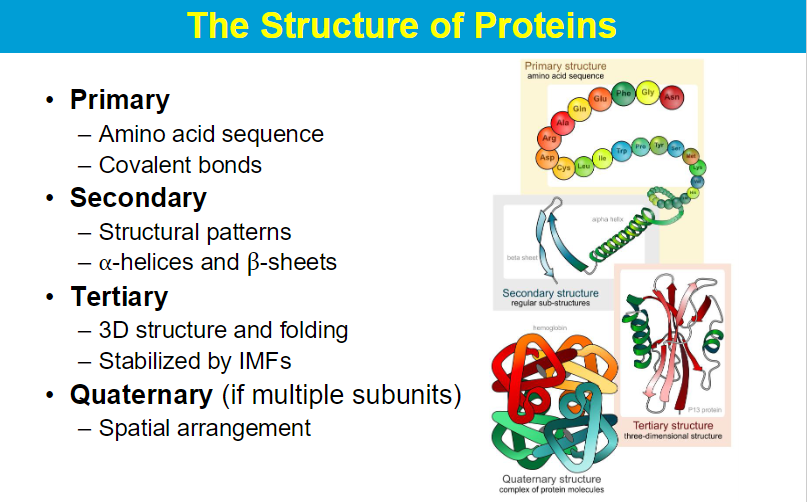
Proteins from Primary to Quaternary Structure
protein size varies*****
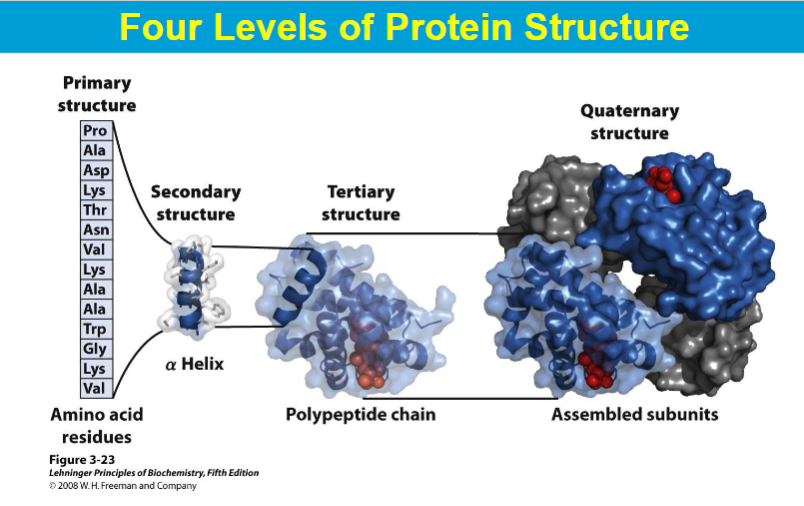
primary structure
structure of the protein is partially dictated by the properties of the peptide bond
the bond is a resonance hybrid
making it rigid and nearly planar
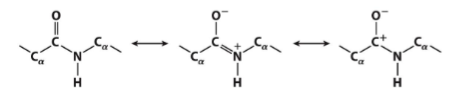
absence of rotation around the C—N bond because of its partial
double-bond character (from resonance)
The sequence and arrangement of amino acids gives the polypeptide a chemical character (i.e., charged, polar, hydrophobic, etc.).
Some polypeptides bind specific targets, which can be used to “fish them out” of a complex mixture. → purification
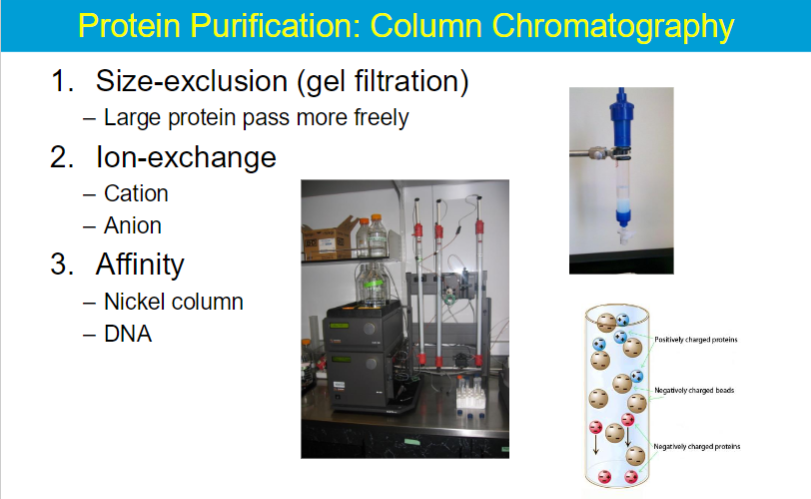
column chromatography
The column is packed with a stationary phase, such as silica gel, aluminum oxide, or macro-porous adsorbent resin. A mobile phase is then passed through the column, separating the compounds based on how they move through the stationary phase.
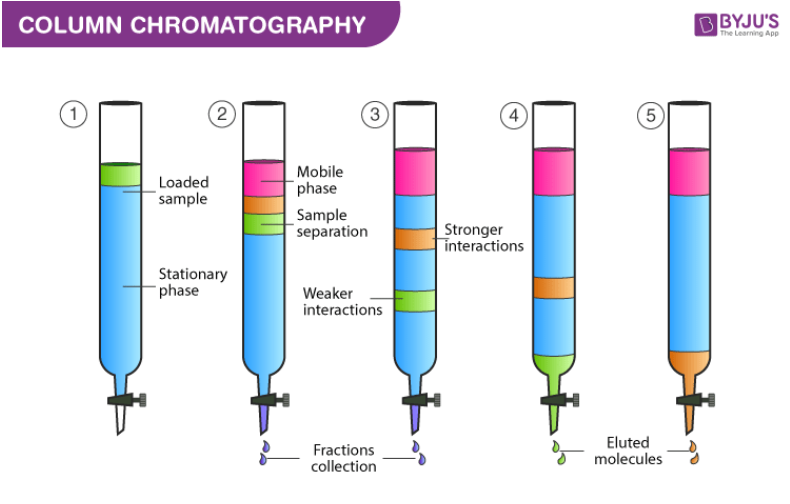
Size-exclusion or gel filtration chromatography separates on the basis of size.
The column matrix is composed of cross-linked polymers with pores of selected sizes. Smaller molecules can enter pores in the polymer beads from which larger molecules would be excluded. Smaller molecules therefore have a larger three-dimensional space in which to diffuse, making their path through the column longer. Larger molecules migrate faster because they pass directly through the column, unhindered by the bead pores
so larger molecules eluted before smaller molecules
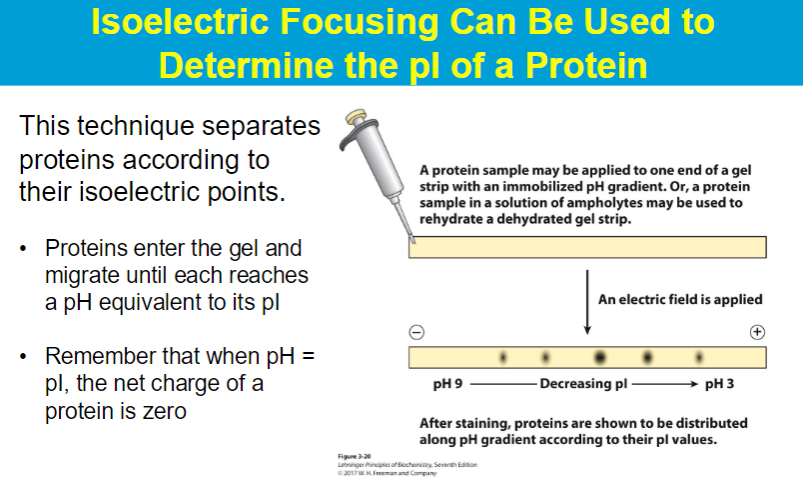
Ion-exchange chromatography separates proteins on the basis of their charges.
ex. if stationary is neg → then positive molecules bind but negative molecules are repelled
so same charge is eluted first
Affinity chromatography separates proteins with specific, high affinity for some ligand (attached to an inert support) from other proteins with no such affinity
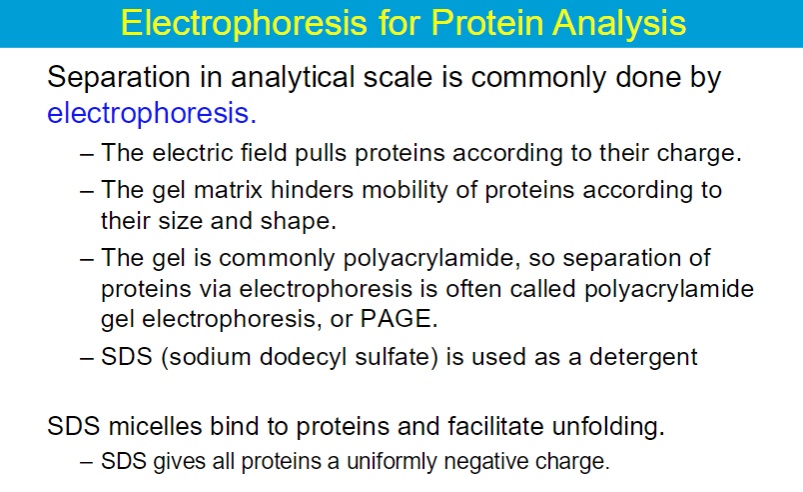
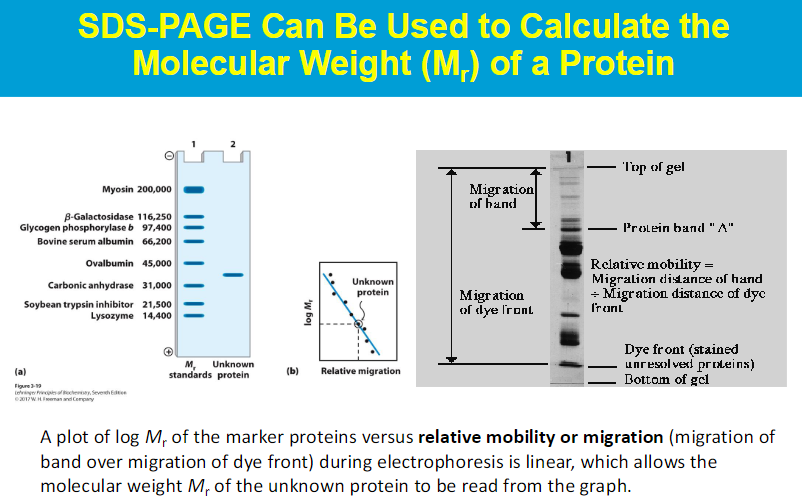
basically SDS is used so that every protein has the same charge, so the mobility of the proteins through the gel is only based on size
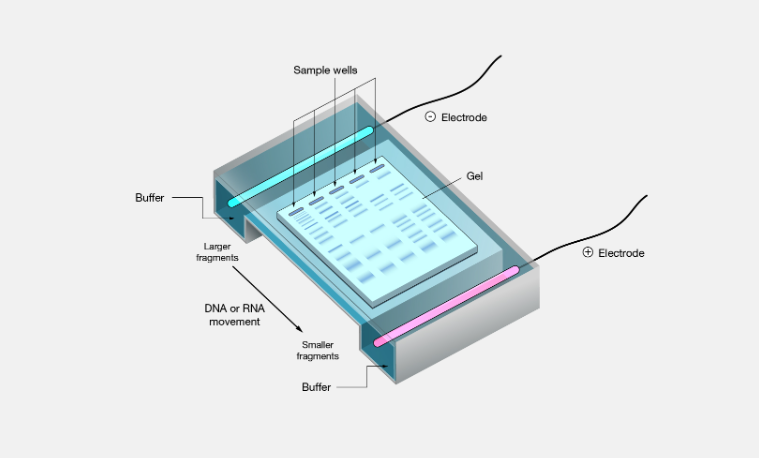
the native shape of proteins does not matter
rate of movement only depends on sizes; small proteins will move faster
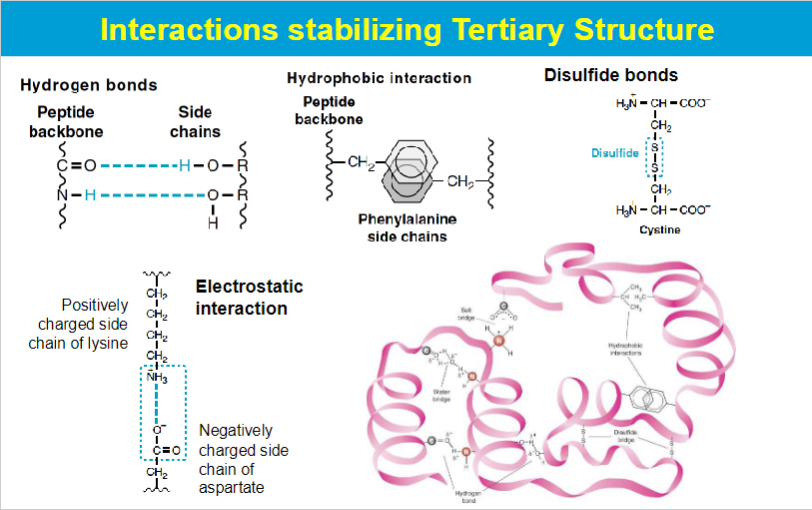
favorable interactions in proteins
hydrophobic effect
aggregation of hydrophobic amino acids (away from water) → this results in the release of structured water molecules (solvation layer). This release increases the net entropy of the system, favoring protein folding and contributing to the stability of the protein's three-dimensional structure.
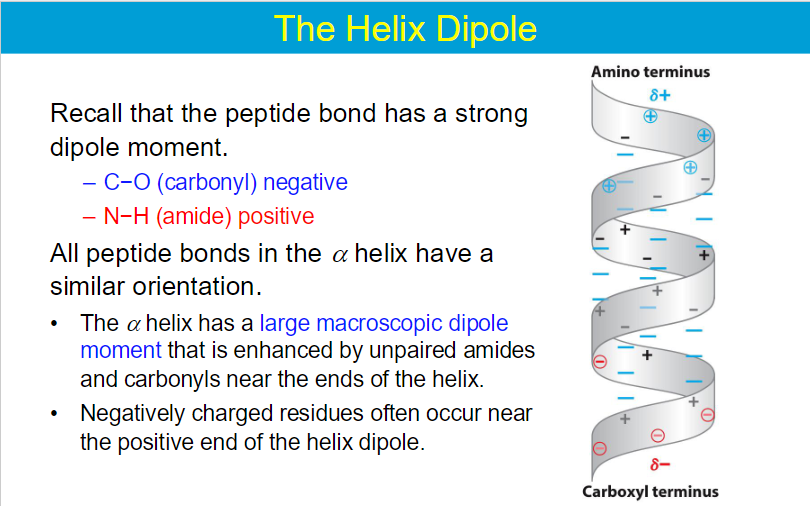
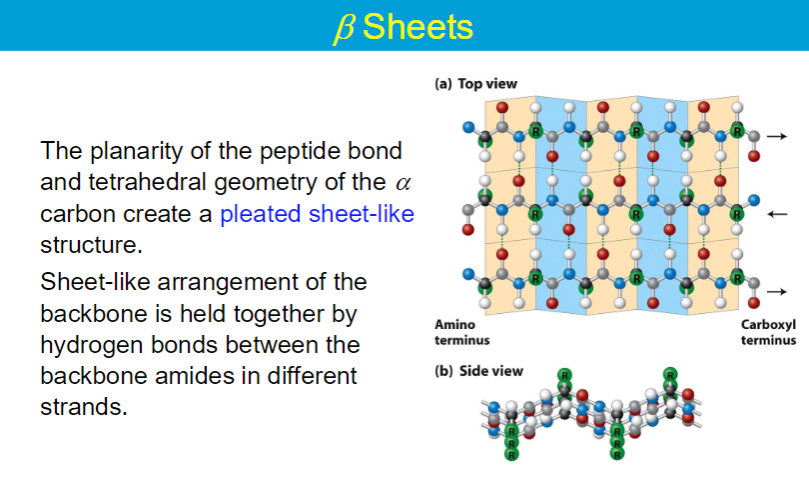
hydrogen bonds: interactions of N-H and C=O in secondary structures
electrostatic interactions:
long-range strong interactions between charged groups
salt bridges
london dispersion
medium-range weak attraction contribute significantly to the stability in the interior of the protein
a molecule's "stability" refers to its tendency to maintain its natural, folded shape ("native conformation"), and the energy difference between a folded and unfolded state of a molecule is typically small, ranging from 20-65 kJ/mol
?? bro idk what this means bruh
When unfolded
Pro: Hydrogen bonds with water
Con: Exposed hydrophobic residues
When folded
Pro: Disulfide bonds, hydrogen bonds, hydrophobic interactions, ion-ion (electrostatic)
Con: Unfavorable entropy
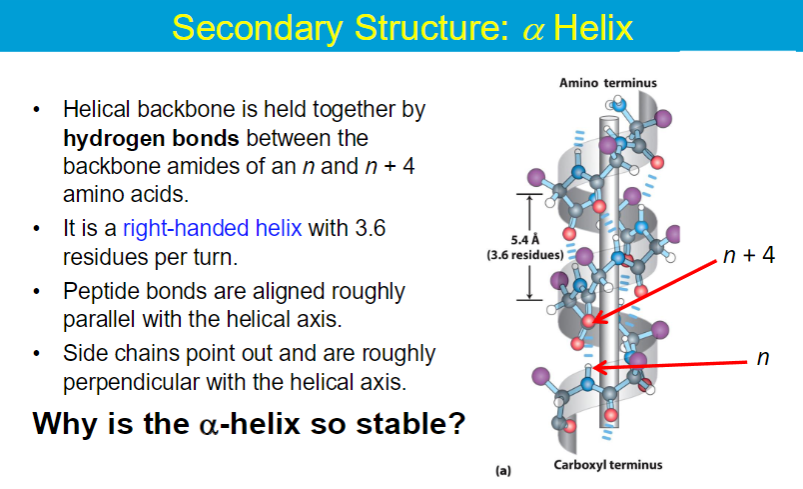
Secondary Structures
α - helix, β - helix
Irregular arrangement of the polypeptide chain is called the
random coil.
small hydrophobic amino acids like alanine (Ala) and leucine (Leu) are considered strong helix formers
meaning they have a high propensity to form alpha helices within a protein structure due to their compact side chains that readily fit into the helical geometry
Proline (Pro) acts as a helix breaker because its unique structure restricts rotation around the N-Cα (phi angle) bond
Glycine (Gly) acts as a helix breaker due to its small side chain which allows for greater conformational flexibility, supporting structures other than a helix.
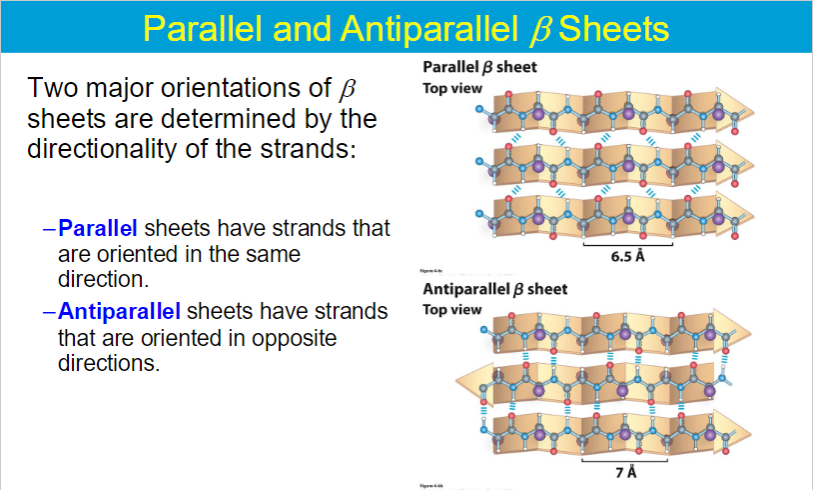
Antiparallel beta sheets have stronger hydrogen bonds compared to parallel beta sheets; this is because the hydrogen bonds in an antiparallel arrangement are more optimally aligned and directly opposite each other, leading to greater stability
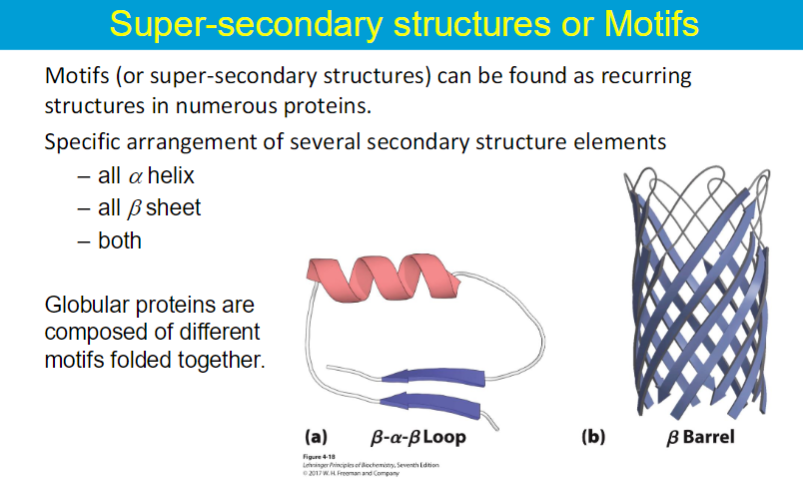
A protein domain is a compact, three-dimensional structure that folds independently from the rest of the protein.
can fold and function as an independent entity from the whole protein
Proteins may consist of several domains
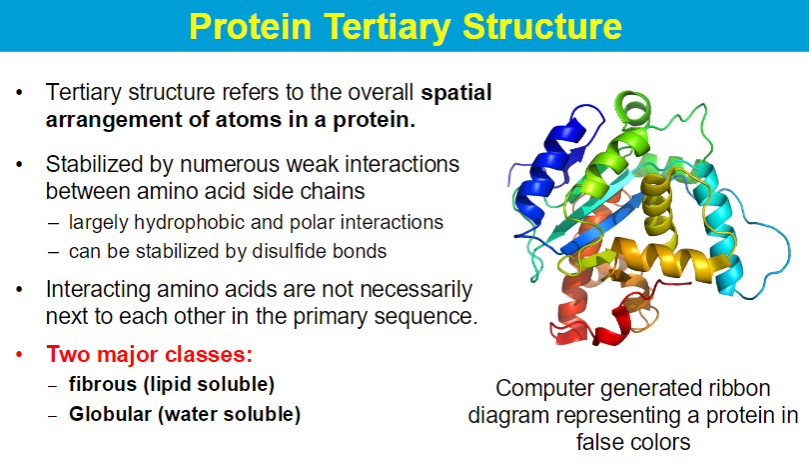
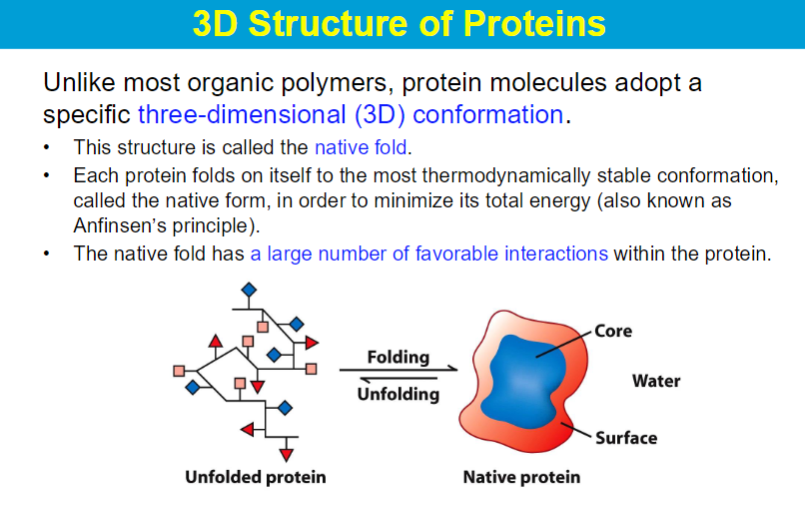
native state has the lowest delta G (lowest energy state)
Globular proteins are proteins that have a compact, spherical shape. They are usually soluble in water and play important roles in cellular processes such as enzymes, hormones, and antibodies.
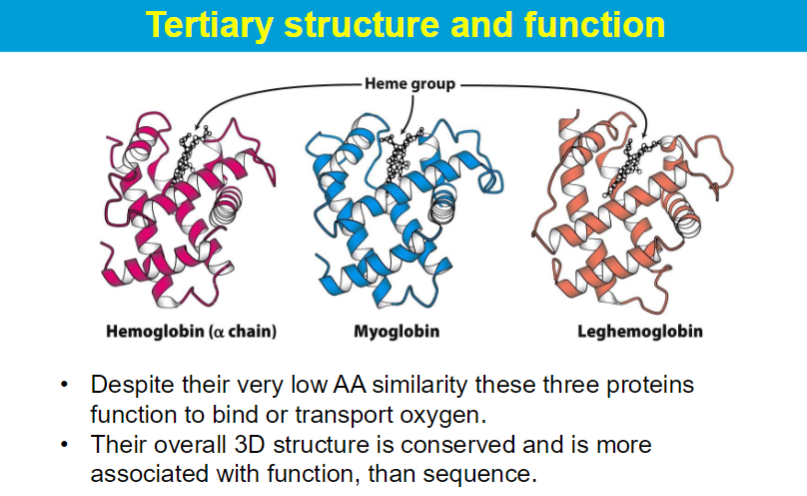
Hydrophobic amino acid residues cluster away from the surface in globular proteins, so much of the protein’s interior is a tightly packed combination of hydrocarbon and aromatic R groups with very few water molecules.
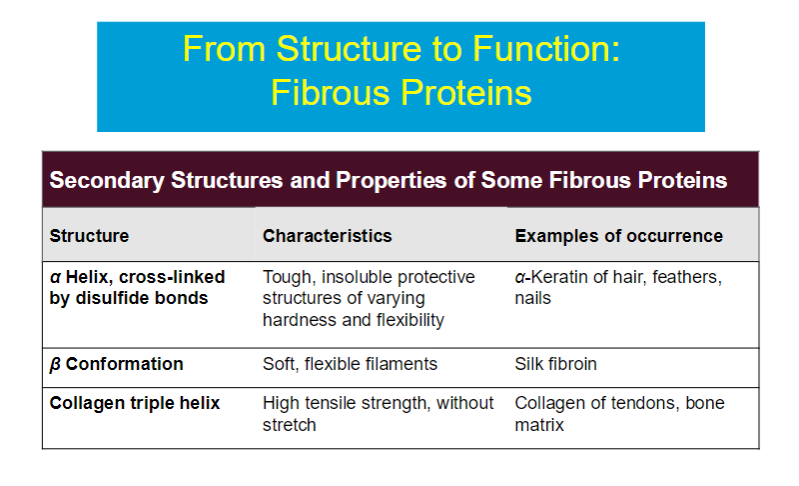
Fibrous proteins are long and narrow or form long, thin structures, and made up of repetitive units, often alpha helices or beta sheets

Used for webs, egg sacks, and wrapping the prey
Extremely strong material
stronger than steel
can stretch a lot before breaking
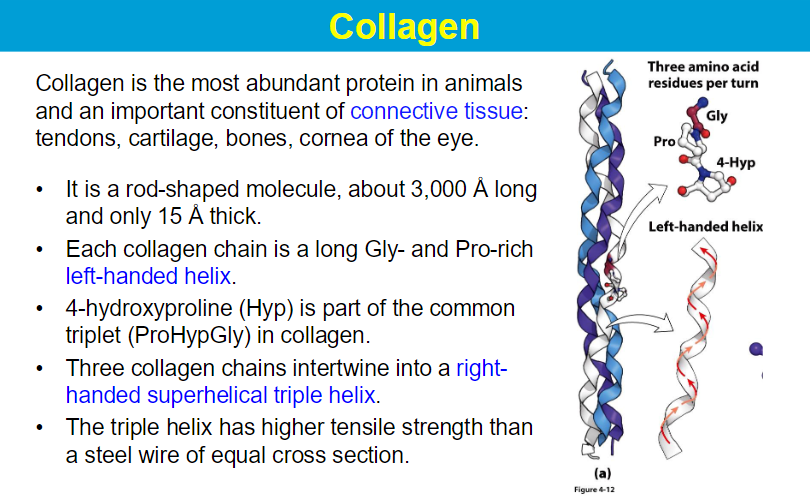
Many triple-helices assemble into a collagen fibril
Collagen superstructures are formed by cross-linking of collagen triple-helices to form collagen fibrils.
Crosslinks are covalent bonds between Lys or His amino acid residues.
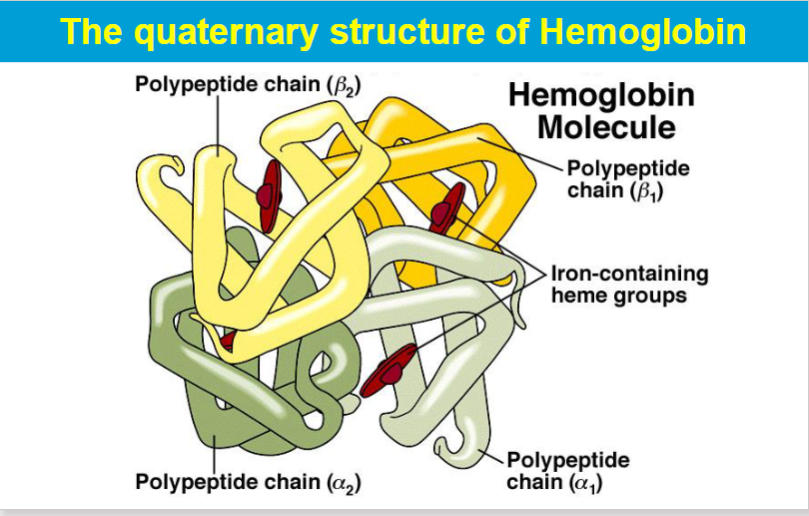
quatenary structure
complex unit composed of 2 or more tertiary structure subunits
subunits= monomers
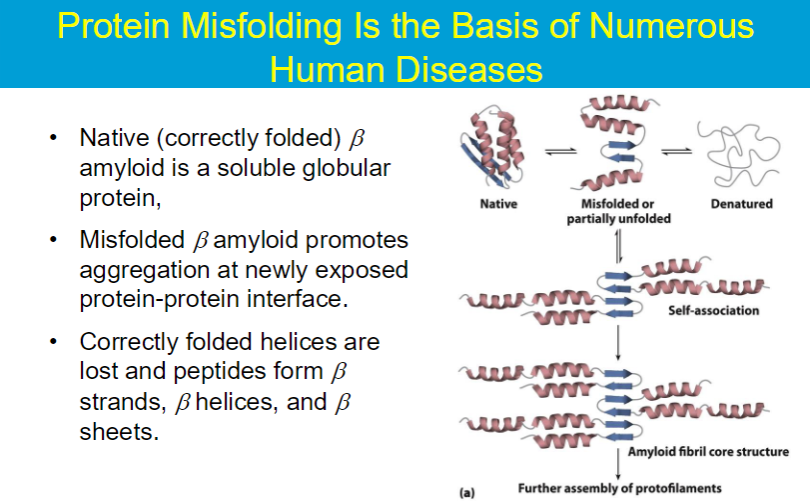
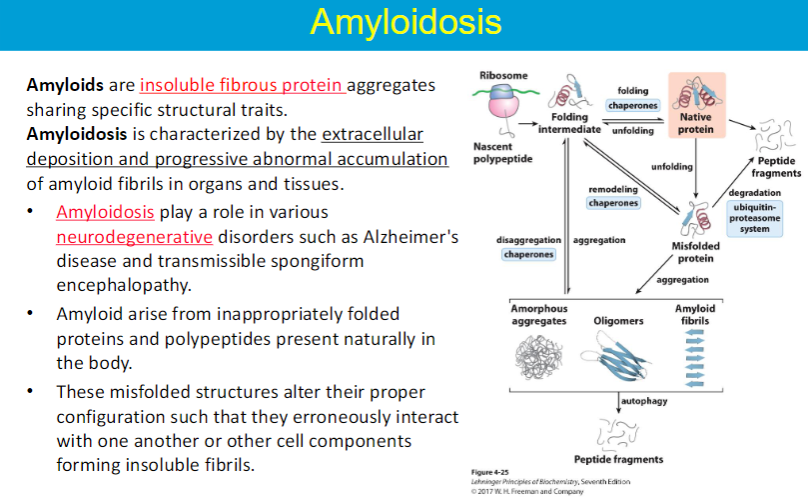
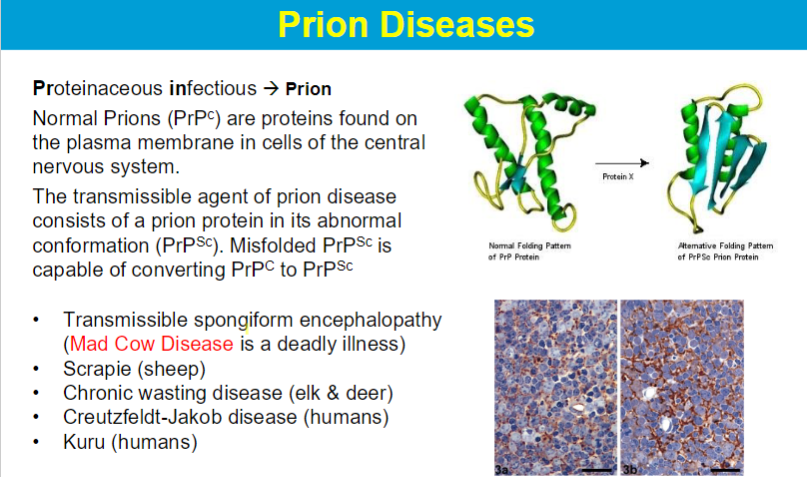
Loss of structural integrity with accompanying loss of activity is called denaturation.
Proteins can be denatured by:
heat or cold
pH extremes
organic solvents
chaotropic agents: urea and guanidinium hydrochloride
molecules that disrupt the hydrogen bonding network between water molecules