Biochem Sept. 8th
Gibbs free energy and spontaneity
ΔG is the difference between the Gibbs free energy of products and reactants:
\Delta G = G{\text{products}} - G{\text{reactants}}Spontaneous (exergonic) reactions: \Delta G < 0
Energy is released; useful for work.
Non-spontaneous (endergonic) reactions: \Delta G > 0
Requires input of energy to proceed.
Common intuition from the transcript:
If products have lower free energy than reactants, the process releases energy (negative $\Delta G$).
A positive $\Delta G$ means the products are higher in free energy than the reactants, so energy input is required.
Example from the transcript:
A → B with \Delta G = +15\ \text{kcal}: uphill, not spontaneous, energy input required.
B → C is downhill with negative $\Delta G$, so it releases energy that can drive other steps.
In metabolic pathways, reactions are often coupled so that energy release from one drives an energy-requiring step, making the overall process spontaneous.
Coupled reactions and net Gibbs free energy
Concept: Cells couple energy-releasing reactions with energy-requiring ones so that the net Gibbs free energy is negative.
Formal expression for coupling:
\Delta G{\text{total}} = \Delta G{A\to B} + \Delta G_{B\to C}If the coupled pair yields \Delta G_{\text{total}} < 0, the overall process is spontaneous.
Real-world context: Many reactions in metabolism are linked in rapid succession or occur in the same time frame to ensure the overall negative free energy change.
Central dogma and the genetic basis of protein synthesis
DNA stores genetic information in cells.
Transcription copies the information from DNA into complementary RNA molecules.
Translation uses RNA as a template to synthesize proteins.
Overall information flow:
DNA (genetic information) → RNA (transcription) → Protein (translation).
Mutations and DNA damage can alter the sequence of amino acids in proteins, changing protein structure and function, which underlies many diseases.
Example from transcript:
Mutations changing amino acid residues (e.g., serine to another residue) can alter protein function or cause misfolding/unfolding.
Pre-class video note (topic two): In the second half of topic two there are many examples about ionization of amino acids and how charges depend on pH; the video is meant to help you prepare for class on Wednesday.
Proteins: building blocks and overall architecture
Proteins are polymers of amino acids.
The basic building blocks are 20 different amino acids linked together in sequence to form proteins.
Protein sequence determines structure and function; mutations in the sequence can alter structure and function.
The sequence is read from the N-terminus to the C-terminus during synthesis.
The first amino acid is usually methionine (Met), initiating the chain.
The chain ends with a C-terminal residue that still has a free carboxyl group.
Proteins must fold into well-defined three-dimensional shapes to perform their functions.
The side chains (R groups) of amino acids determine the protein’s chemical properties and folding behavior.
The backbone (ignoring side chains) is similar across residues and consists of repeating units that include the amide bond:
peptide linkage forms between the carboxyl carbon of one amino acid and the amino nitrogen of the next amino acid: -\mathrm{NH}-\mathrm{C{\alpha}}-\mathrm{C(=O)-} with side chain R attached to the $\mathrm{C{\alpha}}$.
Backbone termini:
N-terminus: free amino group on the first residue
C-terminus: free carboxyl group on the last residue
Condensation (dehydration) reaction forms a peptide bond:
\text{Amino acid}1 -\mathrm{COOH} + \text{Amino acid}2
-\mathrm{NH2} \rightarrow \text{Amino acid}1-\mathrm{CO-NH}-\text{Amino acid}2 + \mathrm{H2O}After incorporation into a protein, each amino acid is called a residue; its position (e.g., residue 4) defines its place in the chain.
Protein properties depend on the side chain, not just the backbone.
Proteins can be sensitive to mutations: a single amino acid change in a long protein (e.g., a 600-residue protein) can cause malfunction or toxicity.
Pre-class and in-class emphasis:
Learning objectives include drawing the basic structure of amino acids, recognizing all 20 amino acids, and understanding how pH affects charge.
Students should be able to recognize amino acid structures and know that the charge state depends on pH and pKa values.
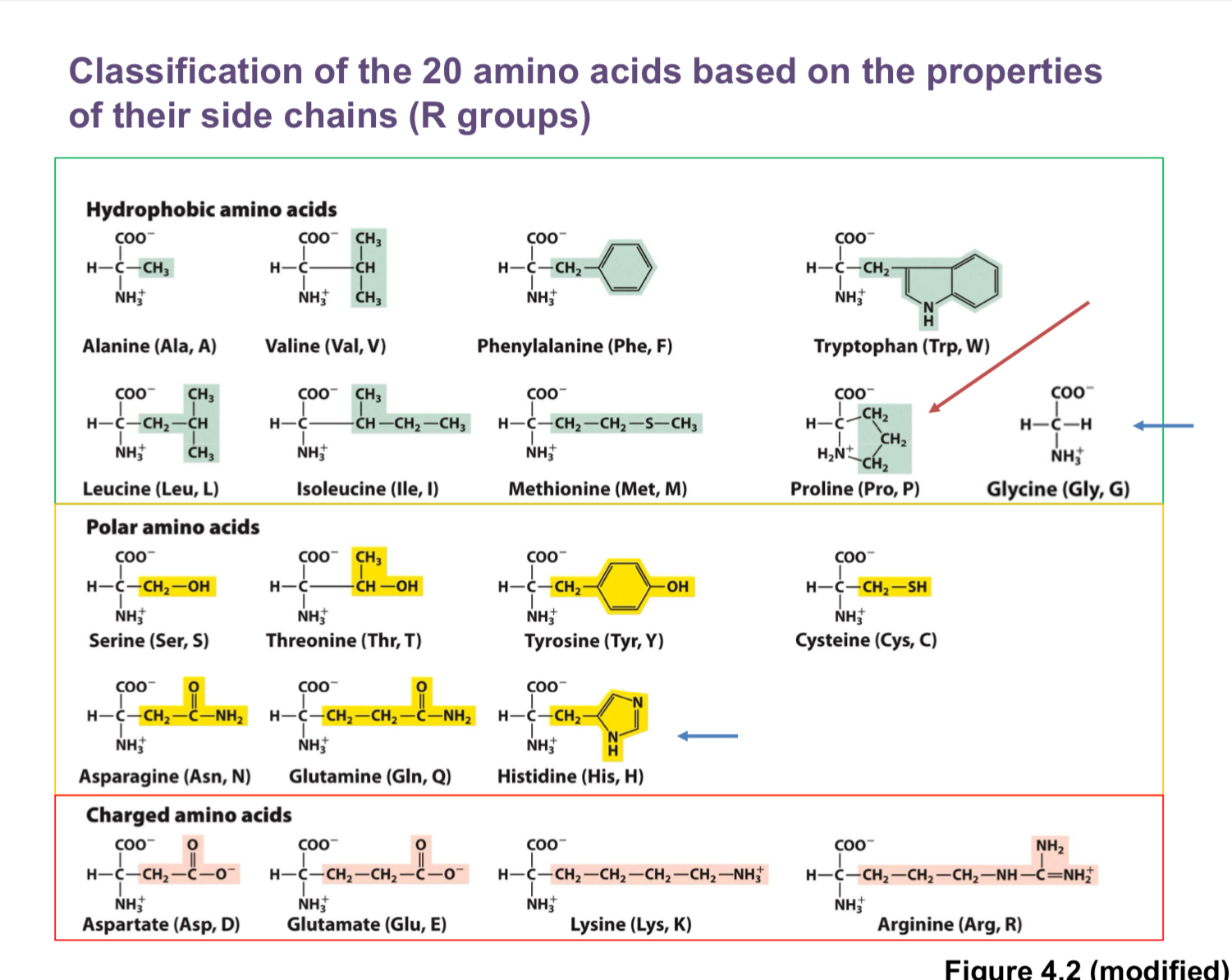
The 20 amino acids: structure, properties, and organization
Each amino acid has:
An α-amino group (—NH2 or —NH3+ depending on protonation)
A carboxyl group (—COOH or —COO− depending on protonation)
A central (α) carbon attached to a hydrogen and a side chain (R group)
The α-carbon is a stereocenter for all amino acids except glycine.
Glycine (G, GLY):
Smallest amino acid; side chain is a single hydrogen
Not chiral (lacks a chiral center)
Often used to increase flexibility in proteins
Proline (P, PRO):
Unique cyclic side chain that bonds back to the amino group
R group forms a covalent bond with the amino nitrogen, limiting conformational flexibility
Often treated specially in structure analysis
Glycine and the L-form:
In biology, proteins are built from L-amino acids; glycine is an exception in that it is not chiral
Hydrophobic (nonpolar) amino acids (tend to be buried in protein cores):
Alanine (A, ALA): small hydrophobic side chain (methyl)
Valine (V, VAL): branched
Leucine (L, LEU): larger aliphatic
Isoleucine (I, ILE): branched
Methionine (M, MET): contains sulfur; thioether; hydrophobic
Phenylalanine (F, PHE): aromatic ring
Tryptophan (W, TRP): largest aromatic side chain; very hydrophobic; least frequent
Note: Tryptophan is large and tends to be in the protein core; could be membrane-associated but often internal
Aromatic and polar varieties:
Tyrosine (Y, TYR): aromatic ring with hydroxyl (OH); can participate in hydrogen bonding; more polar than phenylalanine
Polar (uncharged) amino acids:
Serine (S, SER): has an OH group; can form hydrogen bonds with water
Threonine (T, THR): similar to serine but with an extra carbon; OH group
Asparagine (N, ASN): amide side chain
Glutamine (Q, GLN): amide side chain
Sulfur-containing amino acids:
Methionine (M, MET): discussed above in hydrophobic group
Cysteine (C, CYS): contains a thiol (SH); can form disulfide bonds with another cysteine
Disulfide bonds:
Covalent bonds between two cysteine residues (S−S) formed in oxidizing environments; contribute to protein stability
Histidine (H, HIS):
Polar; imidazole side chain with multiple nitrogens
Can be protonated or deprotonated near physiological pH; charges can flip near pH ~6–7, making it important in enzyme active sites
Positively charged (basic) amino acids:
Lysine (K, LYS): positively charged at physiological pH
Arginine (R, ARG): positively charged; contains guanidinium group, strongly basic
Negatively charged (acidic) amino acids:
Aspartate (D, ASP): negatively charged at physiological pH
Glutamate (E, GLU): negatively charged at physiological pH
The four-group framework for quick memorization (as taught in the transcript):
Hydrophobic (nonpolar)
Polar (uncharged)
Positively charged (basic)
Negatively charged (acidic)
Special notes mentioned in the transcript:
Glycine is the smallest amino acid and is not chiral
Proline’s unique ring system affects protein structure
Tryptophan is large and relatively rare; often found in protein interiors
Cysteine’s SH group enables disulfide bonding
Histidine’s ionization near neutral pH makes it a common catalytic residue
Methionine and cysteine both contain sulfur; methionine is nonpolar and thioether-linked, cysteine has sulfhydryl group
Asparagine and glutamine have amide side chains
Phenylalanine and tyrosine are aromatic; tyrosine adds a polar OH group
The side chain determines much of a residue’s role in folding, stability, and interactions
Practical examples and correlations from the transcript:
MSG = monosodium glutamate (glutamate salt)
Dopamine can be synthesized from phenylalanine or tyrosine; it is a neurotransmitter involved in reward pathways; its structure shows an aromatic ring similar to phenylalanine/tyrosine
Aspartame is a dipeptide composed of aspartic acid and phenylalanine with a methyl ester group; described as a sugar substitute that is much sweeter than sucrose
The lecture emphasizes recognizing the 20 amino acid structures and using one-letter codes in exams (G, A, V, L, I, M, F, W, P, Y, S, T, N, Q, C, H, K, R, D, E)
Learning objectives for this topic (summary of what you should be able to do)
Draw the basic structure of amino acids
Identify the important functional groups in amino acids
Describe the four (or four-plus-two) classes of amino acids based on side chains
Recognize the structure of all 20 amino acids
Understand that the charge of amino acid residues can change with pH
Explain how to calculate pH and pKa values and determine the charge on amino acid groups given pH and pKa
Appreciate how sequence and side chains influence protein folding and function
Understand the concept of amino acid residues, N-terminus vs C-terminus, and how peptide bonds link amino acids into proteins
Recognize that mutations in amino acids can alter protein function, stability, or cause disease
Be familiar with common examples and non-exam but illustrative molecules (e.g., MSG, dopamine, aspartame)
Quick references and memorization tips from the transcript
Glycine (G, GLY): smallest; not chiral
Proline (P, PRO): side chain forms a ring back to the amino group
Tryptophan (W, TRP): large, hydrophobic, least frequent; often buried in protein cores
Methionine (M, MET) and Cysteine (C, CYS): sulfur-containing; cysteine forms disulfide bonds via SH groups
Histidine (H, HIS): ionizable near neutral pH; useful in catalytic roles
Acidic residues (D, E) vs basic residues (K, R, H) and their ionization behavior with pH
Aromatic residues (F, Y, W) and their roles in structure and function
Hydrophobic residues help drive protein folding into a stable core
Polar uncharged residues (S, T, N, Q, Y) can form hydrogen bonds with water and other groups
The four-class framework and how it maps to the four major categories (with Gly and Pro exceptions noted)
Sequence orientation matters: the first residue is at the N-terminus; the last residue at the C-terminus
nomenclature:
Residue vs amino acid distinction: residue is the amino acid within a protein
One-letter code vs three-letter code usage; most exams provide a structure and ask you to identify the residue
Closing notes
The content connects basic chemistry (acid-base concepts) to biology (protein structure and function) and to physiology (metabolic energy changes, energy coupling in cells).
Remember to review ionization of amino acids at different pH values and how to estimate charges using pKa values, as emphasized for topic two.
The material sets up the central dogma and frames why understanding amino acids and proteins is foundational for later topics in metabolism and disease.