lecture 8: G protein coupled receptors
signalling recap
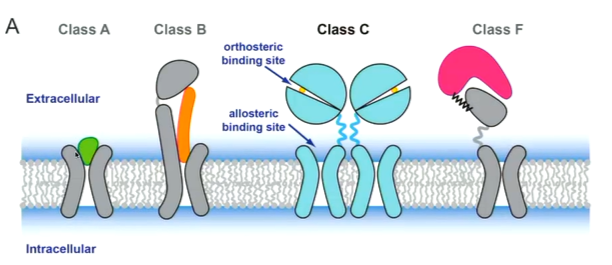
GPCRs are a large family of membrane proteins that have 7 transmembrane helices and are involved in signalling. This makes them very good drug targets. They can be subdivided:
A - rhodopsin like
B - secretin like
C - metabotropic glutamate receptor mGluR like
F - frizzled
there is another system that subdivides the GPCRs differently into glutamate, rhodopsin, adhesion, frizzled and secretin. The rhodopsin like receptors are most common type. Class A structures are the simplest with a 7 TM helix and the ligand binds to the outside. Class B bind peptides and have a larger section at the N terminus. Class C are more complicated with a dimer and instead, ligands bind to the orthosteric binding site. It also has an allosteric binding siye. Class F are involved in protein protein interactions.
As the name suggests, they signal through G proteins. The signalling molecule binds to the receptor and the G protein interacts with the interaction and this causes the subunit to dissociate. The alpha subunit can activate enzymes such as adenylate cyclase which can convert ATP to cAMP to cause a downstream affect. In a resting state, GPCRs have some basal activity and this can be modulate by ligands. Agonists binds to the protein and switches the conformation of the protein to the active state. The inverse agonist changes the conformation to an inactive protein. the antagonist will not change the conformation to prevent other molecules binding.
G proteins are guanine nucleotide binding proteins and are heterotrimeric proteins with an alpha subunit that binds GTP as well as a beta and a gamma subunit. The alpha subunit is built of two domains of a Ras like domain and the nucleotide binds ta the nucleotide binds at the interface. The beta subunit falls into the beta propeller like structure the gamma subunit is one helix chain helix motif.
The GDP bound g protein is inactive and the binding of the agonist to the GPCR will change the conformation of the protein and this allows the G protein to bind which also changes conformation. This means the alpha subunit opens to release GDP and GTP can return The GPCR is known as a GTP exchange factor. When this happens, the subunits interactions are broken and so the alpha subunit is released to interact with other enzymes such as adenylate cyclase and the beta domain can interact with enzymes or transporters such as the calcium channel. This remains unit the GTP is hydrolysed.
There are many different types of subunits that do different things. You ca have an inhibitory protein - Gai - that inhibits enzymes such as adenylate cyclase. Other subunits work through phospholipase C.
history
rhodopsin
rhodopsin was the first crystal structure of a GPCR that was solved and also have a 7 TM helix structure. It is the photoreceptor of the retinal rod cell and acts through the G protein transducin. It binds covalently to the ligand and the 11 cis retinal binds to the lysine residue which activates the protein. This is a strong inverse agonist. It was noticed that there is a DERY motify and an ionic lock. This motif is seen a lot in GPCR structures what happens is the arginine interacts with the glutamate to bridge helix 3 and 6 to hold it in an inactive conformation.
11 cis retinal keeps the GOCR in an inactive state and when the protein is activated, it absorbs a protein and changes the conformation of the retinal to an all trans retinal which activates the protein. All trans retinal acts as an agonist.
They solved the structure of the rhodopsin without the ligand - opsin. This resembled the activated form of the GPCR. tHERE IS MOVEMENT OF tM 5 and 6. tOPOLOGICALLY, tm 5 AND 6 IS WHERE THE INTRACELLULAR LOOP 3 IS FOUND WHICH IS KNOWN TO INTERACT WITH THE g PROTEIN. . the movement of 5 and 6 corresponds to where the G protein should be binding.
when they look at the ionic lock, with opsin, the arginine does not interact with the glutamate and instead interacts with the tyrosine so is no longer holding the helices together. this is corresponding to what was thought to happen. To work out how it interacts with the G protein, they used a peptide corresponding to the part that is known to bind to the rhodopsin. They made a complex of the peptide and the protein and solved the structure of opsin with the peptide. They could see roughly how the g protein interacts with the opsin. Arg 135 interacts with the peptide when the ionic lock brakes.
This was the initial idea of activation of GPCRs. Since then, the structure with the transretinal has been solved. Now, they have cryo-em structure of human rhodopsin bound to an inhibitory G protein.
b-2 adrenergic receptor
This is a receptor of noradrenaline involved in the regulation of cardiovascular and pulmonary function. Two papers published in relation to finding the structure of a GPCRs with a non covalently bound ligand. Both papers, stemmed from the same person but collaborations with different groups and produced different structures one with a Fab fragment at 3.4 angstrom and one with an engineered GPCR at 2.4 angstroms.
there were some obstacles that were overcome such as the production of the protein which were produced in insect cells. There was also stabilization of domains by using an inverse agonist.
T (flexible at TM3 where GPCR binds). T4 lysozyme was found to have its N and C terminus to be the same distance as TM 5 and 6 in the protein. All the contacts in the crystal lattice were therefore through the T4 lysozyme. They observed similar structure to rhodopsin with the major difference being the ligand binding site being more open than in rhodopsin. This makes sense as the ligand also has to detach here. This is the structure of the inactive structure. When activated, they added agonist and saw that structures were more difficult to get as they are conformationally flexible. They stabilized this using a nanobody that mimics the GPCR. They found a probe that bound to TM6 and this was fluorescent and so fluorescence depended on the environment. They had this probe and tried different nanobodies and selected one that gave the structure that could resemble the structure of the active structure. The made the assumption that the nanobody structure that gave the structure with the G protein bound. There are conformational changes on activation. When compared to the structure with the pseudo activated structure with the inactivated structure, it can be seen that there are more changes to the cytoplasmic phase such as the outward displacement of TM 5 and 6 and an inward displacement of TM 3 and 7.
The ligand binding site is seen to change as packing interactions change which changes the conformations. If you compare to the activated and the pseudo activated structure of rhodopsin, it is seen they are similar.
solving the structure of the G proteins as difficult as they had to prepare the complex and this is difficult as it is a membrane protein, G protein is unstable as well as the complex. Techniques to overcome this is using a high affinity agonist to keep it in the activated conformation. They added both G protein and GPCR and mixed the, They also added a pyrophosphate to hydrolyse GDP released from complex. As well as this, they also found the most stabalising detergent.
Crystallizing the protein is difficult as the detergent can mask the protein and Gas was flexible in the absence of the nucleotide. To overcome this, they added t4 lysozyme to make it bigger at the N terminus as well as nanobodies and used LCP method for crystallization.
The complex observed showed that the active and inactive conformations had changes in TM5 and 6 and changes observed with the nanobody were similar when they got the full complex. There were differences as well such as where the G protein bound as the interaction between the G protein and GPCR are different. The ideas from the nanobody were similar. They learnt that interaction is only with the Gas subunit. The G protein and Gas helix 5 binds to a groove in the GPCR and this change the conformation of A5 and results in a structure which opens up one of the domains to release the GDP to allow activation of the subunit.
There are many structures of GPCRs and G proteins or other ligand or proteins. The structures of GPCRs with X ray are increasing but so are cryo-em
GABAb receptor
GABAb is a family C GPCR and is a heterodimer. It is the receptor for the neurotransmitter GABA. GAB has two different receptors -GABAa and GABAb where GABA a is an ion channel and GABAb is a GPCR. There is an obligate dimer and the “venus fly trap” domain bind to the agonist or antagonist at the GB1 domain. The G protein only binds to GB2. The protein was purified in mammalian cells and solved the inactive structure by Cryo-em and saw the C terminal end was floppy. this tail at the C terminus was removed to produce a new protein and the active structure was solved. The inactive structure was bound to an agonist. The active state structure was also there with the agonist bound with a positive allosteric modulator which pushes the conformation to the active form. Local changes in the VFT domain changed the structure and conformation of the TM region. This is transmitted through a stalk region which transmits to the GB2 domain. The proposed model of activation is different to that in class A GPCRs as here TM6 confirmation is the same but is important in dimer conformation as it moves due to the change in conformation which allows the G protein to bind. TM6 doesn’t really change but the interaction changes.