Recombinant DNA Technology
Vocabulary
Recombinant DNA Technology: Technology used to recombine DNA in new sequences, uses restriction enzymes and vectors to join DNA from two biological sources
Biotechnology: The application of recombinant DNA Technology
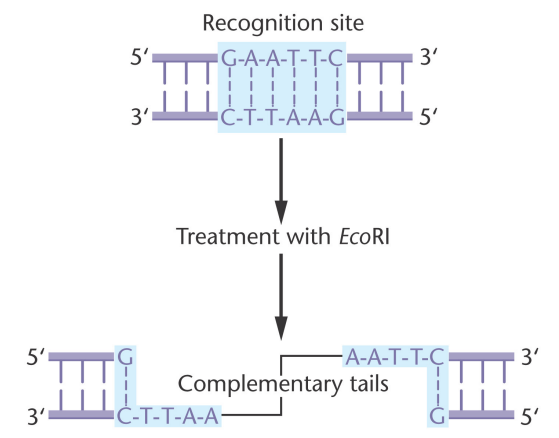
Restriction enzymes: Isolated by Kathleen Danna and Daniel Nathans, come from bacteria and can cleave viral DNA at specific nucleotide sequences. Now known that they create double stranded cuts and can do this to any kind of DNA.
Vector: Something that carries DNA from one organism to another. These provide protection from the host cell, and can make replications of the DNA separate from the host cell
Clone DNA: DNA that has the same sequence as DNA from another source, a host source.
Cloning: The process of making a duplicate of one cell into another, this is done naturally through singled cell mitosis and by hermaphroditic organisms like C. elegans. Monozygotic twins are a pseudo-example of this, same with plants and Dolly the sheep. This process is done in labs often
Embryo splitting: The process of an embryo being bisected very early to produce two half-embryos and then implanted into a surrogate mom, like with monozygotic twins
Nuclear transfer: One cell has the nucleus removed and a new nucleus is implanted from a donor cell, done to an egg. It is then provided nutrients to go through embryogenesis to fully develop, can be done with somatic cells to undifferentiate them
Restriction site: The sequence where a restriction enzyme will bind and make a double stranded break
Sticky ends: A complimentary overhang created by a restriction enzyme
Blunt ends: Created by restriction enzymes when they make double stranded breaks
Palindromic sequence: A sequence that reads the same forward and backwards
Plasmid: An extrachromosomal double-stranded DNA molecule, replicates independently from the chromosome within bacterial cells, often exist in multiple copies in the cytoplasm. In cloning, can be engineered to have a number of convenient restriction sites, and a marker gene to select for presence in host cell
Transformation: How plasmids are introduced into bacteria, genetically alters cells through direct uptake of incorporation of exogenous genetic material. Either uses heat shock of electroporation which has holes in the cell wall which plasmids pass through
Selectable marker genes: Something that will show if the process of DNA hybridization was successful, does things like provide resistance to antibiotics, overall indicates if transformation was successful
Blue-white selection (reporter gene): Used to identify cells containing recombinant and nonrecombinant DNA, a plasmid contains the lacZ gene, which encodes beta-galactosidase. If lacZ is nonfunctional, the process was successful
LacZ: A gene only expressed in the presence of lactose, turned on all the time via IPTG which disrupts the hydrolysis of this gene. Instead IPTG will hydrolyze something else which will turn a colony of bacteria white
Bacterial plasmid: The main vector, first developed, limited by their ability to only accept 15kb to 25kb
Lambda Phage vectors: A vector that comes from a double-stranded DNA virus, up to 45kb of cloned DNA and still able to infect cells and replicate, kills the host cell when it replicates but leaves a plaque with the new DNA
BACs: A vector, can carry 100kb to 300kb, can only make 1 or 2 replicates
YACs: Vector for 230kb to 2Mb, have telomeres, ORI, and centromeres like a natural chromosome
Expression vector: A kind of vector that expresses given DNA, undergo transcription and translation of genes, available for prokaryotes and eukaryotes. Restriction sites are downstream of promoter sequence
Rhizobium radiobacter: A vector used in plant cells, infects plant cells and produces tumors, contains the plasmid Ti (tumor-inducing plasmid) which will permanently integrate into the plant genome
T-DNA: Ti plasmid replacement in a lab, has a selected gene that will be integrated into a plant’s genome
Saccharomyces cerevisiae: Vectors for eukaryotes, easily manipulated with a fully sequences genome, undergoes posttranslational modifications, considered most safe
DNA Library: A collection of cloned DNA for reference, changes based on lab/organization/project
Genomic library: A DNA library with at least one copy of all sequences of a genome in interest, constructed by cutting genomic DNA with restriction enzymes and ligating fragments into vectors
Complementary DNA (cDNA): Contains complementary DNA copies made from mRNAs present in cell populations, represents genes active transcriptionally at the time cells were collected for mRNA isolation
Oligo-DT: A short primer of 10-20 T bases, provides a substrate for reverse transcriptase to make a DNA template by binding to the poly-A tail
RNAase: Digests RNA, particularly in the process of making cDNA
Probe: Used to screen a library and recover clones of specific genes, can be any DNA/RNA sequence complementary to the target gene being identified
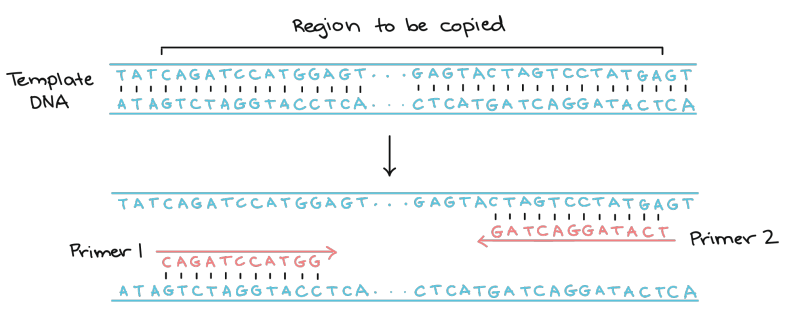
Polymerase chain reaction (PCR): Developed in 1986 by Kerry Mullins, allows for the amplification of specific DNA sequences in vitro, its a rapid method of DNA cloning, close to cloning the host cell. There are many cycles of varying temperatures to denature, anneal, and extend DNA
Taq Polymerase: Used in PCR, a heat stable DNA Polymerase, mimics the machinery of a cell undergoing division
DNA Primer: A short single stranded DNA sequence (20 bases), one complementary 5’ end and another complementary 3’ end of the template
RT-PCR: PCR with reverse transcriptases, cDNA produced and then PCR is done, done with genes that are RNA not DNA
qPCR: Uses due containing probe of DNA-binding dye to label templates. Light emitted by these marked dyes correlate to the amount of amplified DNA product. Examples are SYBR Green and TaqMan. Detects and quantifies amounts
Restriction mapping: Characterizes the sequence and the structure of a segment of DNA by identifying the specific locations of the restriction enzyme cut sites. Cuts DNA with various restriction enzymes and then undergoes electrophoresis
Southern Blotting: A method that detects a specific DNA sequence in a mixed DNA sample, used in detecting DNA from a cDNA library, analysis of long DNA stretches hard to analyze by PCR, and blood/tissue samples. DNA is cut by a restriction enzyme, and segments are separated by gel electrophoresis and transferred to a membrane substrate. It is then denatured into ssDNA, and hybridized with a probe which is then washed off. It is exposed to X-ray film or a camera, and film is developed via computer analysis, patterns are analyzed
Northern Blotting: Used to determine whether a gene is actively being expressed in a given cell/tissue, as to study patterns of gene expression in embryonic/cancerous/disordered tissue
Fluorescent in situ hybridization (FISH): Involves hybridizing probe directly to chromosome or RNA without blotting, carried out with isolated chromosomes on slide of in situ in tissue sections or entire organisms. Helpful when embryos are used for various studies in developmental genetics, can identify which cell types in an embryo express different genes during specific stages of development
Western Blotting: Used for analyzing proteins
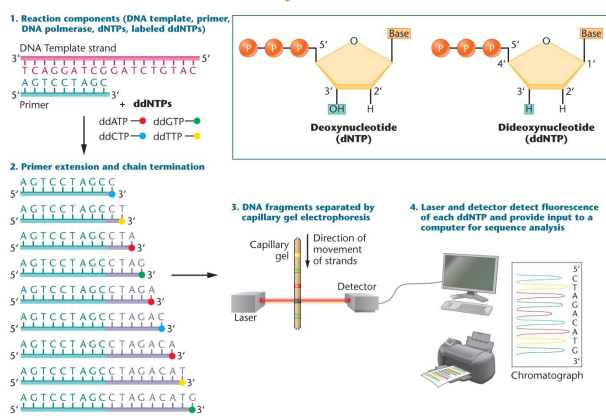
Dideoxynucleotide chain-termination sequencing (Sanger): Most common method of DNA sequencing, uses fluorescent labeled ddntps to stop DNA synthesis. There are dntps and ddntps which are incorporated into a DNA sequence. This is run through electrophoresis and there is eventually a strand ending at each, a computer reads each and provides a genetic sequence
Dideoxynucleotide (ddntp): A dntp with a hydrogen at the 3’ carbon instead of an -OH
3rd Generation sequencing: Automated, uses fluorescent dNTPs, sequences a single molecule of ssDNA. When the phosphate is cleaved, it will shine a color which will be detected by a computer
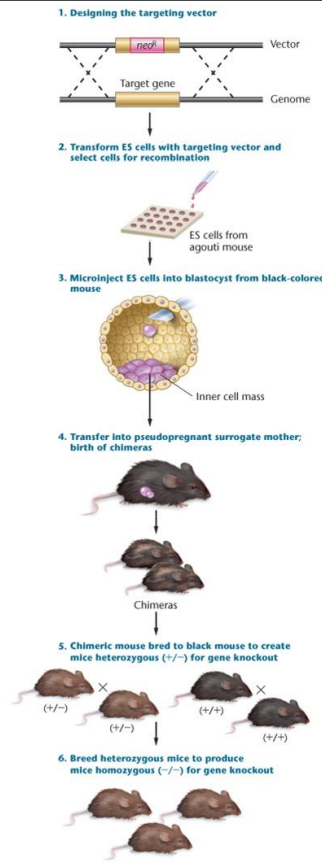
Knockout model: Used to show when specific gene changes, used for in-organism examples, widely used in mice and other model organisms. A target vector introduces DNA through homologous recombination, the new gene is non-functional and causes the mouse to die. Recombinants that grow in neomycin get the ES implanted into a mother placenta
Embryonic stem cells (ES): Used in knockout models, the kind of cell that gets transformed by the vector
Recombinase: Recognizes new parts of DNA in an ES, catalyzes homologous recombination in a small percentage of cells
Neomycin: Where new cells are grown, specifically the ES in knockout models
Chimera: The mouse given birth to with half the mother’s DNA and half the DNA of the recombinant
Conditional knockouts: A knockout organism where the gene isn’t always expressed at all points in life, or is only expressed in specific cells
Transgenic organisms: When genes are transferred between unrelated species, can have a single gene being overexpressed
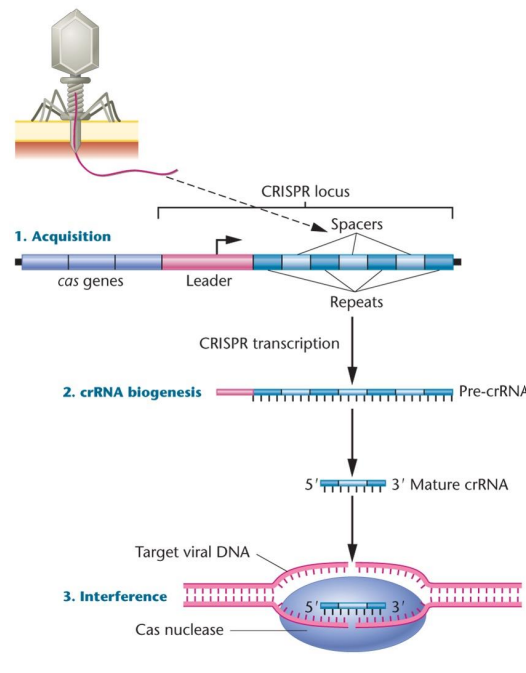
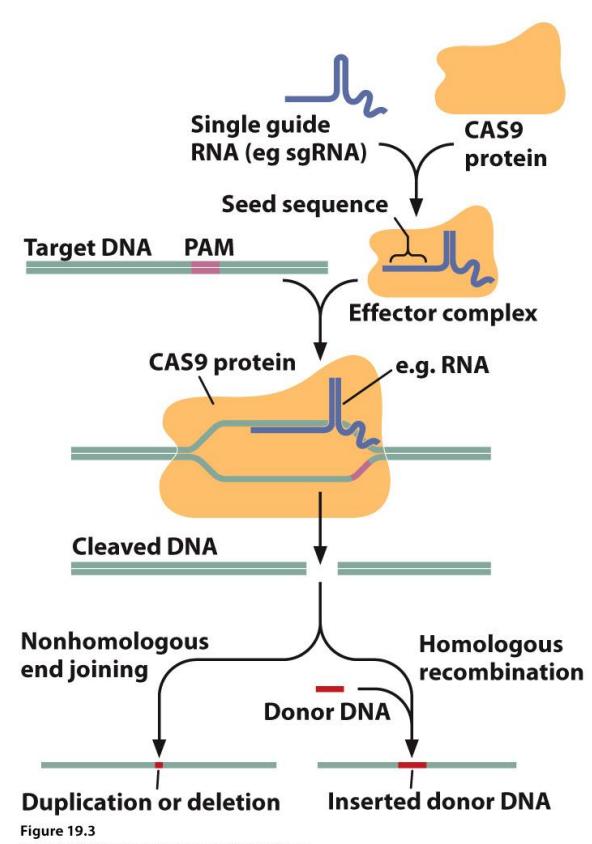
CRISPR-Cas9: Doesn’t introduce any new genetic information, allows for genetic modification and rearrangement of bases within an organism. There are palindromic repeats, and parts of actual sequencing between. An adaptive immune system
To combine DNA, it is treated with a restriction enzyme that will create sticky ends, that will then associate with other similar sticky ends on other DNA molecules with complimentary ends, which will then be sealed by DNA Ligase. Then, the new recombinant DNA is introduced to a bacterial host cell via transformation
To clone a genome, oligo-DT is used, followed by reverse transcriptase to synthesize a DNA strand, followed by RNAase to dissolve the template strand, and then a DNA polymerase to complete the strand
Vectors have the following properties:
Can replicate cloned DNA fragments in a host cell
Can replicate independently of a host cell
Have several restriction enzyme sites to allow insertion of DNA fragments
Carry a selectable gene marker to distinguish host cells that have taken them up from those that have not or a reporter gene that encodes a protein which produces a visible effect
PCR requires 2 primers, which anneal to the denatured DNA and is then built off of by Taq. Steps are as follows:
Denaturation at 92-95 degrees
Primer annealing/hybridization at 45-65 degrees
Extension at 65-75 degrees
The steps are repeated many times using a thermocycler to amplify DNA exponentially. DNA is always double stranded. New strands as well as old strands serve as templates for the next cycle.
Limitations of PCR include its reliance on knowledge of target DNA sequences in order to make a primer, the fact that minor contamination from other sources can cause problems (like skin cells from researchers) and that PCR cannot amplify long segments of DNA
PCR applications include cloning, DNA sequencing, detection of targeted sequences, use with allele specific probes to study/detect genetic disorders, forensic analysis, comparative evolution and ecological studies between species, and locate/determine the nature of genetic conditions very fast
Instead of cross breeding Chimera/F1 mixes, DNA can be implanted into the mother to get knockout organisms. It isn’t as efficient, but is more accurate.