Unit 3: Cellular Energetics
Cell Energy
Energy: the capacity to cause change
Metabolism: the chemical reactions occurring within a living organism
a metabolic pathway begins with a specific molecule and ends with a product
Catabolic pathways (Catabolism): release energy by breaking down complex molecules into simpler compounds
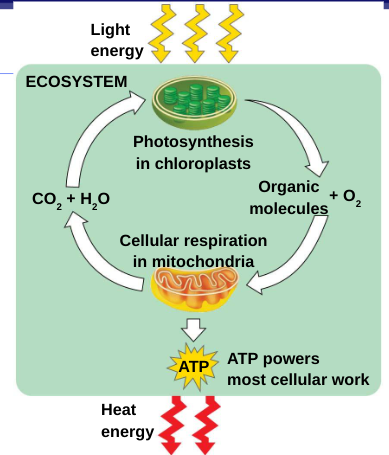
cellular respiration, the breakdown of glucose and other organic fuels to carbon dioxide and water
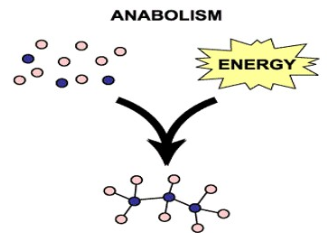
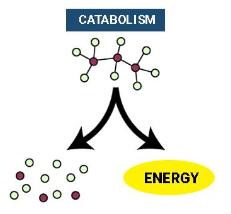 Anabolic pathways (anabolism): consume energy to build complex molecules from simpler ones
Anabolic pathways (anabolism): consume energy to build complex molecules from simpler ones
the synthesis of proteins from amino acids
photosynthesis
 A living system’s free energy (ΔG) is energy that can do work when temperature and pressure are uniform, as in a living cell
A living system’s free energy (ΔG) is energy that can do work when temperature and pressure are uniform, as in a living cell
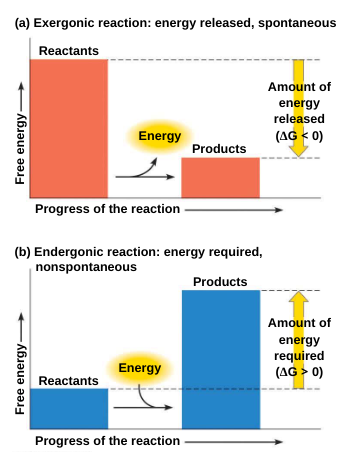
The change in free energy (ΔG) during a chemical reaction is the difference between the free energy of the final state and the free energy of the initial state
ΔG = G (final state) - G (initial state)
Only processes with a negative ΔG are spontaneous
Spontaneous processes can be harnessed to perform work
Enzyme Functions and Pathways
Steps along the pathway (small picture):
Exergonic reaction: a reaction with a net release of free energy and is spontaneous; ΔG is negative
Endergonic reaction: absorbs free energy from its surroundings and is nonspontaneous; ΔG is positive
 Catalyst: a chemical agent that speeds up a reaction without being consumed by the reaction
Catalyst: a chemical agent that speeds up a reaction without being consumed by the reaction
Enzyme: a catalytic protein (a protein that speeds up a reaction without being consumed by the reaction)
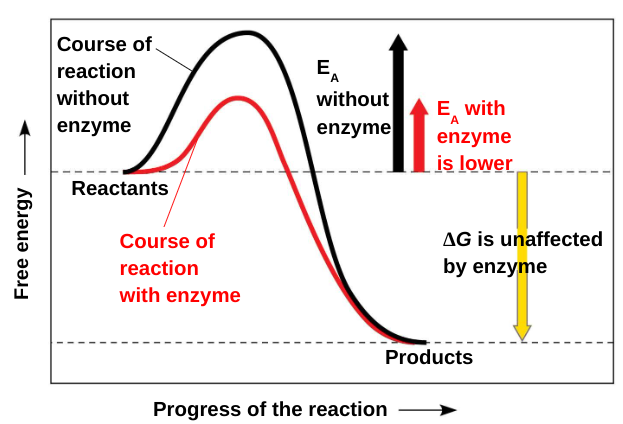
Activation energy: The initial energy needed to start a chemical reaction. Sometimes call the free energy of activation
A catalyst (for example, an enzyme) can speed up a reaction by lowering the activation energy without itself being consumed.
 Substrate: the reactant that an enzyme acts on is called the enzyme’s substrate
Substrate: the reactant that an enzyme acts on is called the enzyme’s substrate
Active site: the region on the enzyme where the substrate binds
The enzyme binds to its substrate, forming an enzyme-substrate complex
Induced fit: enzymes change shape due to chemical interactions with the substrate and the active site
The active site can lower an EA barrier by;
Orienting substrates correctly
Straining substrate bonds
Providing a favorable microenvironment
Covalently bonding to the substrate
The rate of enzyme catalysis can usually be sped up by increasing the substrate concentration in a solution.
When all enzyme molecules in a solution are bonded with substrate, the enzyme is saturated
At enzyme saturation, reaction speed can only be increased by adding more enzyme.
Competitive inhibitors: bind to the active site of an enzyme, competing with the substrate
Noncompetitive inhibitors: bind to another part of an enzyme, causing the enzyme to change shape and making the active site less effective
Allosteric regulation: may either inhibit or stimulate an enzyme’s activity. This occurs when a regulatory molecule binds to the enzyme at one site and affects the protein’s function at the active site. (on/off switch)
Cofactors: non protein enzyme helpers (not from the cell)
Coenzyme: an organic cofactor (enzyme helper)(protein- made in the cell)
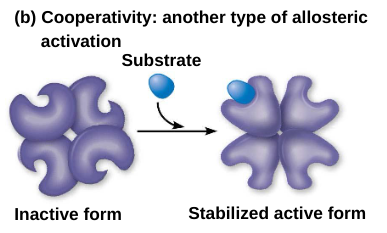
Cooperativity: a form of allosteric regulation that can amplify enzyme activity.
Cooperativity is allosteric because binding by a substrate to one active site affect catalysis in a different active site
In feedback inhibition, the end product of a metabolic pathway shuts down the pathway.
ATP and Cellular Energy
Adenosine Triphosphate:
modified nucleotide
nucleotide = adenine + ribose + phosphate → AMP
AMP + Phosphate → ADP
ADP + phosphate → ATP
ATP → ADP
ATP releases a phosphate group in a spontaneous reaction that can be harnessed to do work
releases energy
ΔG = -7.3 kcal/mol
1 mol of ATP = 7.3 kcals
Organisms can’t store ATP
good energy donor, not good energy storage
too reactive
transfers phosphate too easily
only short term energy storage
carbohydrates and fats are long term energy storage
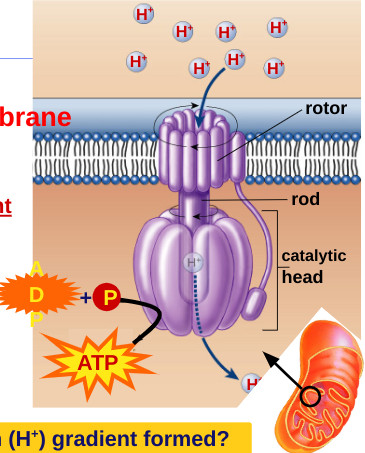
ATP synthase
enzyme channel in mitochondrial membrane
permeable to H+
H+ flow down concentration gradient
flow like water over water wheel
flowing H+ causes change in shape of ATP synthase enzyme
powers bonding of phosphate to ADP: ADP + Phosphate → ATP
 Fermentation (anaerobic): a partial degradation of sugars that occurs without oxygen
Fermentation (anaerobic): a partial degradation of sugars that occurs without oxygen
Aerobic respiration: consumes organic molecules and oxygen and yields ATP
Cellular Respiration
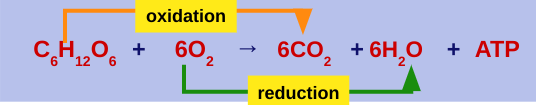
Oxidation-reduction reactions (redox reaction): chemical reactions that transfer electrons between reactants are called redox reactions
Oxidation: a substance loses electrons, or is oxidized (C6H1206 → NADH)
Reduction: a substance gains electrons, or is reduced (the amount of positive charge is reduced) (NADH +02 → NAD+ + H2O)
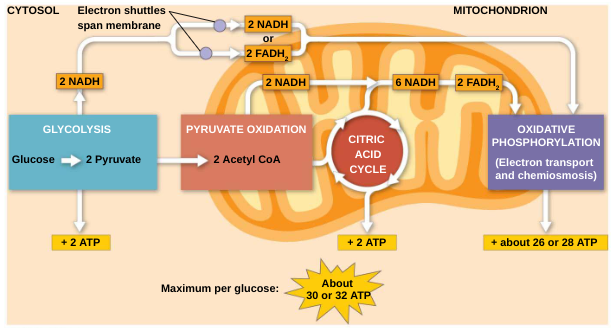
Harvesting of energy from glucose has three stages:
Glycolysis breaks down glucose into two molecules of pyruvate in the cytosol (outside mitochondria)
Pyruvate oxidation and the citric acid cycle (Krebs cycle) completes the breakdown of glucose in the mitochondrial matrix
Oxidative phosphorylation accounts for most of the ATP synthesis and occurs in the inner membrane of the mitochondria (powered by ETC)
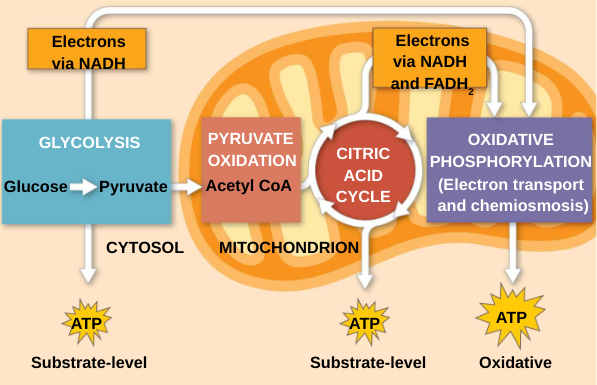
Glycolysis
Glycolysis (“sugar splitting”) breaks down glucose into two molecules of pyruvate
Glycolysis occurs in the cytoplasm and has two major phases — doesn’t happen in mitochondria
The net energy yield is 2 pyruvate, 2 ATP, plus 2 NADH per glucose molecule
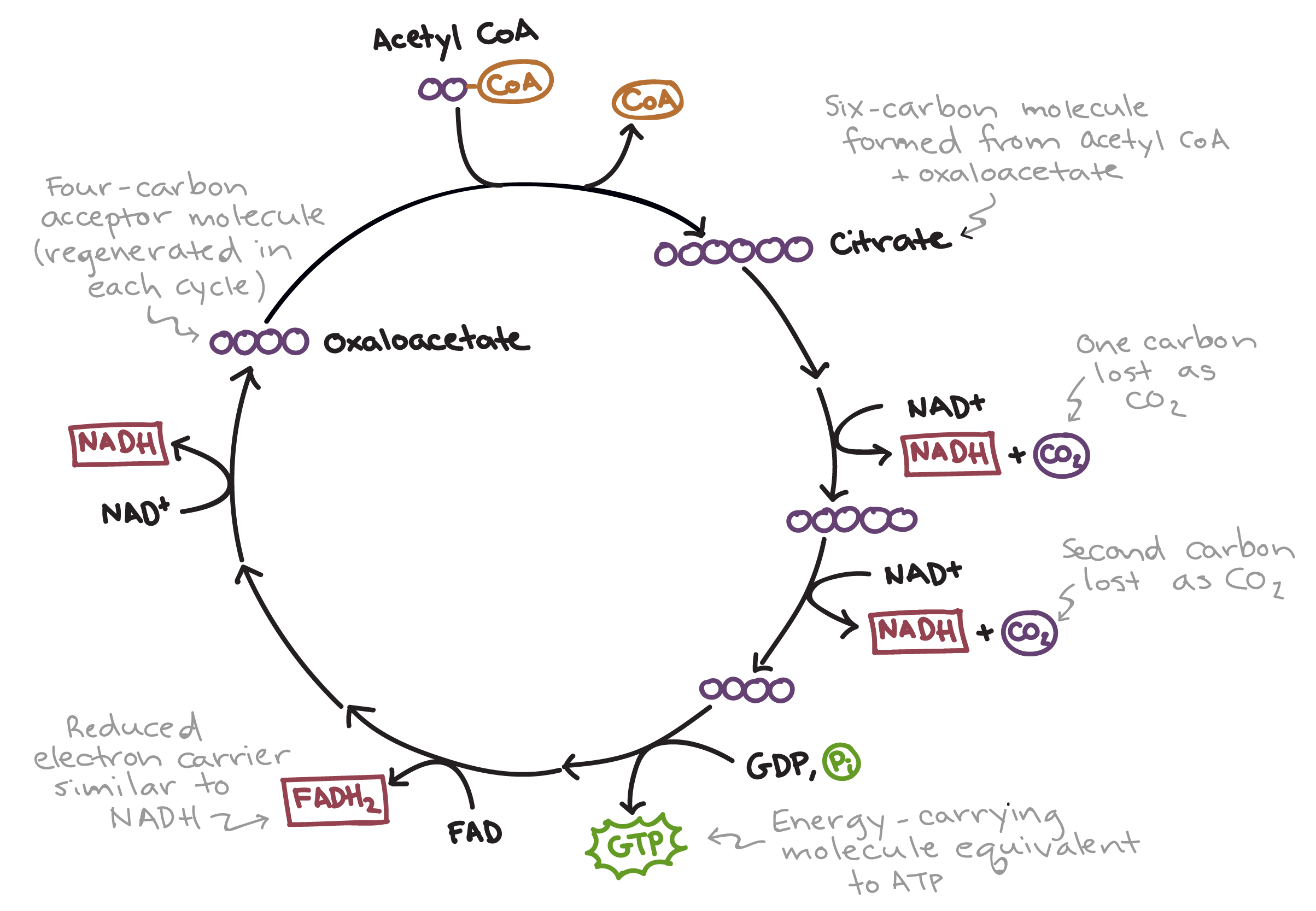
Citric acid cycle
Before the citric acid (Krebs) cycle can begin, pyruvate must be converted to acetyl coenzyme A (acetyl CoA), which links glycolysis to the citric acid cycle + CO2
1 NADH per pyruvate generated here
The citric acid cycle completes the breakdown of pyruvate to CO2
Each pyruvate generates:
1 acetyl CoA
1 ATP
3 NADH
1 FADH2
Total yield per glucose:
2 acetyl CoA
2 ATP
6 NADH [(+2 pyruvate ox.)]
2 FADH2
The citric acid cycle has eight steps, each catalyzed by a specific enzyme
The acetyl group of acetyl CoA joins the cycle by combining with oxaloacetate, forming citrate (citric acid)
The steps of the TCA decompose the citrate back to oxaloacetate, making the process a cycle
The NADH (x3) and FADH2 (x1) produced by the cycle relay electrons extracted from food to the electron transport chain
 NADH and FADH2 (electron carriers) donate electrons to the electron transport chain, which powers ATP synthesis via oxidative phosphorylation.
NADH and FADH2 (electron carriers) donate electrons to the electron transport chain, which powers ATP synthesis via oxidative phosphorylation.
The electron transport chain is located in the inner membrane (cristae) of the mitochondrion.
Electrons drop in free energy as they go down the chain and are finally passed to O2, forming H2O
Electrons are passed through a number of proteins including cytochromes to O2
The electron transport chain generates no ATP directly. It makes the hydrogen ion gradient.
The electron transport chain causes proteins to pump H+ from the mitochondrial matrix to the intermembrane space
H+ then moves back across the membrane, passing through the protein complex, ATP synthase uses the exergonic flow of H+ to drive phosphorylation of ATP.
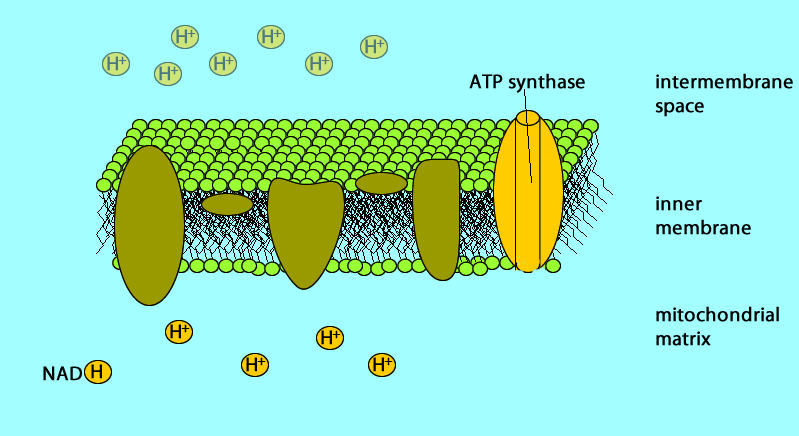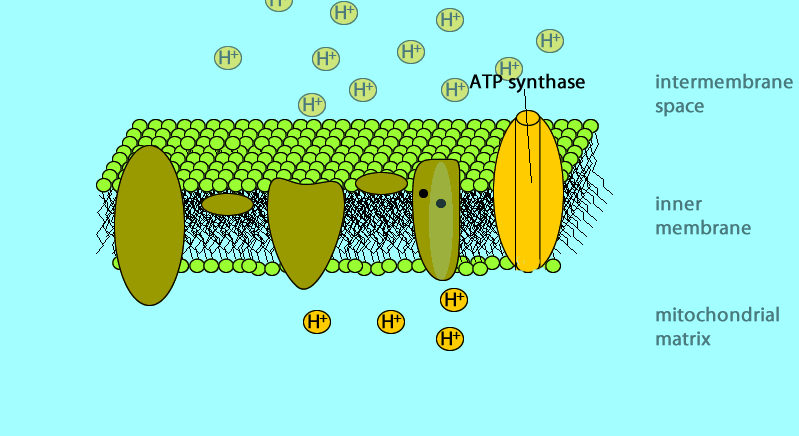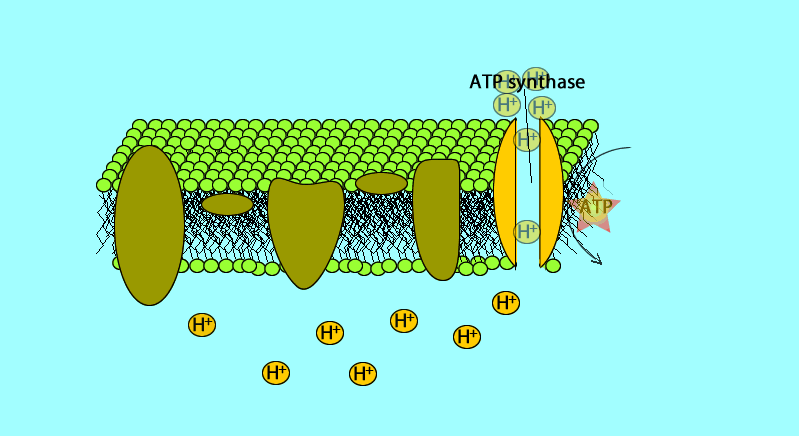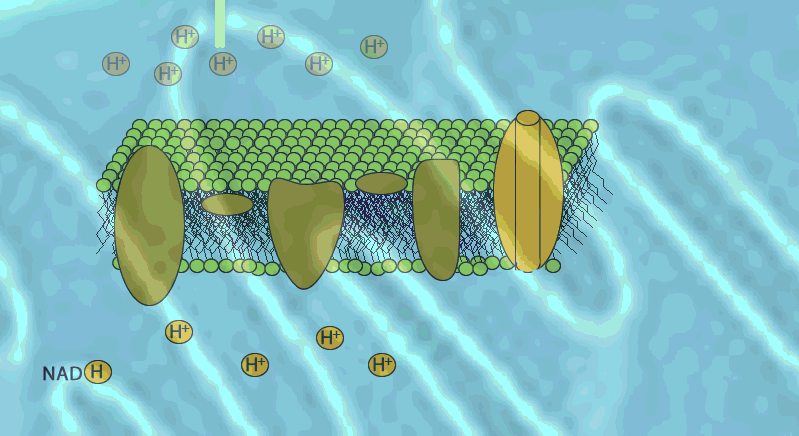
 Glucose → NADH → electron transport chain → proton-motive force (H+) → ATP
Glucose → NADH → electron transport chain → proton-motive force (H+) → ATP
*ATP synthase generates between 26-28 molecules of ATP (total ATP per glucose = 30-32)
Fermentation and Metabolism of Non-carbohydrates
In a lack of oxygen, glycolysis couples with fermentation or anaerobic respiration to produce ATP.
Fermentation uses substrate-level phosphorylation instead of an electron transport chain to generate ATP.
Fermentation consists of glycolysis plus reactions that regenerate NAD+, which can be reused by glycolysis (DOESN’T MAKE ATP)
Two common types are alcohol fermentation and lactic acid fermentation.
In alcohol fermentation, pyruvate is converted to ethanol in two steps:
The first step releases CO2 from pyruvate
The second step reduces the resulting acetaldehyde to ethanol
In lactic acid fermentation, pyruvate is reduced by NADH, forming lactate as an end product, with no release of CO2
Obligate anaerobes carry out only fermentation or anaerobic respiration and cannot survive in the presence of O2
Yeast and many bacteria are facultative anaerobes, meaning that they can survive using either fermentation or cellular respiration.
Glycolysis and the citric acid cycle connect to many other metabolic pathways. Glycolysis accepts a wide range of carbohydrates.
Fats are digested to glycerol (used in glycolysis) and fatty acids. Fatty acids are broken down by beta oxidation and yield acetyl CoA. An oxidized gram of fat produces more than twice as much ATP as an oxidized gram of carbohydrate.
Proteins must be digested to amino acids and amino groups must be removed before amino acids can feed glycolysis or the citric acid cycle.
Photosynthesis
Photosynthesis: the process that converts solar energy into chemical energy
Chloroplasts: the site of photosynthesis in plants.
their green color is from chlorophyll, the green pigment within chloroplasts
CO2 enters and O2 exits the leaf through microscopic pores called stomata
Thylakoids: connected sacs in the chloroplasts; thylakoids may be stacked in columns called grana.
The chlorophyll is in the membranes of thylakoids
Stroma: a dense interior fluid in the chloroplast — site of the Calvin cycle
pigments are substances that absorb visible light
Leaves appear green because chlorophyll reflects and transmits green light
The absorption spectrum of chlorophyll a & b suggests that violet-blue and red light work best for photosynthesis
Chlorophyll a is the main photosynthetic pigment
accessory pigments, such as chlorophyll b, broaden the spectrum used for photosynthesis
Accessory pigments called carotenoids absorb excessive light that would damage chlorophyll
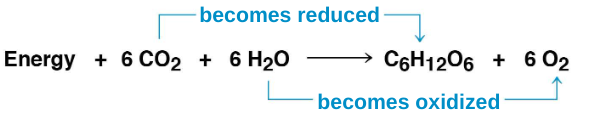
6 CO2 + 12 H20 + Light energy → C6H12O6 + 6 O2 + 6 H2O
Photosynthesis is a redox process in which H2O is oxidized and CO2 is reduced
Photosynthesis is an endergonic process; the energy is provided by light
 Photosynthesis consists of two parts:
Photosynthesis consists of two parts:
The light dependent reactions (in the thylakoids)
split H2O
release O2
Reduce the electron acceptor, NADP+ to NADPH
generate ATP from ADP by adding a phosphate group, photophosphorylation
The Calvin cycle light independent (in the stroma) forms sugar from CO2, using ATP and NADPH
The Calvin cycle begins with carbon fixation, incorporating CO2 into organic molecules, releases PGAL
A photosystem consists of a reaction-center complex (a type of protein complex) surrounded by light-harvesting complexes.
The light-harvesting complexes (pigment molecules bound to proteins) transfer the energy of photons to the reaction center
A primary electron acceptor in the reaction center accepts excited electrons and is reduced as a result
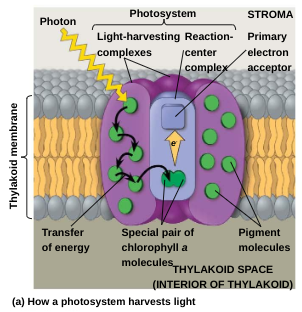 Photosystem II (PS II): functions first and is best at absorbing a wavelength of 680 nm — loses two electrons to ETC
Photosystem II (PS II): functions first and is best at absorbing a wavelength of 680 nm — loses two electrons to ETC
Photosystem I (PS I): functions second is best at absorbing a wavelength of 700 nm — loses two electrons to reduce NADP+ → NADPH
Linear electron flow: involves the flow of electrons through the photosystems and ETC to produce ATP (via H+ gradient and ATP synthase) and NADPH
The Calvin cycle uses the chemical energy of ATP and NADPH to reduce CO2 to sugar
Carbon enters the cycle as CO2 and leaves as a sugar named glyceraldehyde 3-phosphate (G3P) (PGAL)
The Calvin cycle has three phases
Carbon fixation
Phase 1, carbon fixation, involves the incorporation of the CO2 molecules into ribulose bisphosphate (RuBP) using the enzyme rubisco
The product is 3-phosphoglycerate
Reduction
Phase 2, reduction, involves the reduction and phosphorylation of 3-phosphoglycerate to G3P
6 ATP and 6 NADPH are required to produce 6 molecules of G3P, but only one exits the cycle for use by the cell
Regeneration of the CO2 acceptor
Phase 3, regeneration, involves the rearrangement of the five remaining molecules of G3P to regenerate the initial CO2 receptor, RuBP
3 additional ATP are required to power this step
Alternative Pathways for Photosynthesis
On hot, dry days, plants close stomata, which conserves H2O but also limits photosynthesis
The closing of stomata reduces access to CO2 and causes O2 to build up
These conditions favor a more wasteful process called photorespiration
Photorespiration is a metabolic process that uses O2 and releases CO2 — it does not make sugar.
C4 plants minimize the cost of photorespiration by incorporating CO2 into a four-carbon compound
These plants possess an enzyme in the mesophyll cells that have a high affinity for CO2 and can fix carbon even when CO2 concentrations are low.
CAM plants (crassulacean acid metabolism) open their stomata at night, incorporating CO2 into organic acids
stomata close during the dark and CO2 is released from organic acids and used in the Calvin Cycle
ex. cacti, pineapples