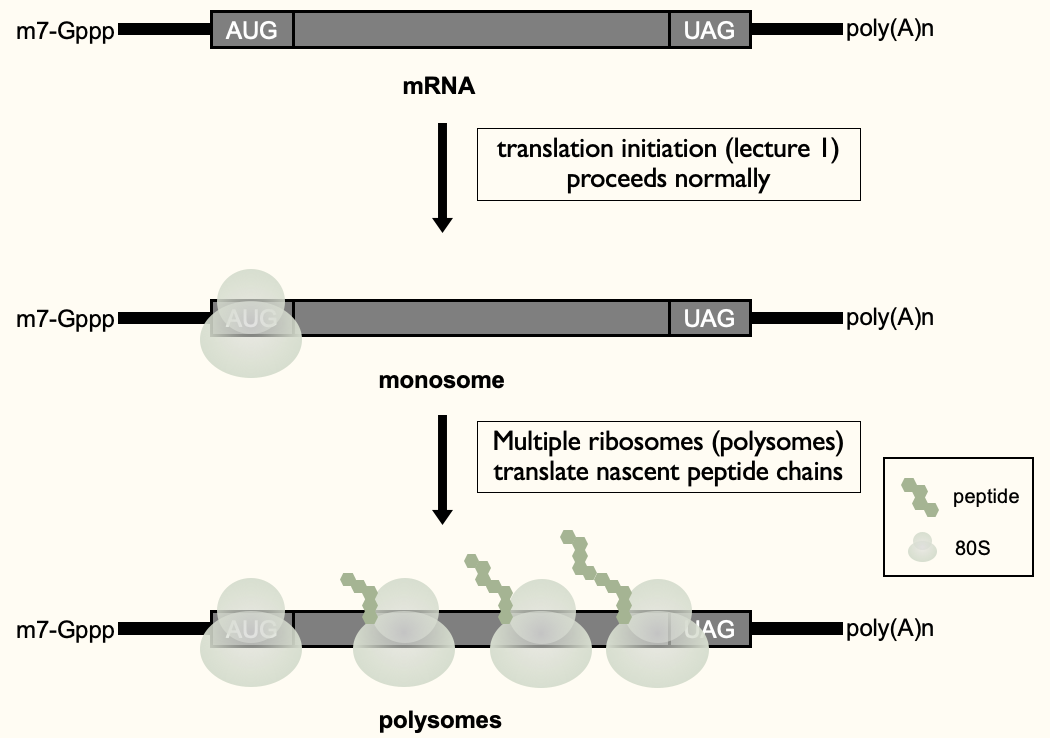
When translation goes right…
Recruited ribosome to 5’ end (monosome) & then further rounds of recruitment of ribosomes to 1 mRNA - to maximise production of proteins from 1 mRNA (polysomes)
Functional protein produced
When translational goes wrong…
**Premature termination codon (PTC)**→ when a termination codon is present in middle of open reading frame - introduced by transcription error in nucleus or mutation in DNA
only makes 1/2 a protein ∴ non-functional & waste of E
toxic truncated proteins → e.g. in transcription factors - 2nd part of protein (not coded) may have contained a regulatory domain which helped keep control of enzyme
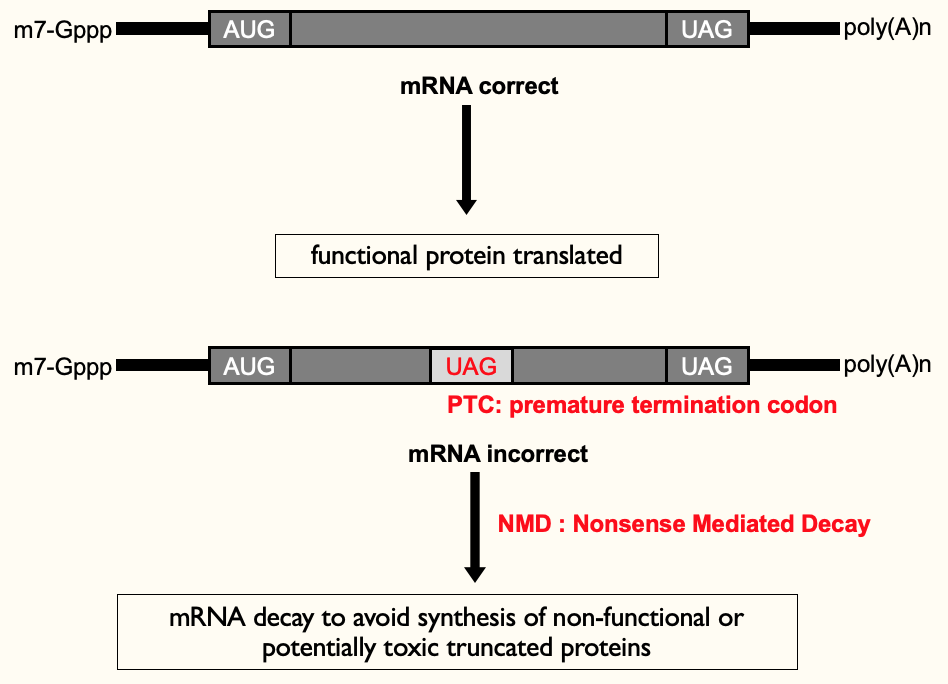
Nonsense Mediated Decay (NMD)
Features in mRNA that help to identify premature termination codon (PTC):
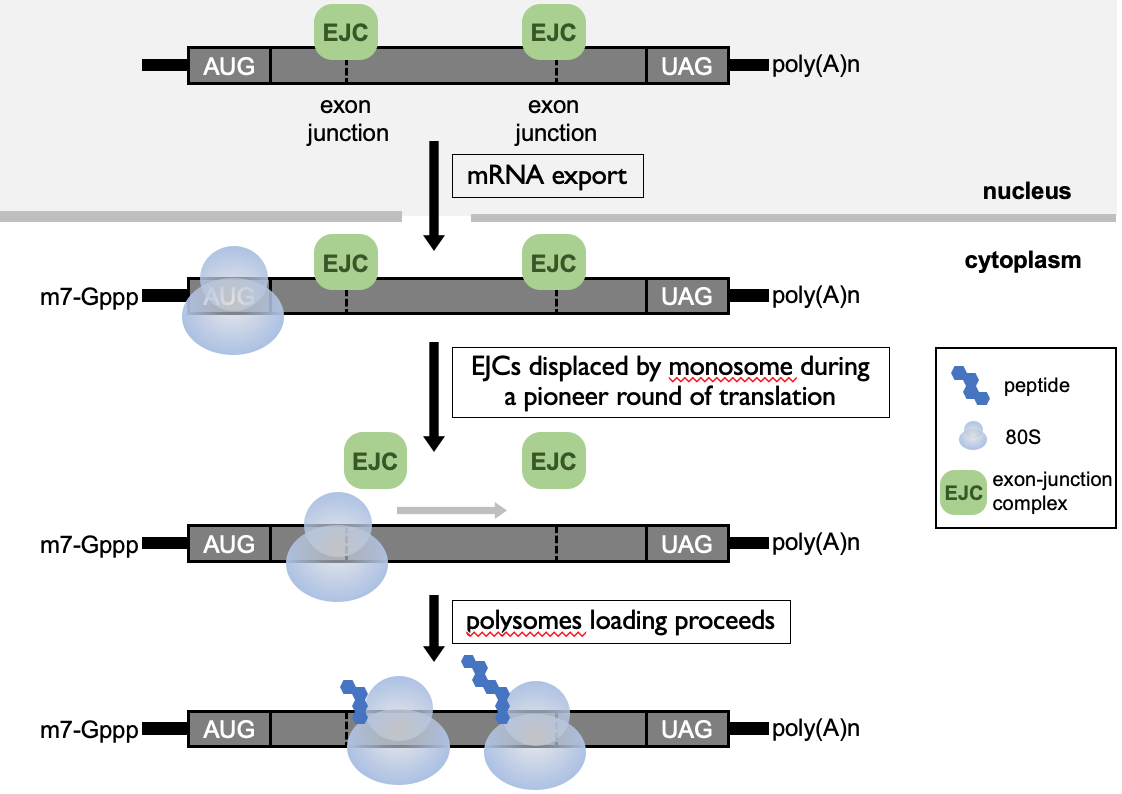
**Exon-junction complex (EJC)**→ at exon-exon boundary
during pioneer round, ribosome is recruited & as it elongates through open reading frame, it displaces the EJCs along the length of mRNA
after pioneer round, no more EJCs & mRNA can be translated efficiently
When PTC is present - translating ribosome starts elongating along 1st portion of open reading frame & knocks off EJCs - then stops at termination codon to terminate
downstream, still EJC present → alarm to ribosome that termination codon isn’t in correct place
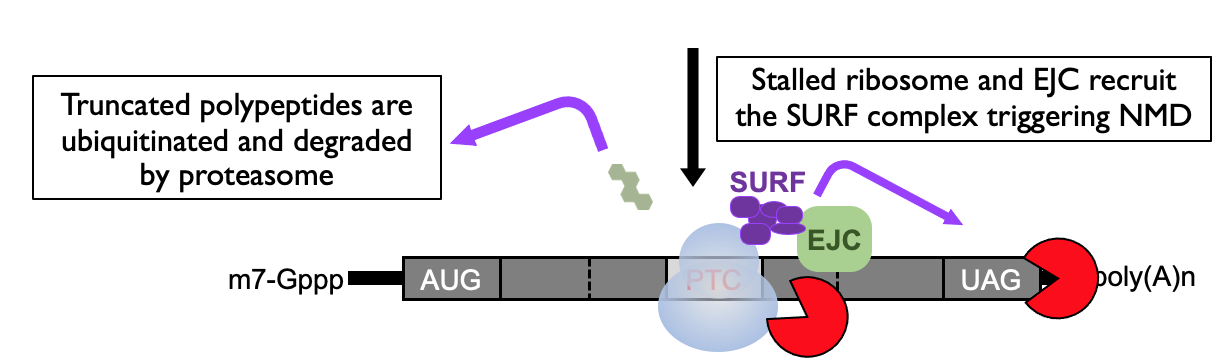
SURF complex formed due to paused ribosome in proximity of EJC → adaptor to bring in RNA decay effectors that dispose of mRNA that is detected as abnormal
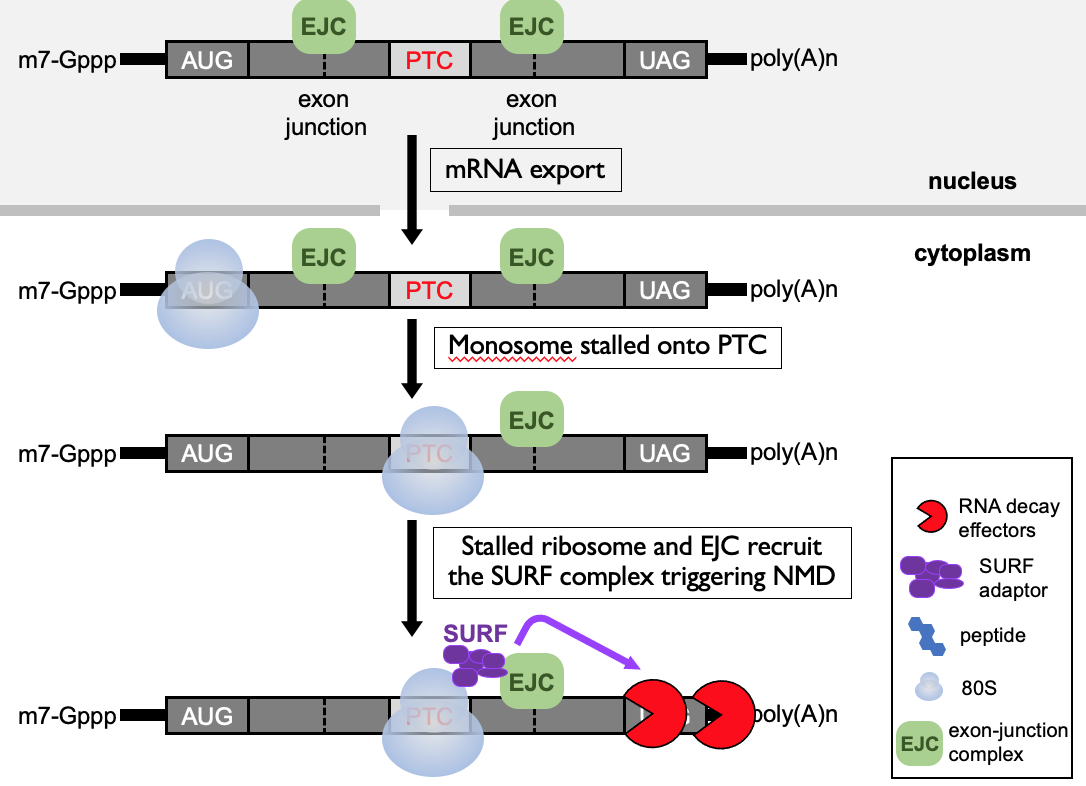
Triggers a safety net to degrade the truncated polypeptide that’s been generated
Decay that’s initiated is an endoribonucleolytic cleavage - cuts in middle of RNA
complex also recruits SMG6
Other quality control pathways
Nonsense-mediated decay (NMD) → (premature termination codons) quality control to eliminate problematic mRNA & limit production of toxic proteins in cytoplasm
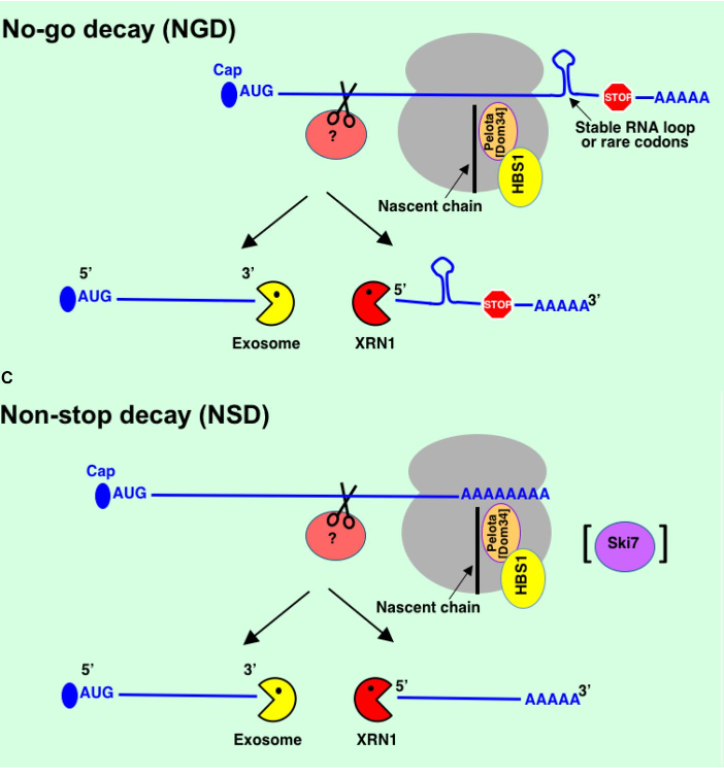
No-go decay (NGD) → ribosome stalled in translation mRNAs e.g. it hits a stable RNA loop, rare codon present in mRNA or damage to RNA
to rescue stuck ribosome - Pelota & HBS1 are recruited
Non-stop decay (NSD) → mRNAs w/o natural stop codons e.g. mistake in transcription, mutation in DNA or transcription termination hasn’t happened fully at end of gene ∴ added poly(A) tail to middle of seq. → ribosome starts translating into poly(A) tail
Specific mRNA translation regulation
Most regulation occurs through 5’ & 3’ UTRs
When global mechanisms have specific consequences (uORFs)
How miRNAs & RBPs can regulate translation from 3’ end
how epitranscriptomic changes can influence translation
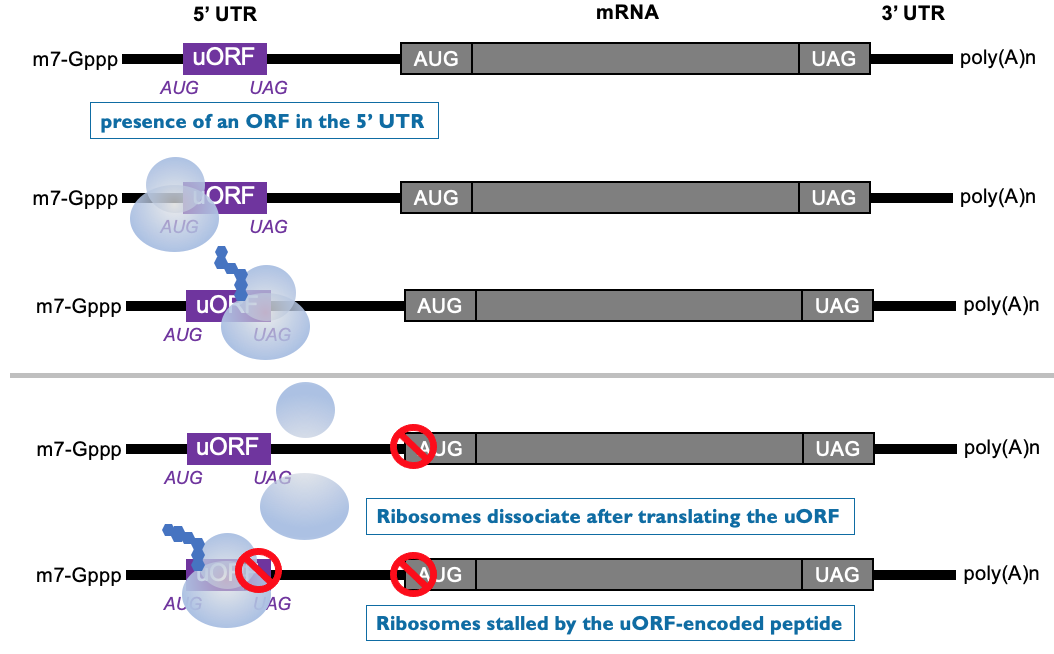
Translational repression by a uORF (upstream Open Reading Frames)
Translation from central ORF is dependant on scanning from 5’ cap of a ribosome & initiating at AUG
Some mRNAs have short ORF in 5’ UTR - encode for small peptides
scanning ribosome initiates at upstream ORF to produce short peptide - at end of translation, ribosome terminates & leaves mRNA
downstream ORF isn’t translated ∴ poorly translated as ribosomes won’t reach downstream AUG
beginning translation at upstream ORF, inhibits initiation at downstream ORF
peptide encoded by uORF can specifically interfere w/ translation at ribosome
Translational control by a uORF (upstream ORF)
mRNAs w/ upstream ORFs are generally encoding products that post-production needs to be v. tightly controlled & strongly inhibited most of the time
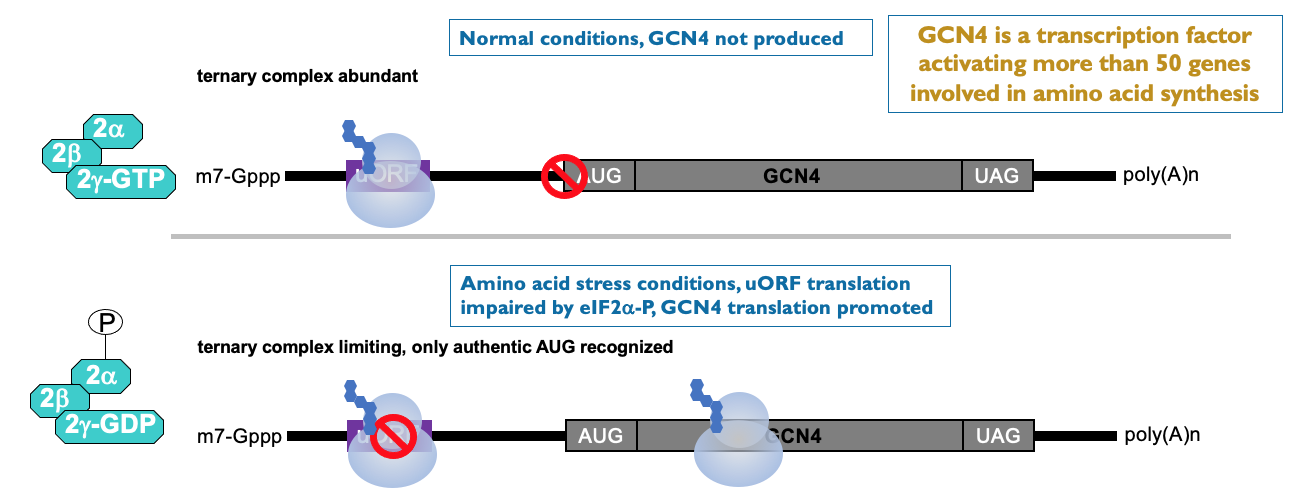
These examples contain ORF & help ↑ translation when eIF2α is phosphorylated:
e.g. GCN4 → transcription factor - activates large no. of genes involved in AA synthesis → most of time - no GCN4 produced as translation inhibited by upstream ORF
low AA & eIF2α kinase has lead to phosphorylation of eIF2 complex → ↓ in ternary complex availability that delivers initiator tRNA to ribosome → ribosome initiated on GCN4 mRNA - scans along UTR but misses upstream ORF due to low availability of initiator tRNA to start translation
translation improves on uORF containing mRNA from low base level
↑ production of GCN4 helps synthesis AA to get out of cell stress situation
e.g. ATF4 → helps produce chaperone proteins to help resolve ER stress → improves translation
GADD34 → helps guide phosphatase to dephosphorylate eIF2α
Translational control from 3’ end - RNA-BPs
RNA binding protein binding to cis-acting element in 3’ UTR can impact on initiation at 5’ cap
Cis-acting element→ element in RNA which acts on same RNA (on itself)
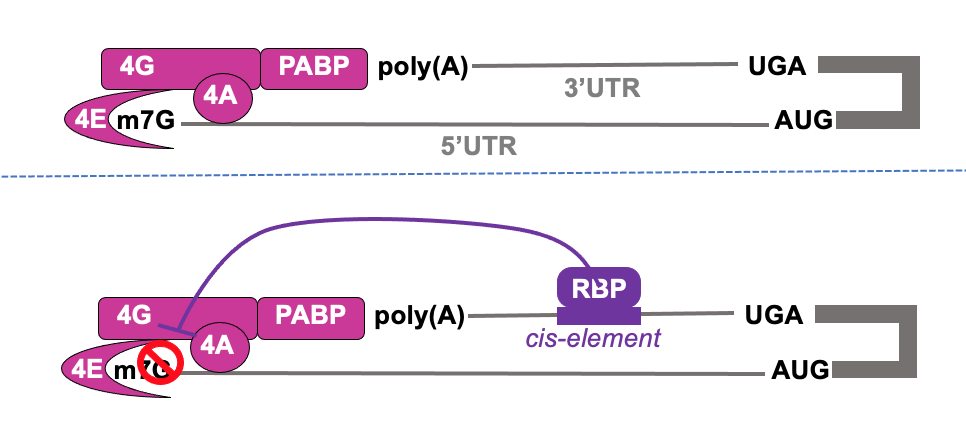
Translational control from 3’ end- e.g. caudal & bicoid
Patterning needs to occur so that v. early on in development, embryo knows which end in head & which is tail
done molecularly by gradients of proteins (morphogens) that influence this patterning - communicate their fate to cells during embryogenesis
e.g. bicoid → gene is selectively expressed at head end (anterior)
transcription factor that’s important for production of other patterning mols. & acts as translational regulator on mRNA called caudal
e.g. Caudal → important for patterning the posterior end of embryo, but it’s mRNA is expressed throughout the embryo - bicoid stops protein being where bicoid is present
Patterning the early drosophila embryo:
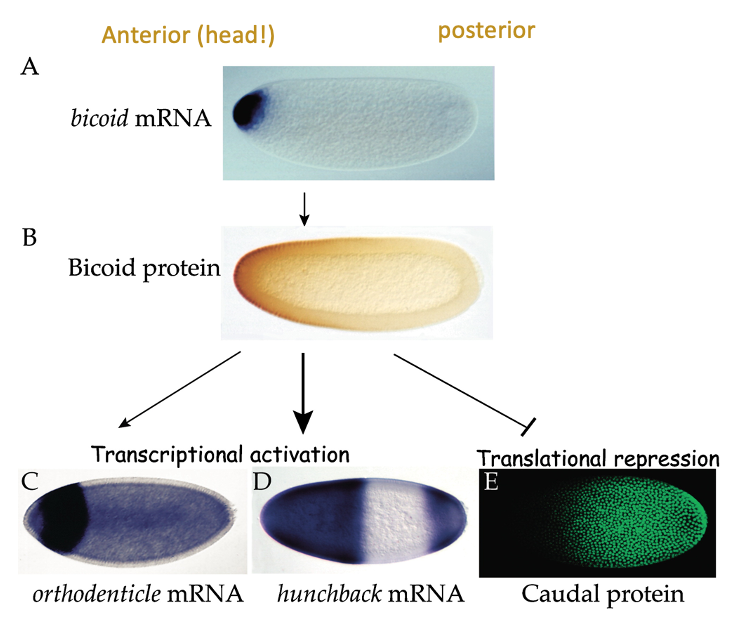
Caudal is an important patterning mol. during embryogenesis
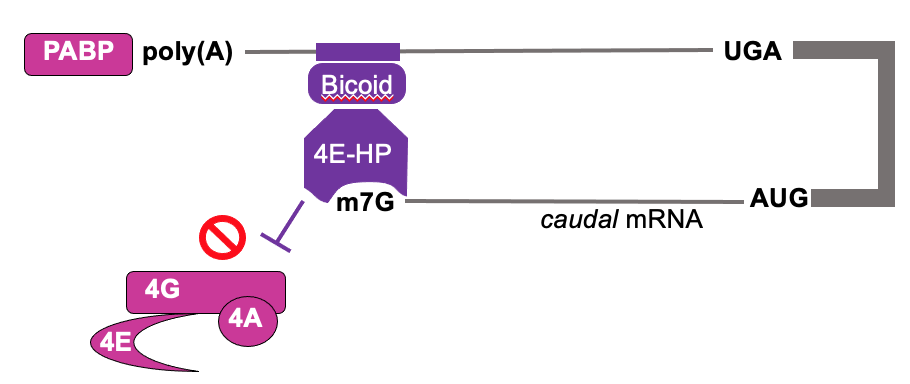
Bicoid (RBP) is RNA binding protein that interacts w/ 3’ UTR of caudal mRNA
Bicoid recruits 4E-HP (4E homologous protein) to bing caudal cap but not initiate translation
4E-HP blocks eIF4F binding & translation of caudal mRNA
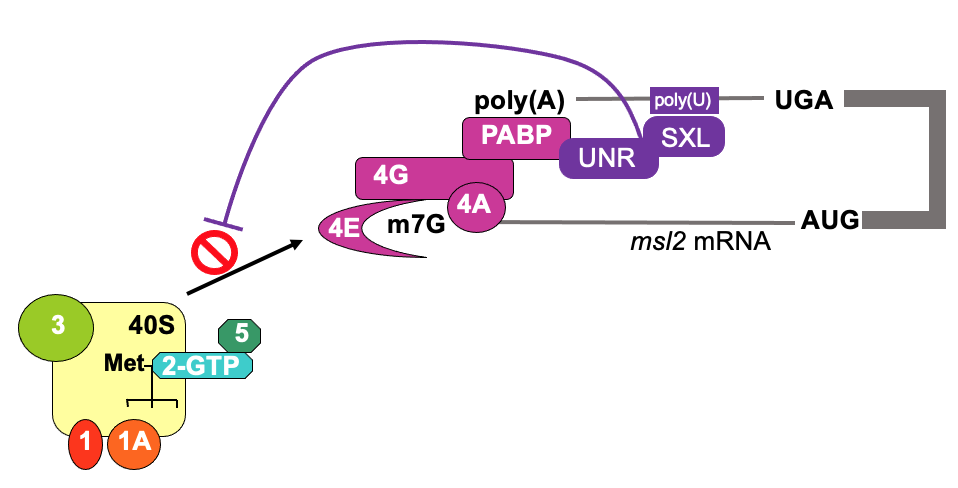
Translational control from 3’ end - e.g. msl2 & SXL
In drosophila, to achieve dosage compensation of X-linked genes, in male flies, the dosage compensation complex (feat. msl-2) is required for hypertranscription (massively upregulate) of single X chromosome
mediated by protein msl-2
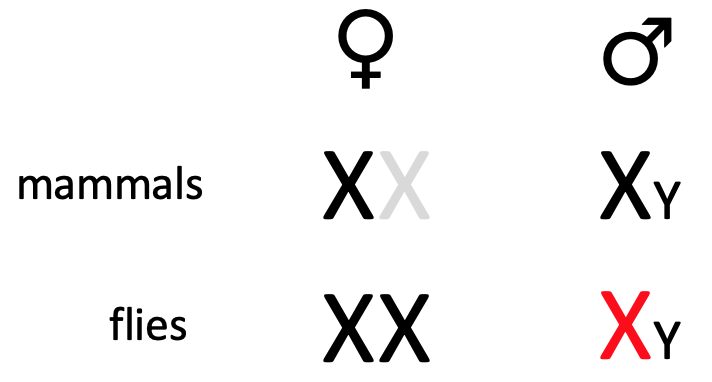
msl2 mRNA is transcribed in both, but translated in male but not female during development - to repress production in female flies
SXL (“sex-lethal”) is only expressed in females & is a binding protein
SXL & UNR bind to 3’ UTR of msl-2 & interact w/ poly(A) binding protein & influences eIF4F complex to stabilise it’s interaction w/ pol(A)
becomes too stable - prevents 4F to recruit 43s ribosome - translation is inhibited on msl2 in female flies
In early embryogenesis, mutation of SXL will compromise establishment of sex
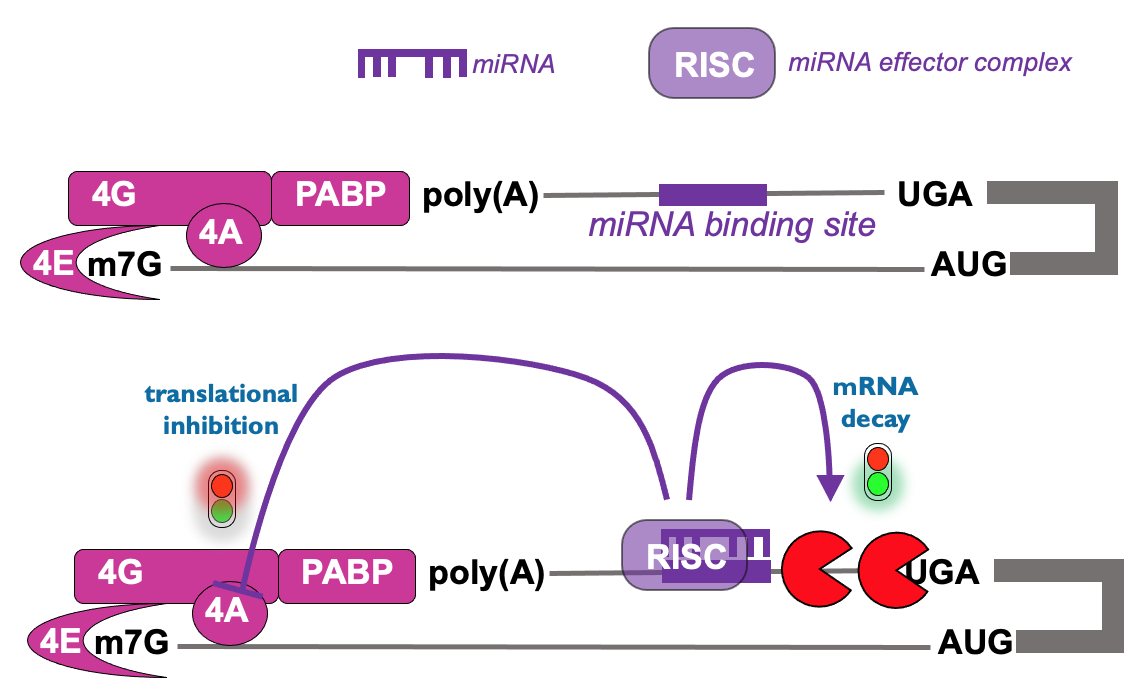
Translational control from 3’ end - microRNAs
Regulatory RNA seq. in 3’ UTR that mediates binding of RNA complex & leads to inhibition of translation initiation → occurs prior to mRNA decay & also occurs on microRNA targets
other example of mech. of control from 3’ UTR
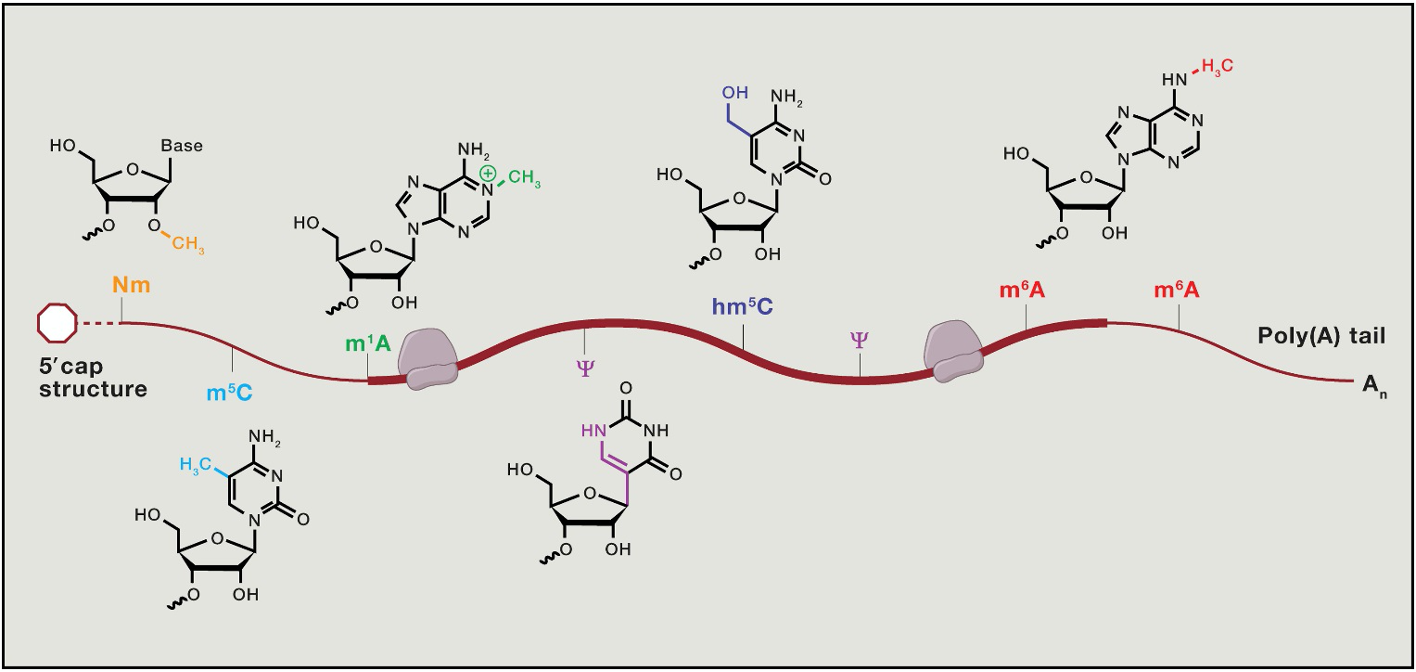
Translational control - beyond the genetic code
RNA bases can be chemically modified in a way that doesn’t Δ the code → the epitranscriptome
influences mRNA fate
e.g. methyl-cytosine or methyl-adenosine
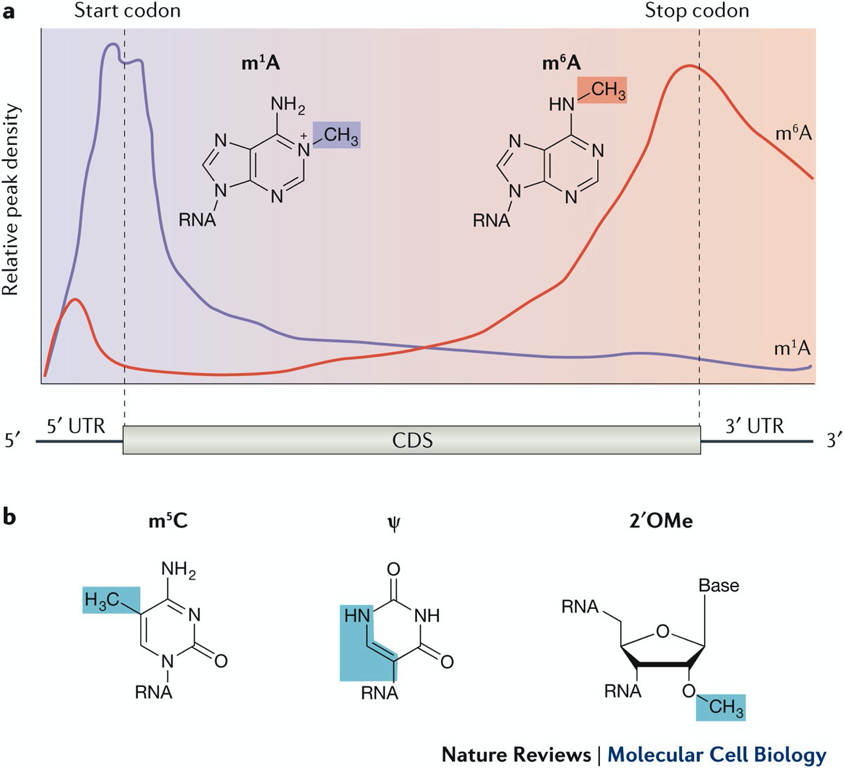
mRNA modifications correlate w/ mRNA features
most freq. modification - m7 cap → methyl 7 on guanosine cap
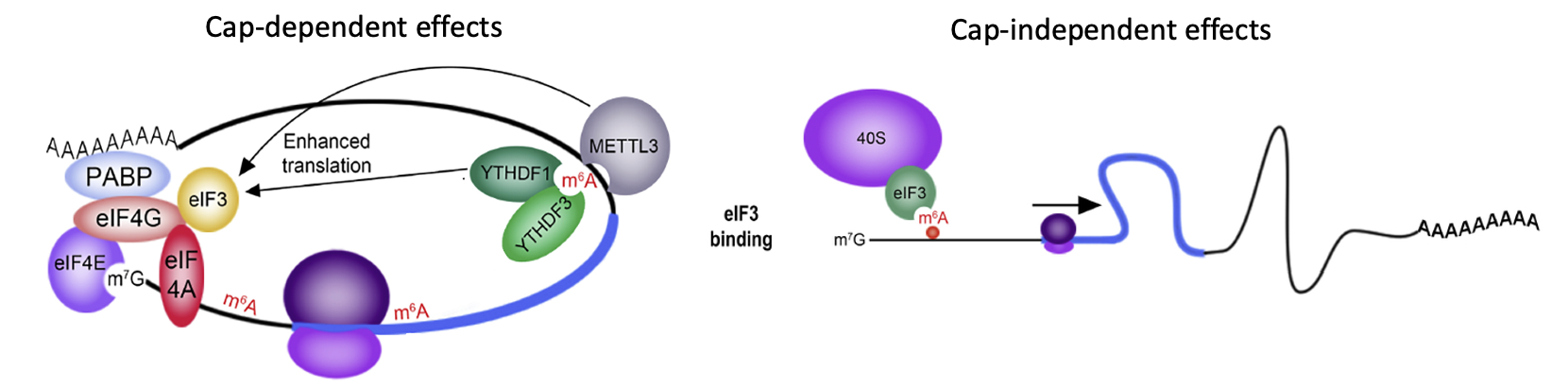
Translational control - N6-methyladenosine (m⁶A)
m⁶A promotes mRNA translation by multiple mechanisms
“reader proteins” can bind m⁶A in 3’ UTR & enhance translation initiation e.g YTHDF1 & 3
METTL3 → helps install m⁶A & adds on m⁶A → promotes translation of message m⁶A is on (cap-dependant translation)
When m⁶A is present on 5’ UTR it can function like IRES → translation factor eIF3 can be recruited by m⁶A directly
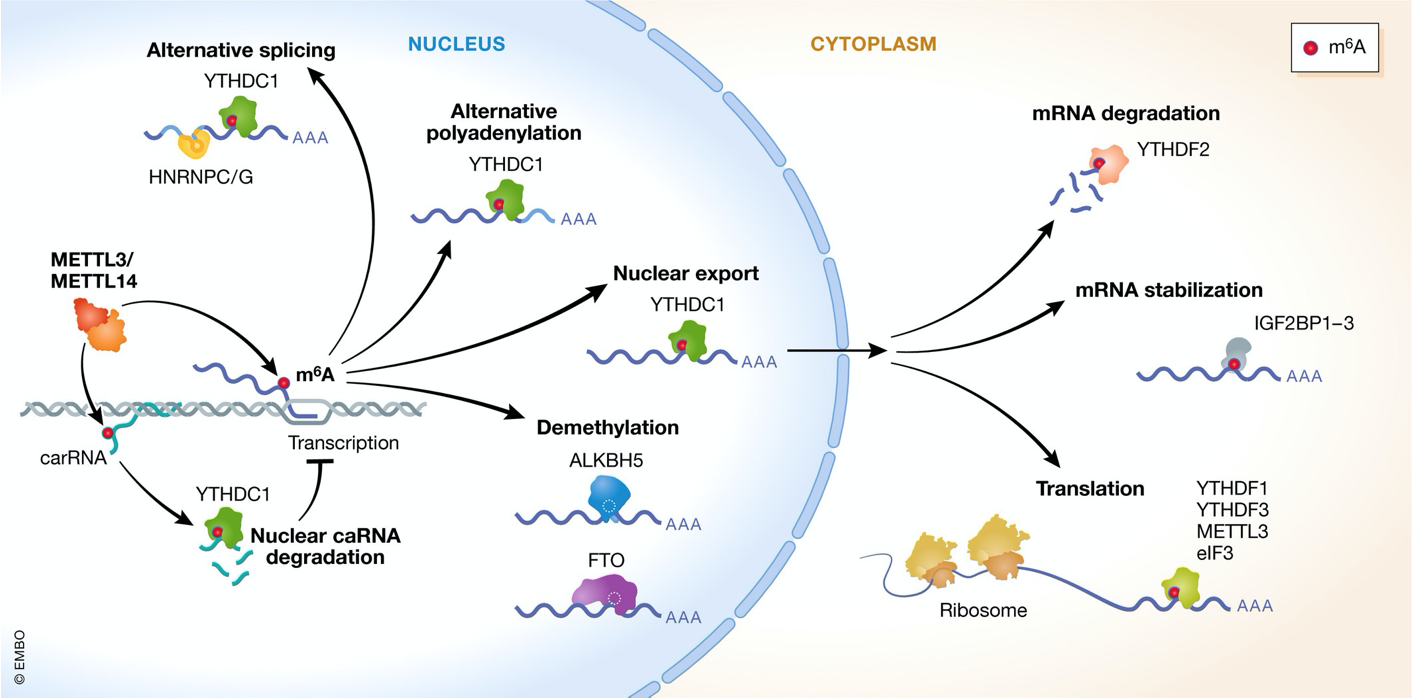
m⁶A controls multiple RNA-regulatory processes
m⁶A implicated in control of RNA stability & degradation
Can impact nuclear export & splicing
Functions are dictated by diff. kinds of BPs that binds the m⁶A modification
can be demethylated later on → turned on or off → epitranscriptomics
Questions & themes to consider
What mechanisms exist to ensure accurate translation of error-free mRNAs?
Which control mechanisms target general translation factors but selectively affect the translation of a subset of mRNAs?
Which mutations or changes in eIFs are associated w/ disease?
Which control pathway normally prevent disease?
How might a circular RNA, lacking a 5’ cap, be translated?
 Knowt
Knowt