ap bio unit 6 review
DNA, RNA, and DNA Replication
Structure of DNA and RNA
- dna and rna: nucleic acids that store genetic information in living organisms; dna is usually used by prokaryotes and eukaryotes; rna is used by some viruses
- prokaryotes: package their dna into circular chromosomes into the nucleoid region
- eukaryotes: package their into linear chromosomes into the nucleus
- plasmids: used by both prokaryotes and eukaryotes to store additional genetic information in the form of small, circular pieces; aka extranuclear dna
| DNA | RNA | |
|---|---|---|
| nucleotide | nitrogenous base, 5-carbon sugar, and phosphate group | nitrogenous base, 5-carbon sugar, and phosphate group |
| nitrogenous bases | adenine, guanine, thymine, and cytosine | adenine, guanine, uracil, and cytosine |
| base-pairing rules | A-T and C-G | A-U and C-G |
| 5-carbon sugar | deoxyribose; has a hydrogen (H) attached to 2’ carbon | ribose; has a hydroxyl (OH-) attached to 2’ carbon |
| strandedness | double-stranded; anti-parallel strands | single-stranded; can fold into 3-D structures in rRNA and in tRNA |
| overall structure | 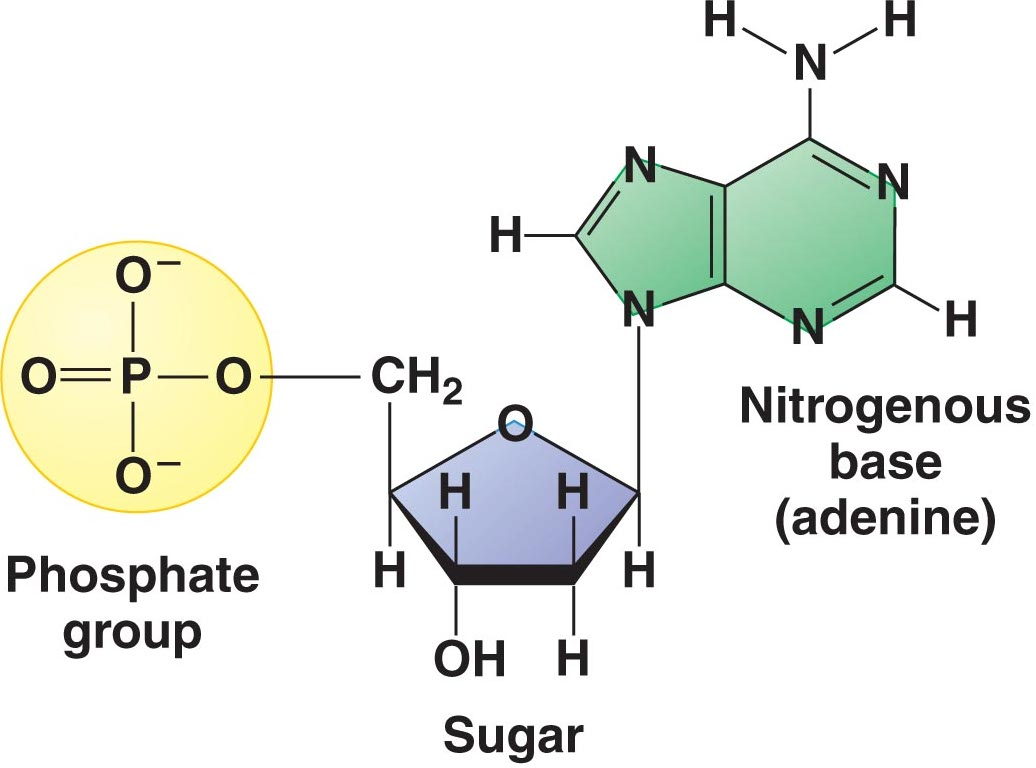 | 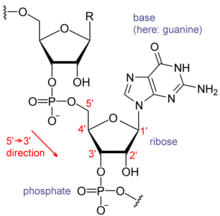 |
| functions | more stable, so it is better for carrying genetic information between generations | has more temporary functions (ex. use of mRNA during transcription) |
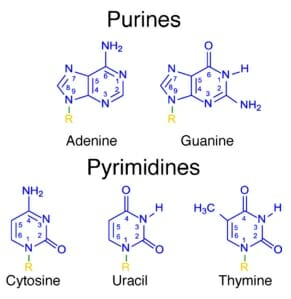
nitrogenous bases: adenine, guanine, cytosine, thymine, and uracil are all nitrogenous bases found in either dna or rna
- purines: two-ringed structure; adenine and guanine
- pyrimidines: single-ringed structure; thymine, cytosine, and uracil
- when adenine pairs with thymine or uracil, it forms 2 hydrogen bonds
- when cytosine pairs with guanine, it forms 3 hydrogen bonds
structure of dna: double helix with anti-parallel strands
- one strand is oriented in the 5’ phosphate group at the end and the other is oriented with the 3’ hydroxyl (OH-) group at the end
- purine of one strand is paired with pyrimidine of other strand
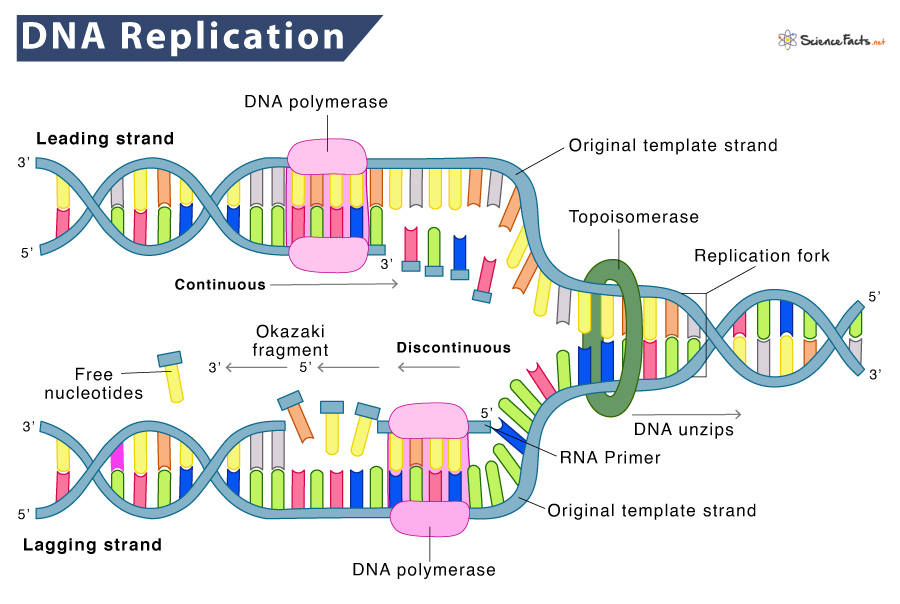
DNA Replication
each of the two strands of dna serves as a template for a new strand during replication
- semiconservative: describes dna because each new double helix is composed of a template strand and a new strand
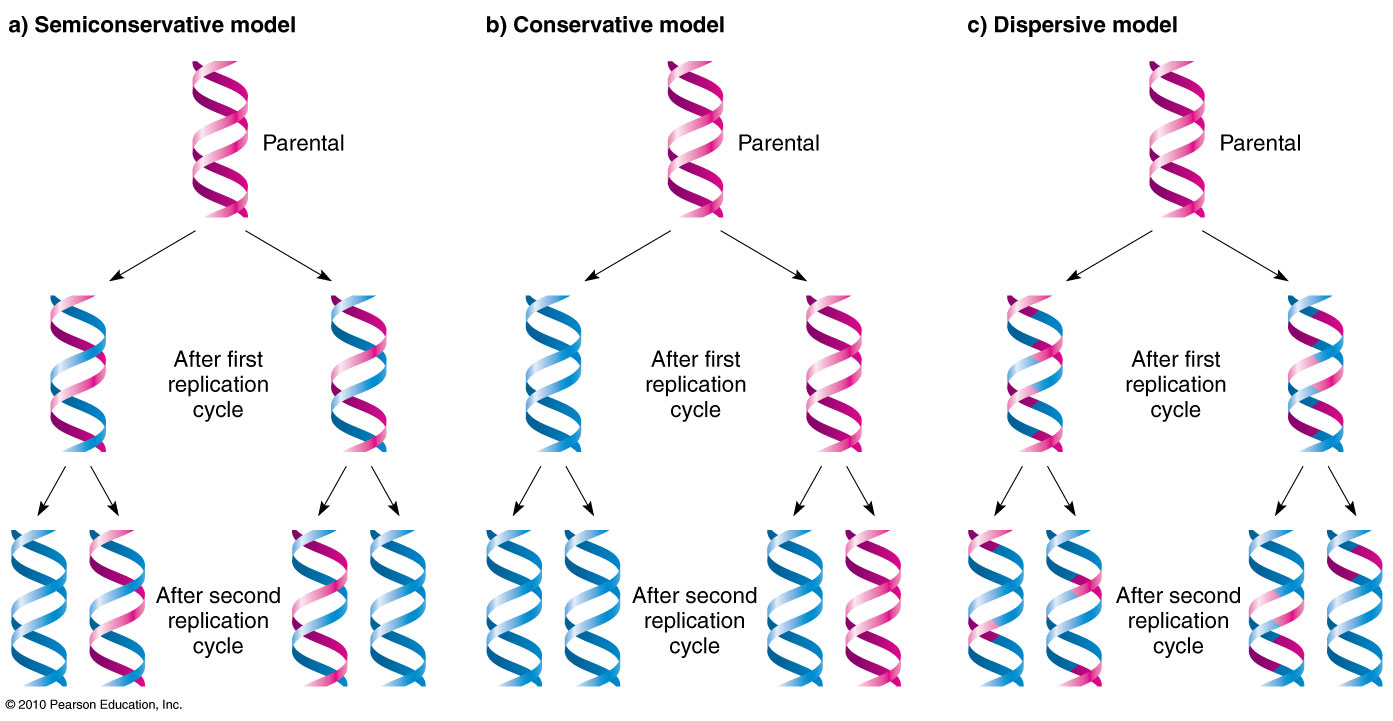
dna replication follows the process below:
- initiation: origin of replication is recognized and the process of helicase (enzyme) unwinding the two dna strands begins
- topoisomerase enzymes: used to make temporary nicks in sugar-phosphate backbone of dna to relieve the pressure from supercoiling as helicase unwinds it
- elongation: rna polymerase is used to read the template strand of dna and synthesize an rna primer using some complementary rna nucleotides
- new dna nucleotides can be added to the rna primer that is synthesized
- dna polymerase: the enzyme that adds new nucleotides to the 3’ hydroxyl group i
- leading strand replication: one new strand (the leading strand) is made as a continuous piece in the 5’ to 3’ direction (it is read in the 3’ to 5’ direction)
- lagging strand replication: formed in the 3' to 5' (direction away from the fork, which is the 5’ to 3’ direction) and is made in small pieces called okazaki fragments
- ligase: the enzyme that brings together the okazaki fragments
- termination: when converging replication forks meet, the process ends
Transcription and Translation
Transcription: Prokaryotes vs. Eukaryotes
- transcription: process in which genetic information in a sequence of dna nucleotides is copied into newly synthesized rna molecules
- rna polymerase: the enzyme responsible for transcribing a dna sequence into an rna molecules
- the function of an rna molecule depends on its structure and sequence:
- mRNA (messenger rna): single-stranded; carries information from dna to the ribosome
- codon: 3 base pair sequence in mRNA that are complementary to the dna strand; used to create specific animo acids during translation
- tRNA (transfer rna): folds into a 3-D structure to act as an adapter molecule during translation
- one end of tRNA will bind to a specific amino acid and the other end contains an anticodon, which will pair up with the appropriate mRNA codon at the ribosome during translation
- rRNA (ribosomal rna): folds into a 3-D structure; rRNA and proteins form ribosomes to perform translation
- 3-D rRNA acts like a ribozyme (catalyzes the reactions during translation)
Process of Transcription
- pre-initiation: rna polymerase binds to a promoter
- promoter region: only serves as a binding site for rna polymerase; doesn’t code for any amino acids
- TATA box: most eukaryotes have this promoter region
- transcription factors: help rna polymerase bind to the promoter region and start transcription
- initiation: rna polymerase adds new rna nucleotides in the 5’ to 3’ direction
- elongation: the dna strand being transcribed by rna polymerase is used to create a complementary strand
- the newly synthesized strand will be used during translation
- termination: results in the release of the newly synthesized mRNA from the elongation complex
- in eukaryotes: termination of transcription involves cleavage of the transcript, followed by a process called polyadenylation
- polyadenylation: modifications to eukaryotic transcribed rna strands to prevent degradation
Modifications to Eukaryotic Pre-mRNA
removal of introns: spliceosomes remove the introns from the pre-mRNA to join the exons together
- intron: noncoding rna sequences in pre-mRNA; may function in regulation of gene expression
- exon: coding sequences of eukaryotic rna
- spliceosome: structures made of small nuclear rna (snRNA) and small nuclear ribonucleoproteins (snRNPs)
- alternative splicing: exons can be joined in different combinations to generate multiple rna transcriptions from the same gene; results in variety of transcriptions, in comparison to the one sequence that can be generated in prokaryotes
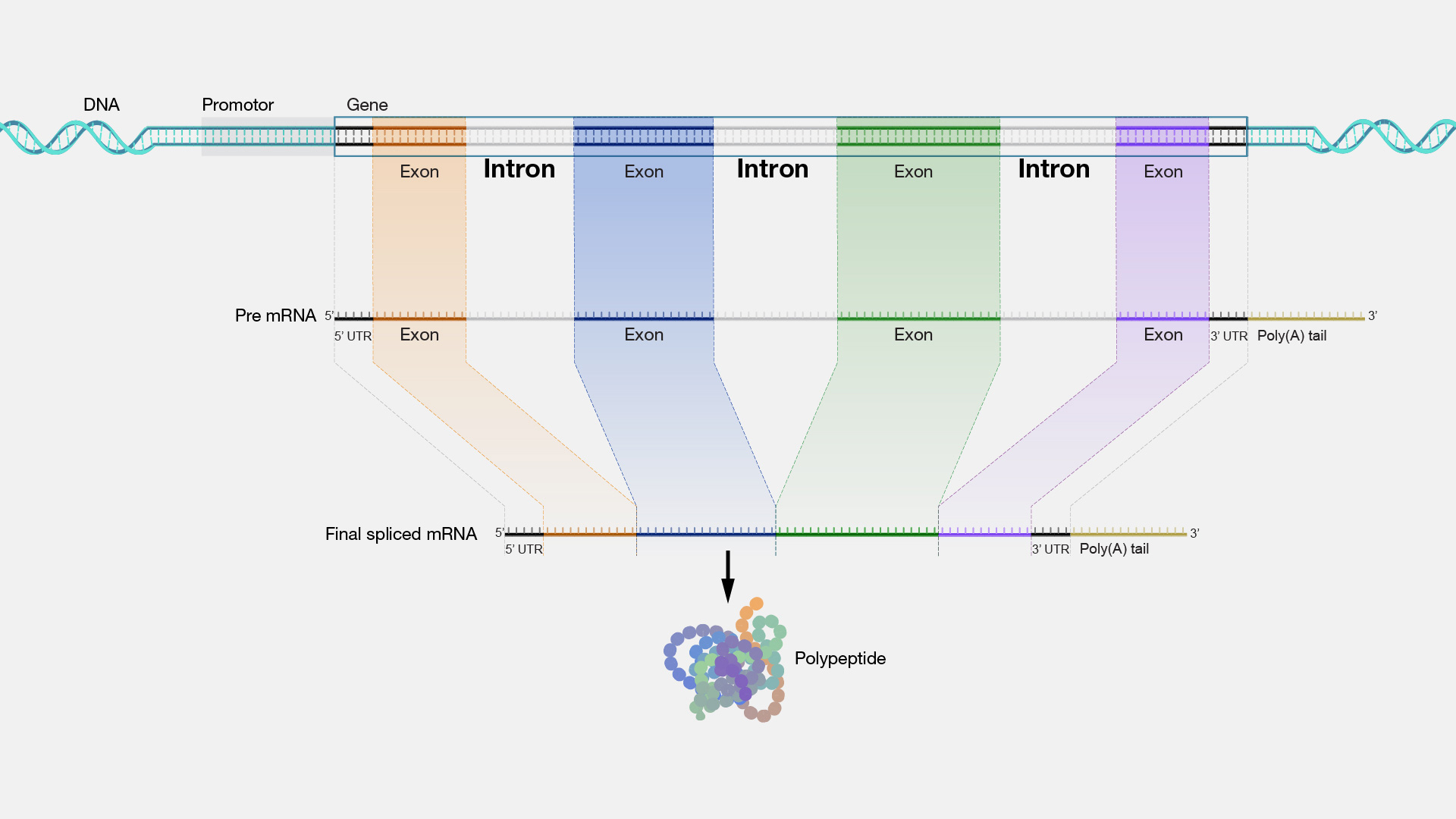
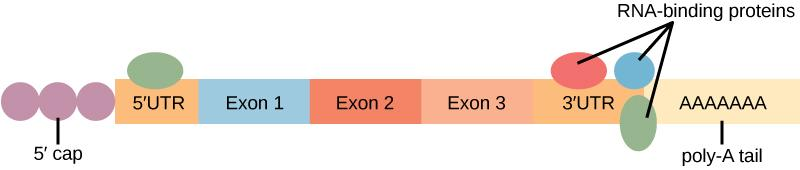
addition of GTP cap: 5’ guanosine triphosphate cap is added to pre-mRNA transcript to prevent degradation; GTP cap also helps with initiation of translation
addition of poly-A tail: enzyme poly-A polymerase adds a string of adenine nucleotides to the 3’ end of the pre-mRNA to prevent degradation
- mRNAs with longer 3’ poly-A tails tend to have longer durations in the cytosol to allow for more copies of the protein (that the mRNA codes for) to be generated
mature mRNA is formed after these modifications
Translation
- translation of mRNA molecules occurs in the ribosomes in prokaryotes and eukaryotes
- ribosomes: found in cytoplasm of both and the rough ER of eukaryotes
- eukaryotes: cytoplasmic ribosomes code for proteins that stay inside the cell; ribosomes on rough ER code for proteins leaving the cell
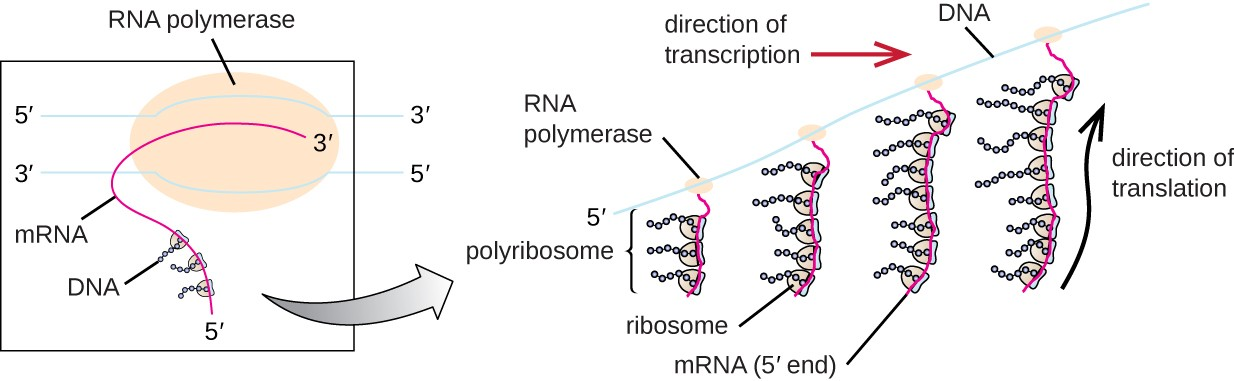
- prokaryotes: no nucleus, so translation can occur as transcription occurs; multiple ribosomes can be simultaneously translating prokaryotic rna to form polyribosomes (aka polysomes)
Process of Translation
initiation: genetic code is read in codons (3 base pairs); translation is initiated when rRNA pairs with start codon (AUG)
- a tRNA with the anticodon (UAC) brings the appropriate amino acid to the ribosome and the anticodon on the tRNA pairs with the codon on the mRNA
- each codon on mRNA codes for one amino acid, but multiple codons can code for the same amino acid
- the redundancy results in “silent” mutations, where there is a different code, but no change in the actual amino acid sequence
- almost all organisms use the same genetic code, which is evidence of common ancestry
elongation: after 1st amino acid is placed, ribosome translocates (moves) to the next codon; a new tRNA with the appropriate anticodon and amino acid will pair with the next codon; ribosome catalyzes the formation of peptide bond between the two amino acids
- once the peptide bond is formed, the 1st tRNA releases its amino acid (which is now connected to the 2nd amino acid) and the 1st tRNA is released from the ribosome
- this continue until a stop codon is reached
termination: stop codons (aka “nonsense codons) are reached and release factors bind to the ribosome; release factors cause the ribosome to disassemble and release the polypeptide chain
- release factors: type of protein used to terminate the translation process
Flow of Information from Nucleus to Cell Membrane
- in eukaryotes, dna follows this process:
- DNA → mRNA → protein at ribosome on rough ER → protein at golgi apparatus → protein in vesicle → protein outside of cell membrane
- dna is transcribed into mRNA in the nucleus
- ribosomes on the rough ER use the information from the mRNA to form proteins
- the proteins will move from the rough ER to the golgi apparatus
- the golgi will modify and package the proteins into vesicles for transport
- the proteins (now in vesicles) are exported from the cells
- vesicles will fuse with the cell membrane to release their protein contents from the cell
INSERT PICTURE!!
- retroviruses: use rna as their primary carrier of genetic material; coontain the enzyme reverse transcriptase, which allows the virus to make a copy of dna from rna
- the virus injects this dna into the host cell, which will then transcribe and copy the dna, spreading the virus
- reverse transcriptase: less accurate than rna polymerase, causing retroviruses to have high mutation rates
Regulation and Mutation
- phenotypes: determined by expression and regulation of genes
- regulatory proteins: proteins that can turn on or off genes by binding to specific nucleotide sequences
- regulatory sequences: nucleotide sequences that regulatory proteins bind to
Regulation of Gene Expression in Prokaryotes
prokaryotes use operons to regulate gene expression
- operon: cluster of genes with a common function under the control of a common promoter
- contain regulatory sequences for regulatory proteins and genes for structural proteins
- promoter: non-coding regulatory sequences that serve as binding sites for rna polymerase
- operators: non-coding regulatory sequences that serve as binding sites for repressor proteins (type of regulatory protein)
- structural gene: coding sequences that contain the genetic code for the proteins required to perform the function of the operon
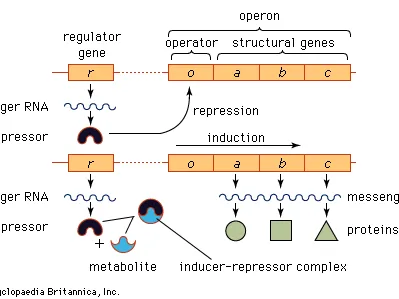
Inducible Operon
usually have a catabolic function (digesting molecules) and are turned off unless the appropriate inducer molecule is present
repressor protein in an inducible operon binds to the operator sequence to block transcription of the operon by rna polymerase
inducer: binds to repressor protein and changes its shape so that it can no longer bind to the operator sequence; allows rna polymerase to being transcribing the operon
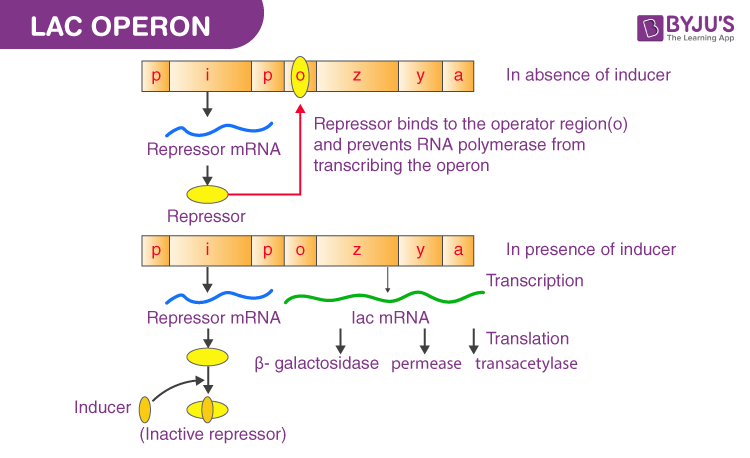
lac operon: used to produce protein required for the digestion of lactose (sugar)
- no lactose present: lac repressor protein binds to operator to block transcription of the operon
- lactose present: lactose (as the inducer) binds to the lac repressor protein to change its shape so that it can not bind to the operator sequence; allows rna polymerase to transcribe the genes for the proteins that digest lactose
cAMP, CAP(catabolite activator protein), and lac operon: binding of repressor proteins can sometimes positively “upregulate” gene expression
- glucose levels are low: cAMP levels in cell increase and more cAMP binds to CAP in the lac operon
- CAP binds to a CAP binding site near the promoter, stimulating transcription
- transcription of lac operon is increased when glucose is absent, allowing for the cell to use energy from lactose more efficiently
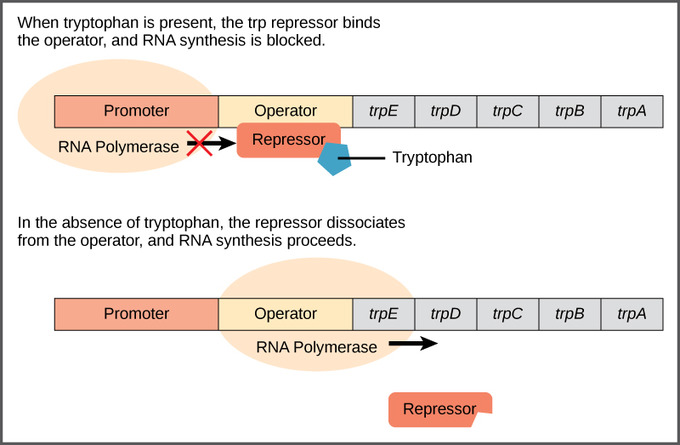
Repressible Operon
usually have an anabolic function (synthesizing molecules) and are turned on unless the product of the operon is abundant in the cell
trp operon: used to produce enzymes necessary for the synthesis of amino acid tryptophan
- tryptophan acts as a co-repressor, and it must be bound to the trp repressor before they can bind to the operator (no tryptophan means no binding to turn off the operon)
- this type of feedback mechanism allows the cell to make the enzymes needed to synthesize tryptophan only when the cell needs it
Inducible vs. Repressible Operons
- key differences include:
- function of operon (catabolic or anabolic)
- whether the repressor protein can bind on its own to the operator or if it needs a co-repressor
Regulation of Gene Expression in Prokaryotes
- eukaryotes can also use interactions between regulatory sequences and regulatory proteins to turn genes on or off and regulate levels of gene expression
- examples of regulatory sequences in eukaryotes:
- promoters: sequences that serve as binding sites for rna polymerase
- regulatory switches: sequences to which activator proteins or repressor proteins may bind
- enhancers: regulatory switches that activator proteins or transcription factors bind to
- silencers: regulatory switches that repressor proteins bind to
- examples of regulatory proteins in eukaryotes:
- repressors: bind to regulatory switches to turn off/suppress gene expression
- activators: bind to regulatory switches to turn on/activate gene expression
- transcription factors: help rna polymerase bind to the promoter region and start transcription them to communicate
- epigenetic changes: reversible modifications to nucleotides of dna sequence (ex. methylation); also affect gene expression in eukaryotes
- methylation: adding a methyl group to a nucleotide
- methylated nucleotides are less likely to be transcribed, so the cell can modify gene expression by changing the level of methylation in various genes
- histone proteins: a protein that provides structural support for a chromosome; can be epigenetically modified by acetylation of the histone proteins
- acetylation: adding an acetyl group to the histone protein; causes dna to be more loosely wound around histone proteins
- makes dna more accessible to rna polymerase, and in turn, more likely to be expressed
- euchromatin: dna that is more loosely wound around histone proteins; more accessible to rna polymerase and more likely to be expressed
- heterochromatin: dna in chromosomes that is tightly wound around histone proteins; less accessible to rna polymerase and results in reduced gene expression
- small interfering rna (siRNA): single-stranded molecules that bind to complementary mRNA molecules to form double stranded rna (dsRNA)
- enzymes can detect and destroy dsRNA, resulting in no translation of the targeted mRNA molecules; reduces gene expression
Gene Expression and Cell Specialization
- differential gene expression: regulation of gene expression results in different genes being expressed in different cells; influences the function of cells and the resulting phenotype
- example: skin cell expressing lower levels of melanin protein would appear lighter than skin cell expressing higher levels of melanin protein
- timing of different gene expressions is critical to the formation of specialized tissues and organs in zygotes
Mutations
- mutations: changes in genetic material of an organism; may result in changes to an organism’s phenotype; can provide genetic variation in populations
- not all mutations are harmful because the organism’s environment will determine whether or not the mutation is harmful or beneficial (or has no effect)
- other mutations do not cause changes to amino acid sequences because of redundancy
- mutations can be caused by:
- environmental factors (ex. chemicals or radiation)
- random errors in dna replication
- mistakes in mitosis or meiosis
- nondisjunction (resulting in aneuploidy)
- aneuploidy in animals: can be fatal, cause sterility, or other disorders
- aneuploidy in plants: results in polyploidy (entire extra set of chromosomes), which can increase the survival of the plant
- horizontal transmission: genetic mutations can be transferred horizontally between members of the same generation; methods include:
- transformation: the uptake of naked foreign dna by a cell
- transduction: the transmission of dna from one organism to another by viruses
- conjugation: transmission of dna through cell-to-cell contact (usually through a connection called a pilus)
- pilus: a hair-like structure associated with bacterial adhesion and related to bacterial colonization and infection
- transposition: the movement of dna between chromosomes or within a chromosome; aka “jumping genes”
Biotechnology
Overview
- biotechnology has many purposes; examples include:
- cancer therapies
- improvements in agricultural yields
- gene therapies for genetic disorders
- de-extinction projects
- extinction projects
- etc.
Bacterial Transformation
- bacterial transformation: introduces foreign dna into bacterial cells; the foreign dna (usually a plasmid) may integrate into the host cell’s chromosome or remain separate from the host cell dna (stays in cytoplasm)
- methods of bacterial transformation:
- heat shock: bacterial cells are mixed with foreign dna and then quickly exposed to cold-hot-cold temps. to create temporary microscopic pores through which the foreign dna can enter the host cell
- selectable marker: foreign plasmid dna needs a marker so that cells that have incorporated the plasmid dna cannot be detected
- selectable markers are usually an antibiotic resistant gene that is not present in non-transformed bacterial cells
- recombinant dna: when plasmid contains a gene from another organism; dna can be cut with restriction endonucleases (aka restriction enzymes) and recombined/connected with dna ligases
- some recombinant plasmids contain the selectable marker and the gene that codes for the desired protein product
- example: insulin, which is now produced in large amounts by bacteria that contain the proper recombinant plasmids
Gel Electrophoresis
- gel electrophoresis: a technique used to separate dna fragment by size and charge
- dna fragments can be formed by treating a dna sample with restriction enzymes to cut the dna at specific base pairs
- in the structure of dna, phosphate groups have a slightly negative charge, leading to am overall negative charge in dna molecules
- a positive cathode is placed at the bottom of the gel plate to attract dna molecules to the bottom; shorter fragments move quicker than longer fragments
- this allows for distinctions to be made between shorter and longer fragments of dna based on how far they traveled in a given amount of time
- the estimated fragment lengths can be run alongside other dna samples to provide a “ruler” to estimate the size of each fragment produced
- each organism has a different pattern of fragments, resulting in a dna “fingerprint”
Polymerase Chain Reaction (PCR)
- PCR: used to amplify specific dna fragments and create millions of copies of a specific dna fragment; involves cycles of dna replication using primers that are specific to the beginning and end of the dna sequence that is being amplified
- three stages of PCR cycles:
- denaturing of dna: separates the two strands of dna; usually done at a high temp.
- annealing of the primers: temp. is lowered slightly from the temp. used to separate the dna strands and primers that are complementary to the targeted dna strand are used to anneal with the dna sequence being amplified
- anneal: to form hydrogen bonds with
- extension of the primers: dna polymerase adds new nucleotides to the primers to create two copies of the targeted dna sequence
- each cycle of PCR doubles the number of copies of the targeted dna sequence
- PCR is used in the sequencing of dna, which is used for:
- forensics
- bioinformatics
- medicine
- evolutionary biology
CRISPR-Cas9
- CRISPR: clustered regularly interspaced short palindromic repeats
- CRISPR-Cas9: a system that functions in nature as an adaptive immune system in bacteria
- when bacteria becomes infected by bacteriophages, it cuts up the viral dna and inserts the fragments into short palindromic repeats into the bacteria’s dna
- when another virus infects the bacteria, it can use the fragments to compare the dna from the new virus to the stored sequences
- CRISPR can be used to edit dna sequences
- Cas9 enzyme uses a fragment of rna to find where to cut dna at a specific part in the sequence
- this can create a “knockout” gene, which is cut and no longer functional; prevents the development of some genetic diseases or allows the scientists to understand the function of that gene