Nucleic acids and DOGMA
Nucleic acids to know
DNA - Deoxyribonucleis acid
RNA - Ribonucleic acid
The structure of nucleic acids
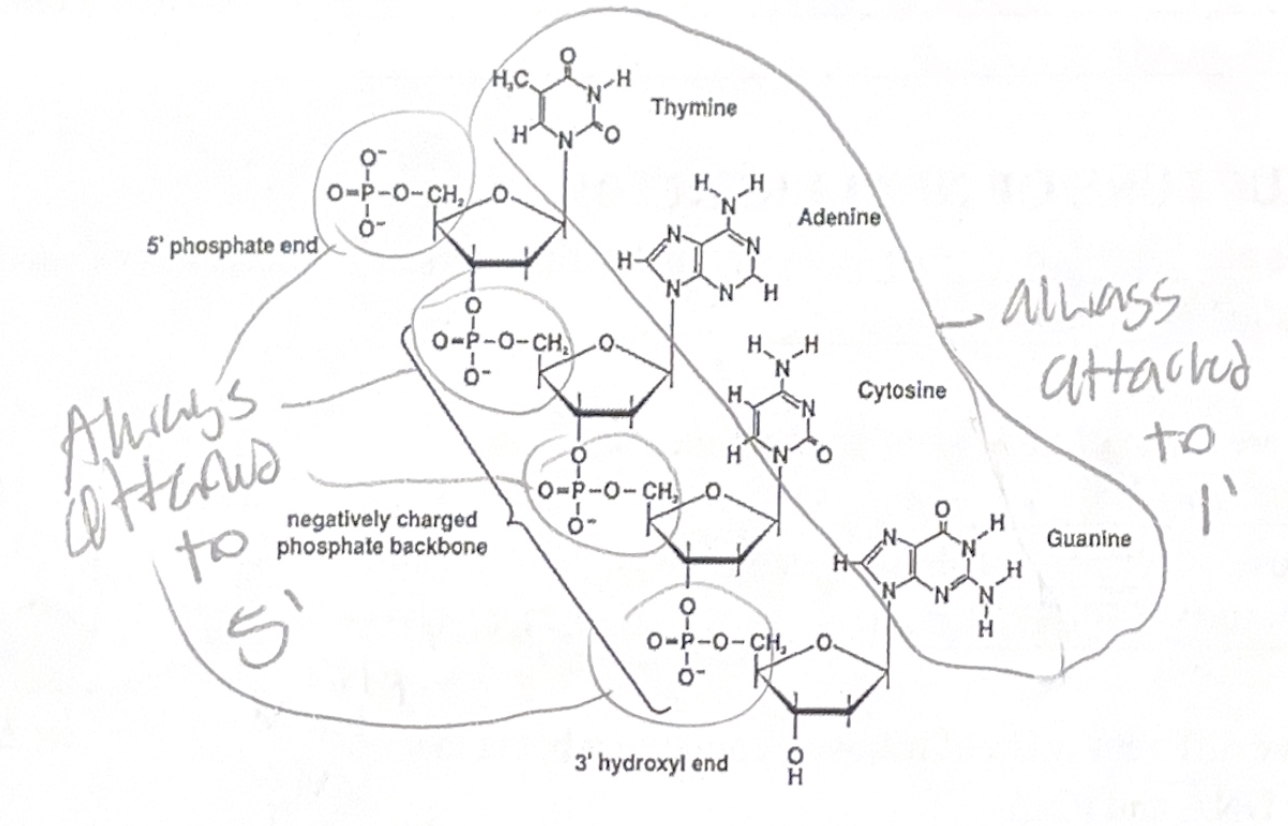
Nucleic acids are polynucleotides, meaning that they are polymers of small molecules called nucleotides.
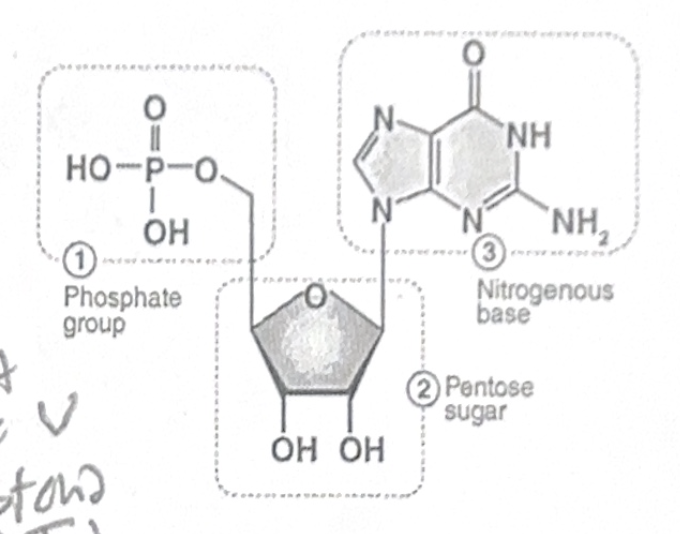
Nucleotides are made of three smaller components:
1. Phosphate group
2. Pentose sugar (ribosome or deoxyribos)
3. Nitrogenous base (A, T (or U for RNA), G, C)
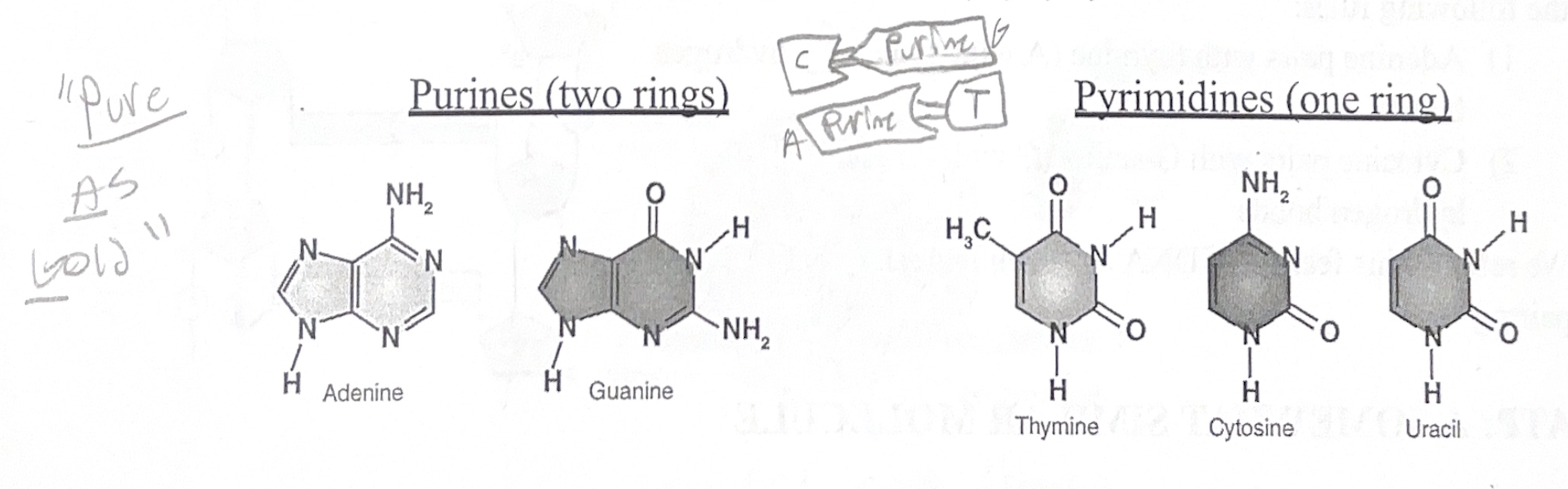
There are five different types of nitrogen-containing bases that are found in DNA and RNA.
In DNA: Adenine, Thymine, Guanine, and Cytosine
In RNA: Adenine, Uracil, Guanine, and Cytosine
These bases are often referred to by their first letters: A, T (or U), C, and G
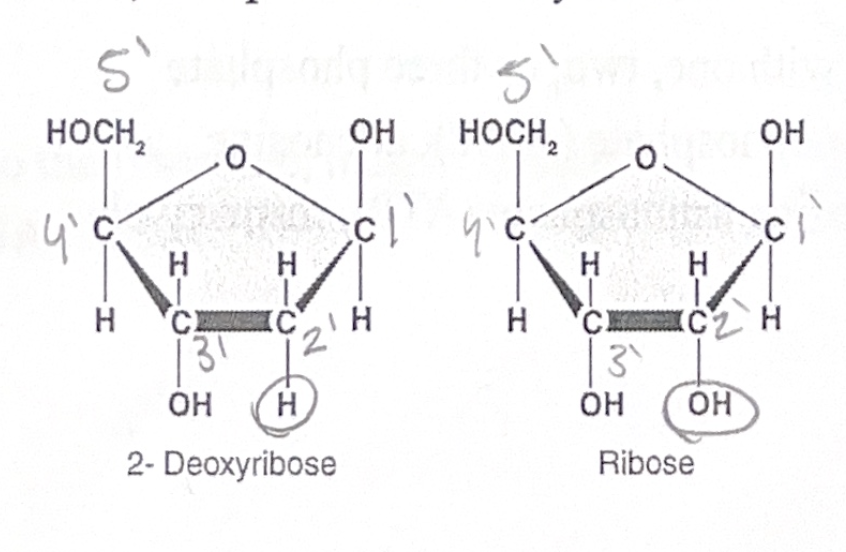
The pentose sugar can either be ribose (in RNA) or deoxyribose (in DNA). These two are very similar, except that the deoxyribose has one fewer oxygen.
Polynucleotides (DNA and RNA)
DNA and RNA are formed by covalent sugar-phosphate bonds called phosphodiester bonds, which link successive nucleotides together.
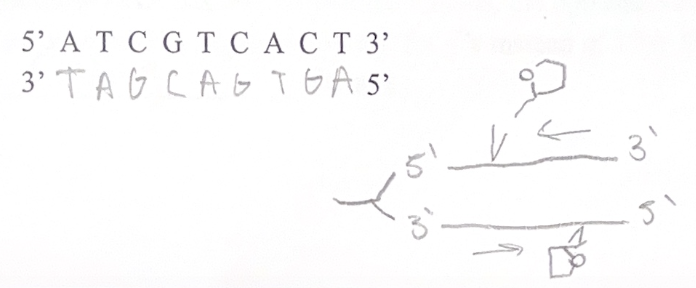
DNA molecules are made of two antiparallel polynucleotide strands. Antiparallel means that each strand running in an opposite direction.
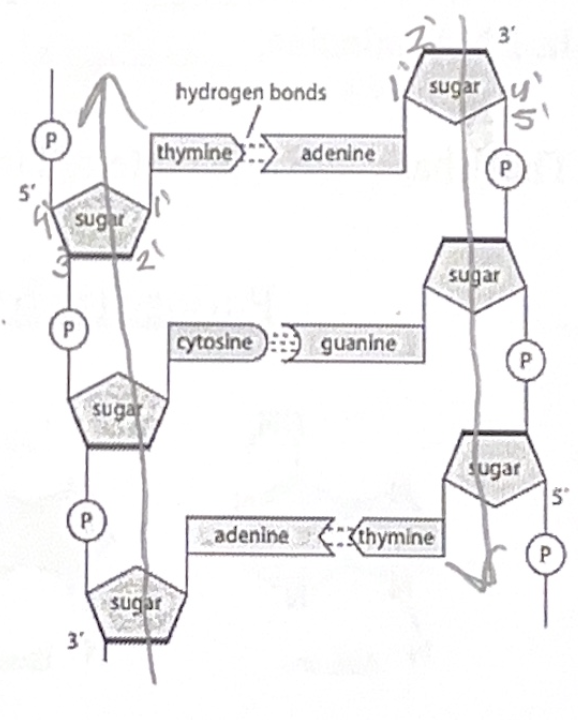
The two strands are held together by hydrogen bonds and follow the following rules:
1. Adenine pairs with Thymine (A with T) via 2 hydrogen bonds
2. Cytosine pairs with Guanine (C with G) via 3 hydrogen bonds
We refer to this feature of DNA as “complementary base pairing”
ATP: a somewhat similar molecule
ATP stands for adenosine triphosphate. Although it is not part of DNA or RNA, its structure is very similar to that of a nucleotide
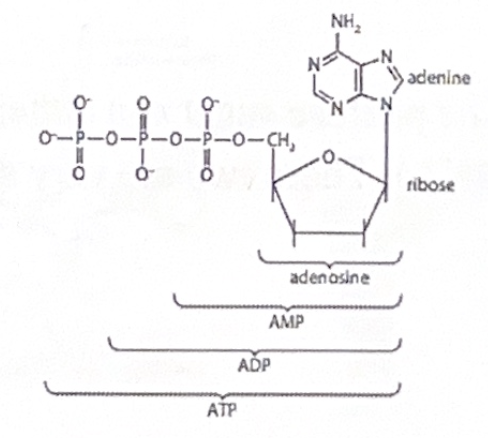
Adenosine can be combined with one, two, or three phosphate groups to give adenosine monophosphate (AMP), adenosine diphosphate (ADP), or adenosine triphosphate (ATP), respectively.
The central DOGMA: transcription
Background
A genome is the complete set of genetic information within an organism.
- The human genome is ~3 billion nucleotides long.
- The parts of the genome which encode polypeptides are called genes
- Genes comprise only ~2-3% of the human genome
- The other ~98% of our genome is considered non-coding and does not directly code for proteins.
The central DOGMA
“I shall…argue that the main function of the genetic material is to control (not necessarily directly) the synthesis of proteins. There is a little direct evidence to support this, but to my mind the psychological drive behind the hypothesis is at the moment independent of such evidence” - Francis Crick, 1957
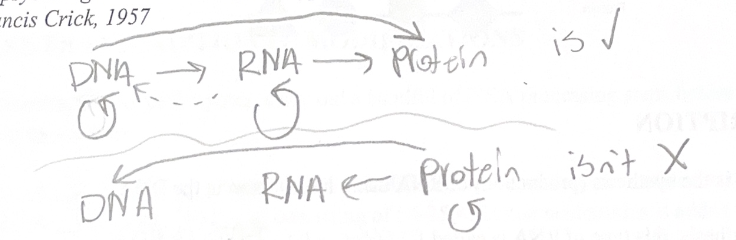
The process of protein synthesis involved two overarching processes:
1. Transcription: DNA to RNA
2. Translation: RNA to Protein
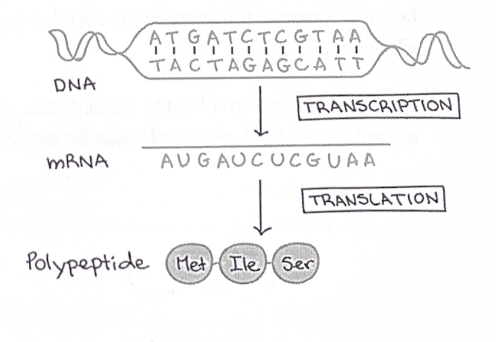
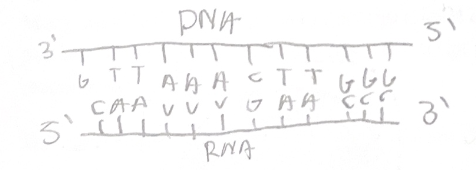
The genetic code
For each gene, only one of the two DNA strands is transcribed. This strand is called the template strand
Because DNA has two antiparallel strands, the opposite strand of DNA will look identical to the encoded mRNA sequence (except for T’s instead of U’s). We call this strand of DNA the coding strand.
The sequence of nucleotides bases in a DNA molecule is a code for the sequence of amino acids in a polypeptide.
This code is broken up into three-letter chunks called codons
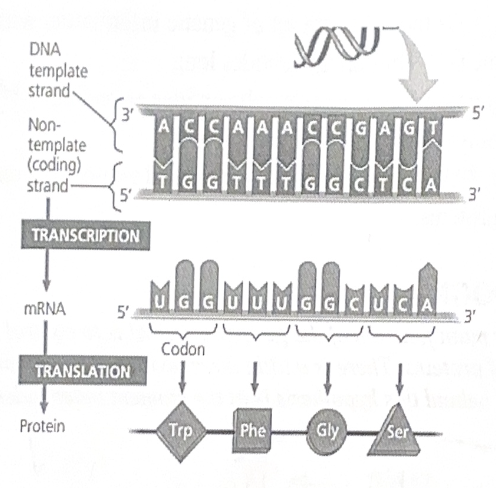
Transcription is the synthesis (production) of RNA using information in the DNA.
In protein synthesis, this type of RNA is called messenger RNA (mRNA), because it carries the instruction encoded in the DNA to the site of protein production, the ribosome.
The main enzyme involved in transcription is RNA polymerase, which assembles the RNA polynucleotide by moving in its 5’ to 3’ direction, adding onto its 3’ end.
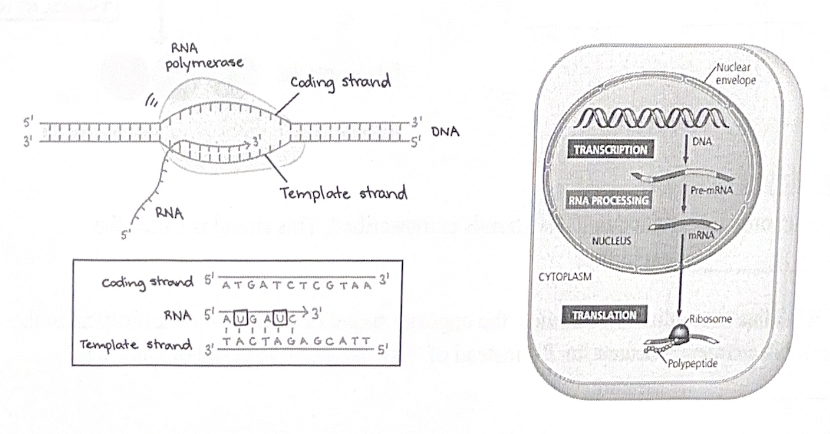
The steps of transcription
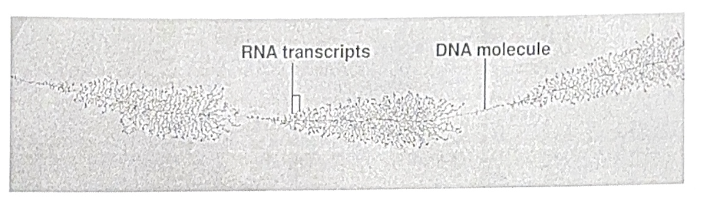
1. Initiation - RNA polymerase binds to a promoter
2. Elongation - RNA polymerase synthesized mRNA (RNA polymerase can only move in its 5’ to 3’ direction, adding onto its 3’ end)
3. Termination - RNA polymerase reaches a terminator and mRNA is released
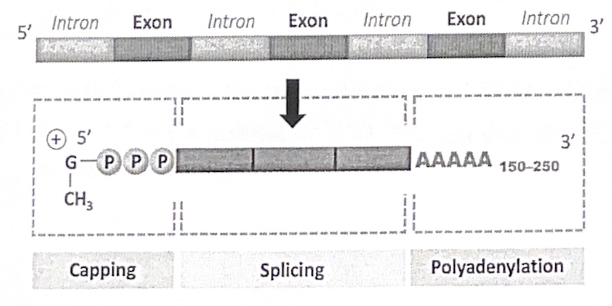
Post-transcriptional modifications
Eukaryotes, but not prokaryotes, carry out a handful of RNA processing steps before the mRNA leaves the nucleus:
1. A 5’ cap consisting of a modified guanine nucleotide is added to the 5’ end
2. A poly-a tail consisting of 20-250 adenine nucleotides is added to the 3’ end
These modifications:
- protect the mRNA
- facilitate mRNA export from the nucleus
- help ribosomes attach to the 5’ end of the mRNA before translation
Many eukaryotic transcripts also undergo a level of RNA splicing, in which large portions of the original (primary) RNA transcript are removed and the remaining portions are reconnected.
The intervening sequences that are removed are called introns
The expressed sequences that remain are called exons
The central DOGMA: translation
Translation is the process by which cells convert the genetic information in mRNA into a sequence of amino acids, forming a polypeptide.
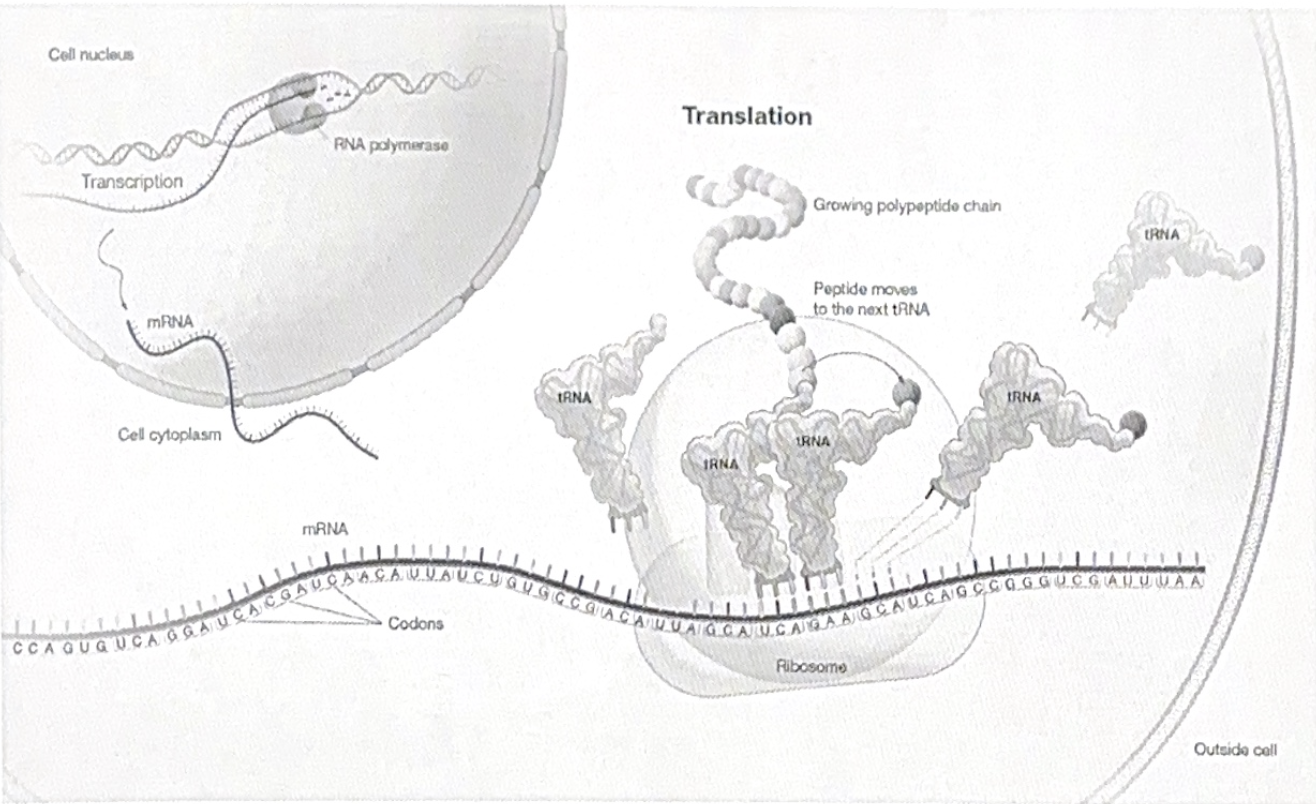
The components of translation: Ribosomes, tRNA, and mRNA
The ribosome is made of ribosomal RNA (rRNA) and protein and has two subunits (large and small).
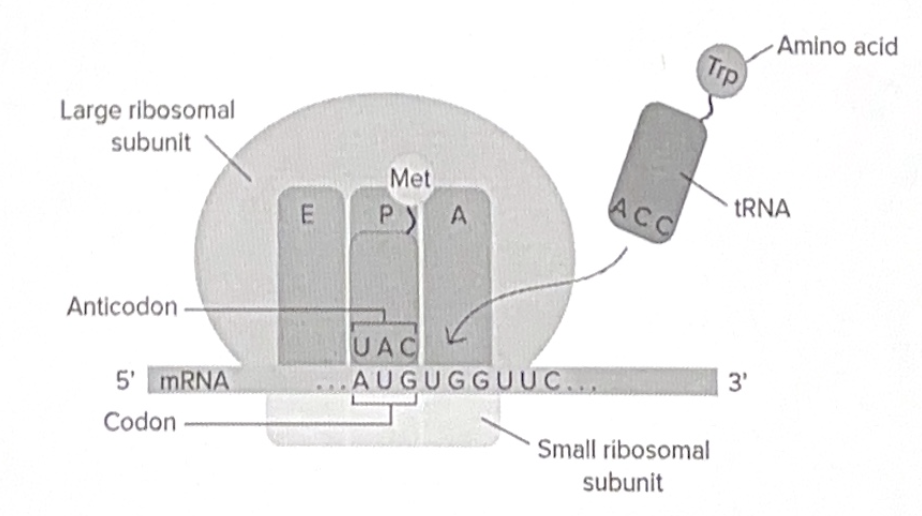
A special type of RNA called transfer RNA (tRNA) carries amino acids to the ribosome. tRNA acts as a molecular “bridge” that matches mRNA codons with their corresponding amino acids.
tRNAs bring amino acids to the ribosome, matching their anticodon with mRNA codons
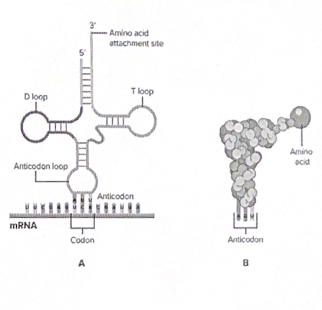
The translation process
Like transcription, translation has there steps:
1. Initiation - small ribosomal subunit binds to mRNA and start the codon
2. Elongation - tRNA brings specific amino acids to the ribosome, matching with codons on mRNA
3. Termination - translation stops when a stop codon (UAA, UAG, UGA) is reached and a release factor binds to the stop codon
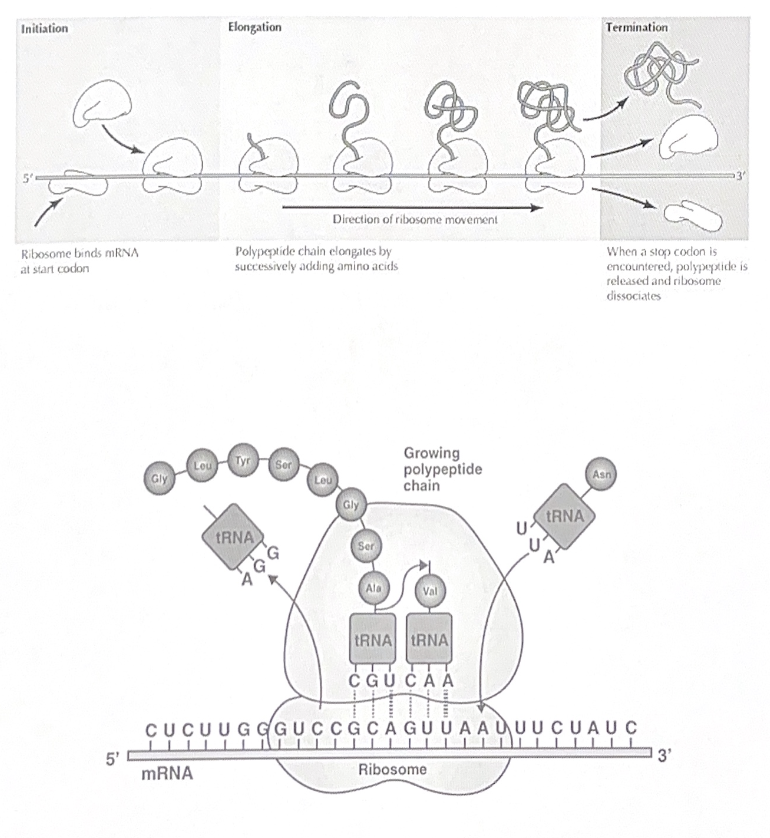
DNA replication
DNA replication occurs during s phase of the cell cycle. It is a “semiconservative” process meaning that each strand is used as a template for a new strand.
The steps of DNA replication are:
1. Helicase unzips the double helix at the replication fork
2. A short RNA ‘primer’ is added by primase
3. DNA polymerase binds to the primer and adds new bases onto the template strand.
- DNA polymerase adds bases to the 3’ end of the growing strand (moving 5’ to 3’)
4. DNA ligase fuses Okazaki fragments on the lagging strand
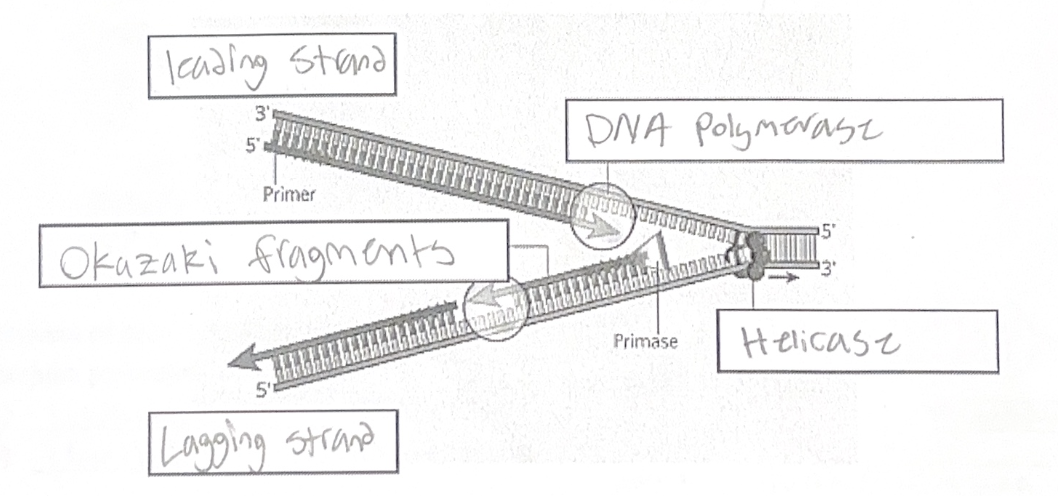
Some rules to remember:
1. DNA polymerase moves from 5’ to 3’
2. Molecules of DNA (and RNA) extend in the 5’ to 3’ direction
3. DNA polymerase moves continuously on the leading strand, but needs to “jump” backwards repeatedly on the lagging strand (which is why we see Okazaki fragments)