W4 L11: Non-coding RNAs
Non-coding RNA = doesn’t code for proteins
1st non-coding RNA
- Alanine tRNA in baker’s yeast
- Cloverleaf structure was solved in 1974
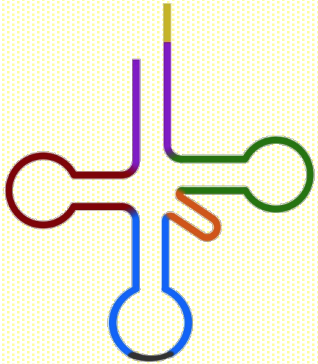
tRNA-Phe
tRNA decode the mRNA sequence during protein synthesis at ribosome
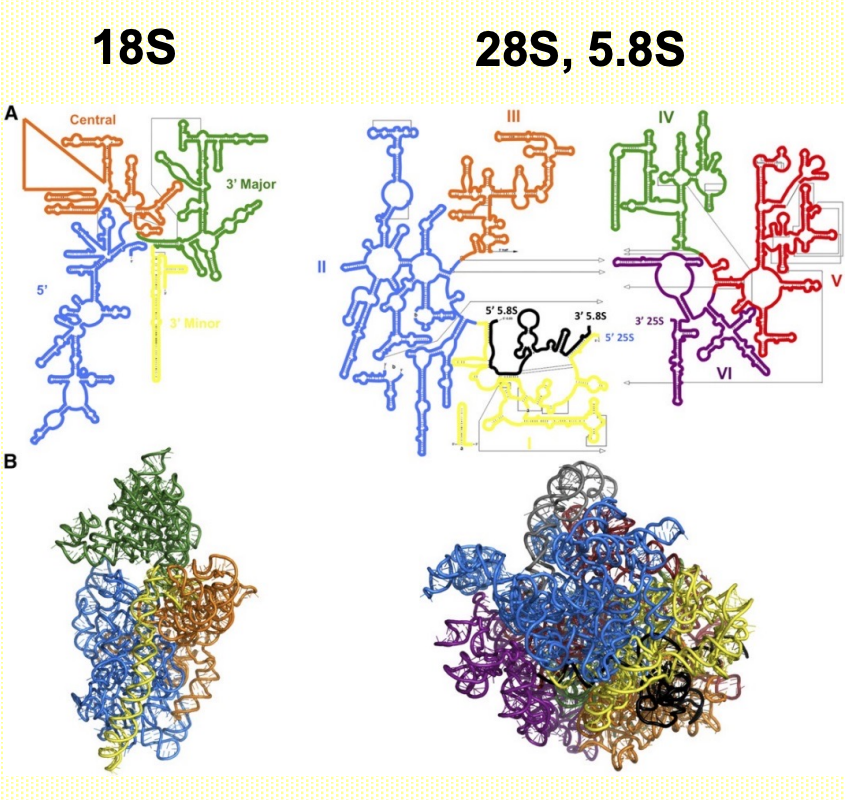
Ribosomal RNA
- 80% of total RNA
- Transcribed by Pol I (except 5S by Pol III)
- Humans: 300-400 rDNA repeats on diff. chromosomes (13-15, 21, 22)
- Eukaryotic ribosomes (2 subunits)
- 40S subunit > 18S rRNA
- 60S subunit > 5S, 5.8S, 28S
- 5.8S, 18S & 28S are made from a single transcript (45S precursor) > rRNA processing in nucleolus
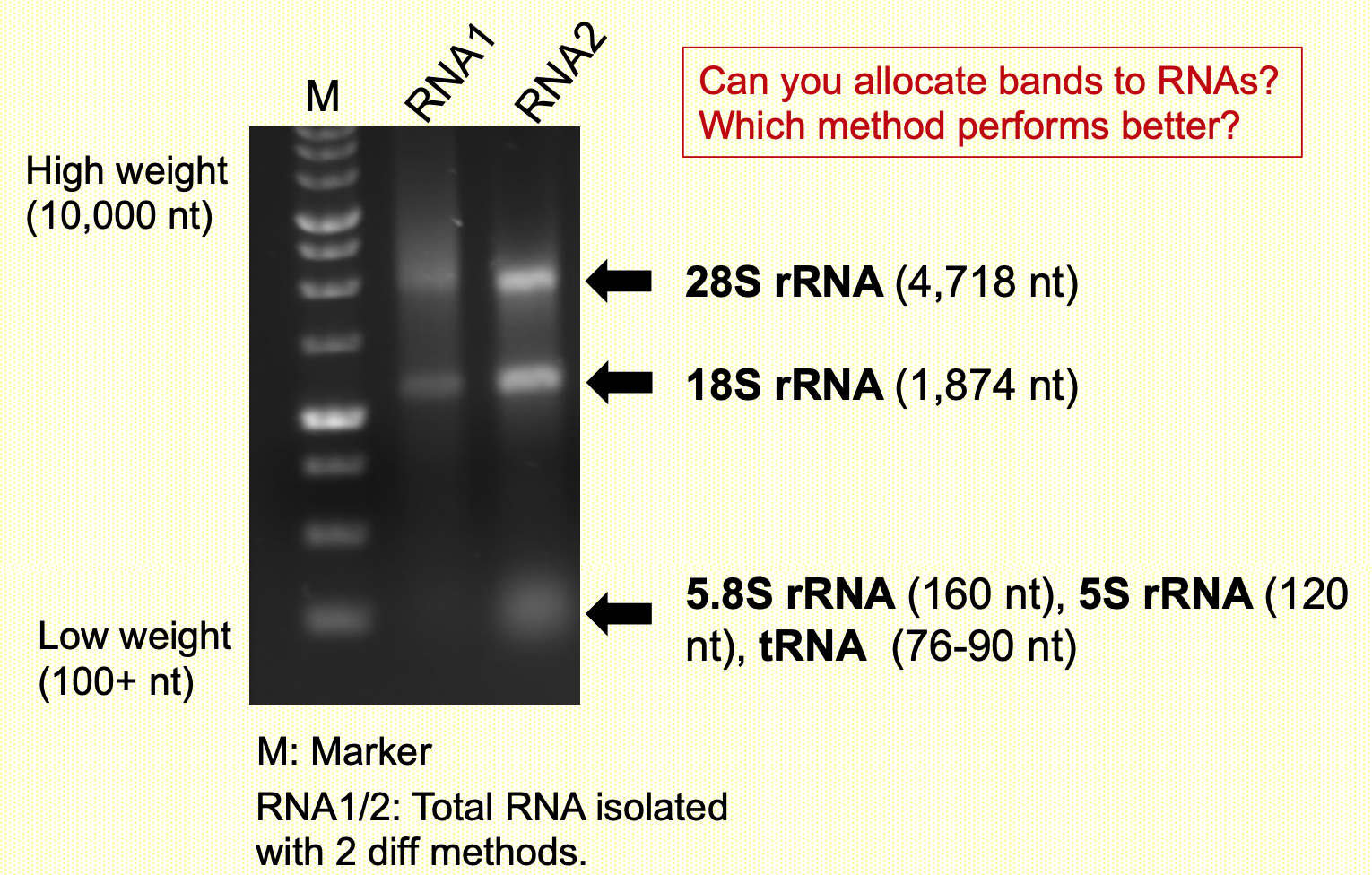
Total RNA isolated from human cells
28s is biggest, 5.8s is smallest
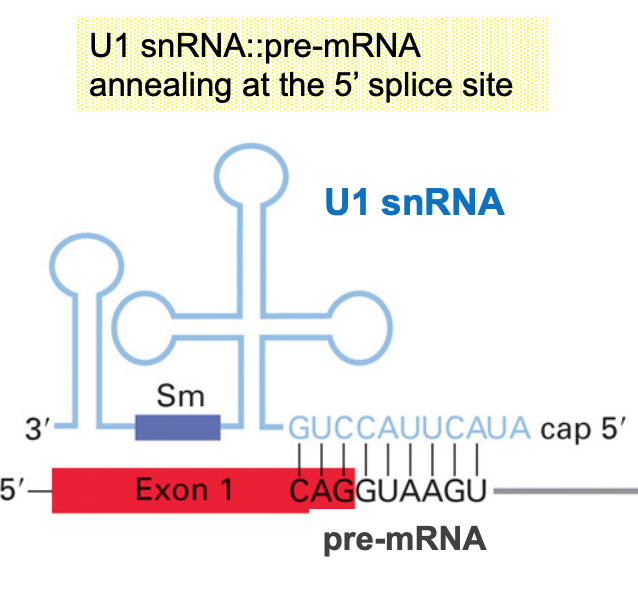
Small nuclear RNAs - snRNAs
U-rich sequence involved in splicing (U1, U2, U4, U5, U6)
Base-pairing w/ pre-mRNAs defines splice-sites
From 107- 210 nts long, each associated w/ 6-10 proteins
- small nuclear ribonucleoprotein particles (snRNPs)
Highly expressed (1 mio snRNPs) & evolutionarily conserved
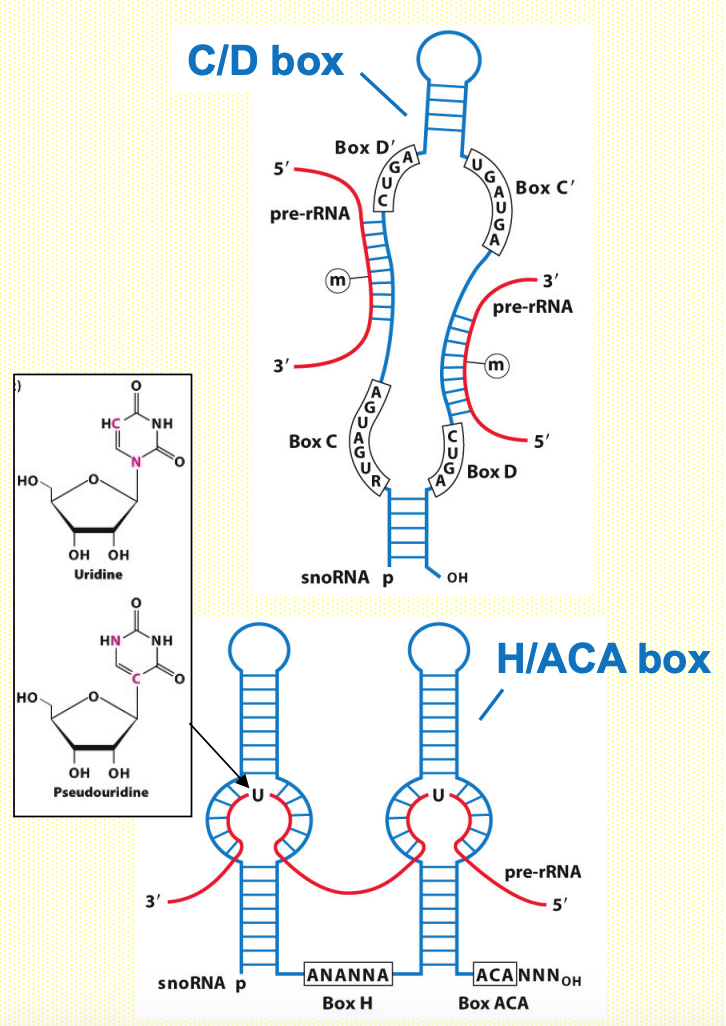
Small nucleolar RNAs - snoRNAs
- 200 diff species in mammals, 60-150 nts length
- Most snoRNAs are encoded w/in introns of Pol II transcribed genes
- Assemble w/ proteins to form small nucleolar ribonucleoproteins (snoRNPs)
- Guide specific RNA modifications in i.e. rRNA by base-pairing
- C/D box > directs 2’-O-ribose methylation by recruiting methyl transferase enzyme
- H/ACA box > recruits an enzyme that converts uridine to pseudouridine
- Main function takes place in nucleolus (>rRNA processing)
- In C/D box → hybridise/ anneal w/ complementary seq. in rRNA → guide methylation of bases in rRNA
- In H/ACA box → modify uridine base & form pseudouridine
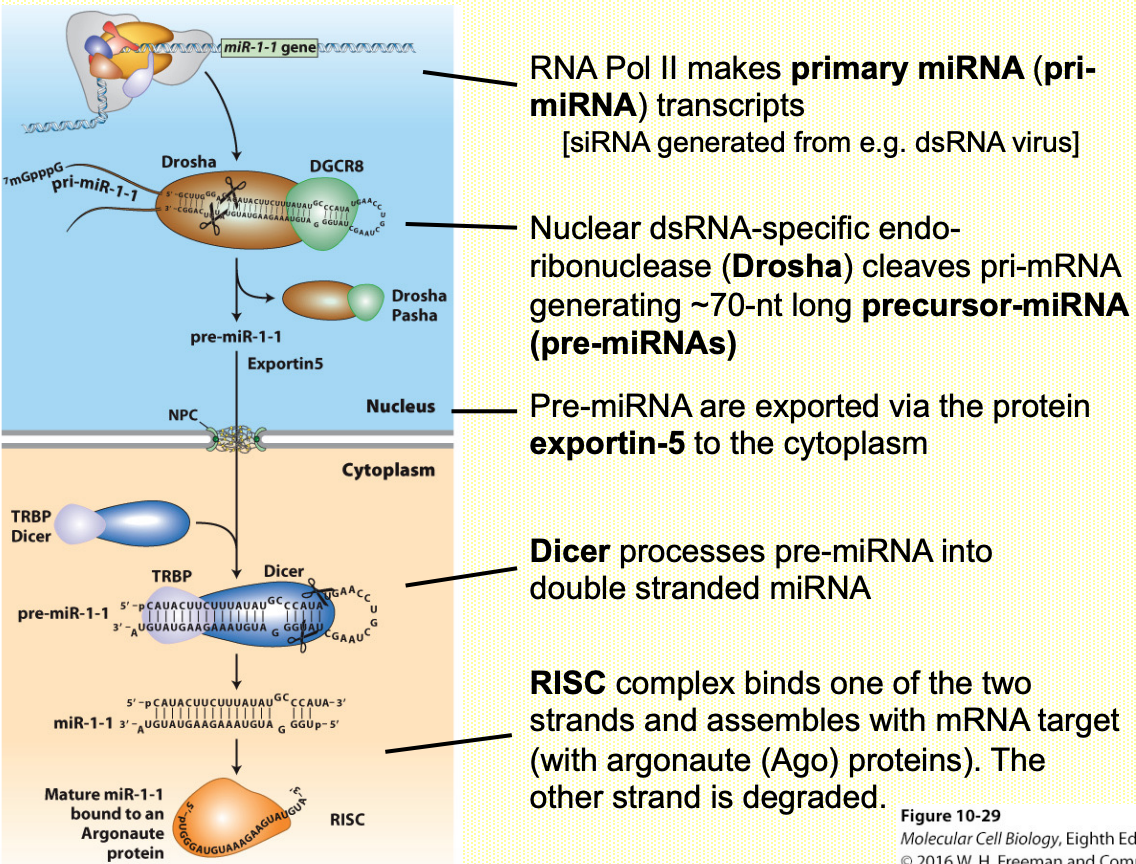
miRNAs & siRNAs
Small non-coding RNAs affect translation or decay of mRNAs in cytoplasm
20-22 nucleotides long RNA molecules
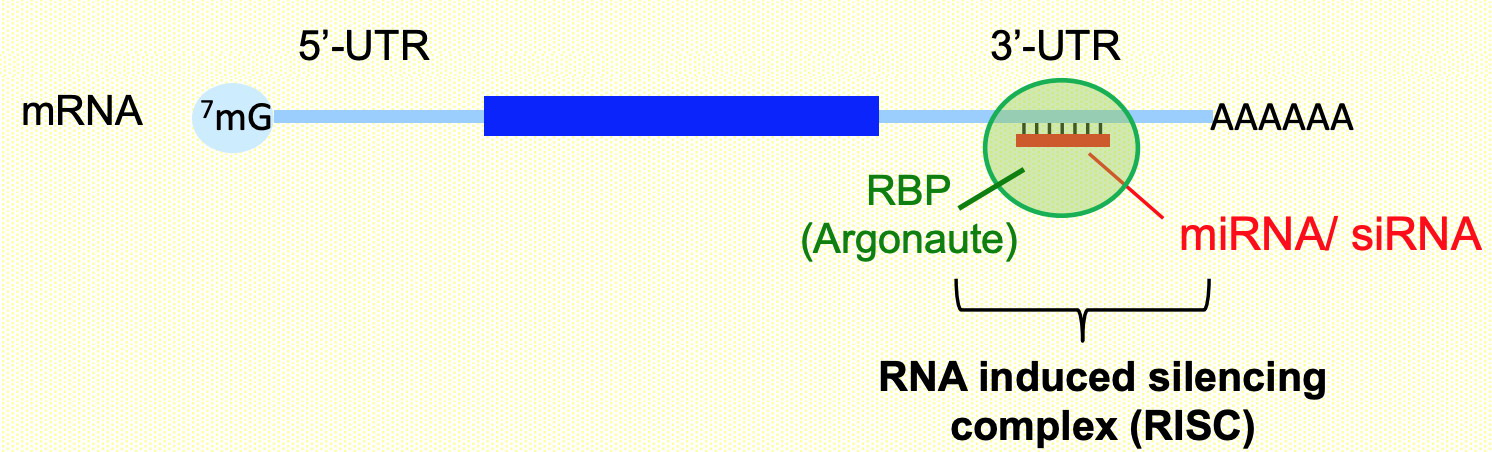
Specifically bind to complementary sequences locates in 3’-UTR regions of mRNAs (in complex w/ RNA-binding proteins (i.e. Ago)
Repress mRNA expression by promoting decay &/or inhibit translation
miRNAs → promoting deadenylation, translation, repression, decay
siRNA → cleavage of mRNA & exosome-mediated degradation
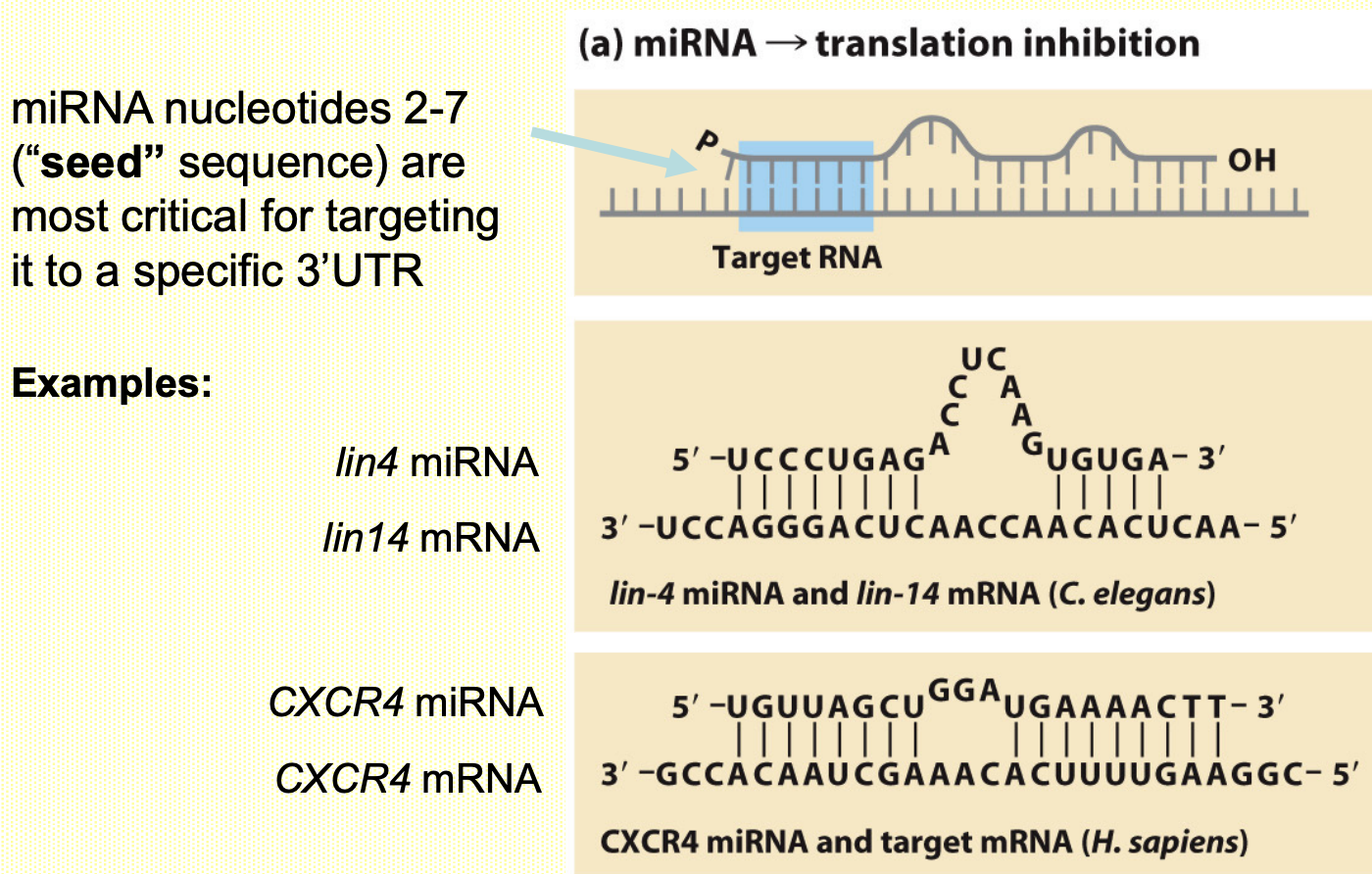
MicroRNAs repress translation by ‘imperfect’ hybridisation w/ target mRNAs in cytoplasm
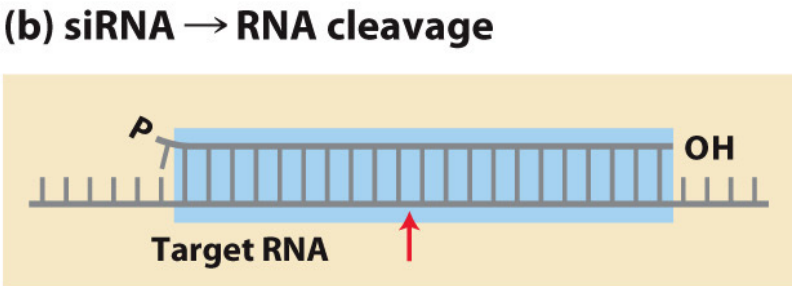
siRNAs cleave mRNAs upon ‘perfect’ hybridisation
Established as defence mechanism against invading dsRNA viruses & unwanted actions of transposons/repetitive elements expressed from genome
“invading” dsRNA processed into siRNA that targets invader mRNAs for degradation
siRNA mediated defence mechanism are crucial in plants, worms & insects - less in mammals where a protein based system to fight viruses has taken over
How are miRNAs/siRNAs generated in cells?
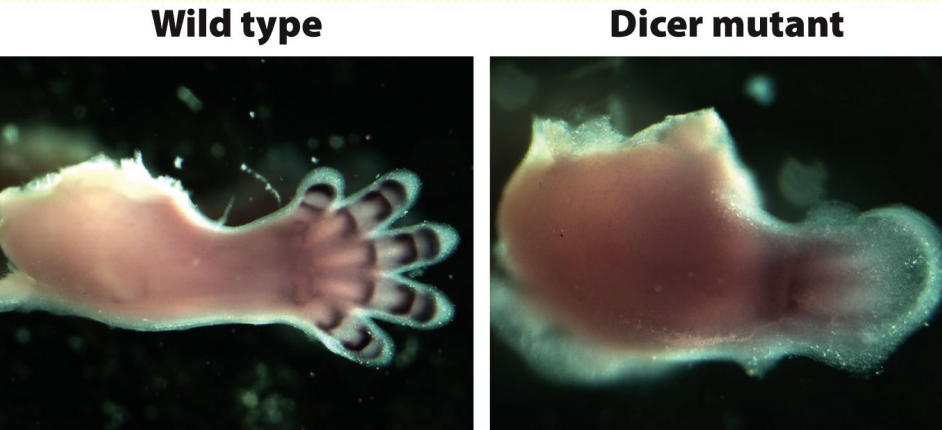
Developmental defects in inducible 𝘋𝘪𝘤𝘦𝘳 gene knock-out
‘Stable’ 𝘋𝘪𝘤𝘦𝘳 gene knock-out eliminates generation of miRNA in mammals & is embryonic lethal
‘Conditional’ 𝘋𝘪𝘤𝘦𝘳 knock-out in limb primordia leads to defects in tissue morphogenesis/ development
miRNA & disease
- ~5,000 diff miRNA regulate expression of almost every gene (>80%)
- miRNA combinatorial control genes w/ crucial functions in cell proliferation, development, inflammation, ageing
- Many miRNAs have been linked to disease (e.g. cancer oncogenes or tumour suppressor)
- 1,100 miRNAa linked to more than 850 diseases
- Represents a novel target for diagnostics (biomarkers) & drug development
- discovery of extracellular miRNAs detectable in blood
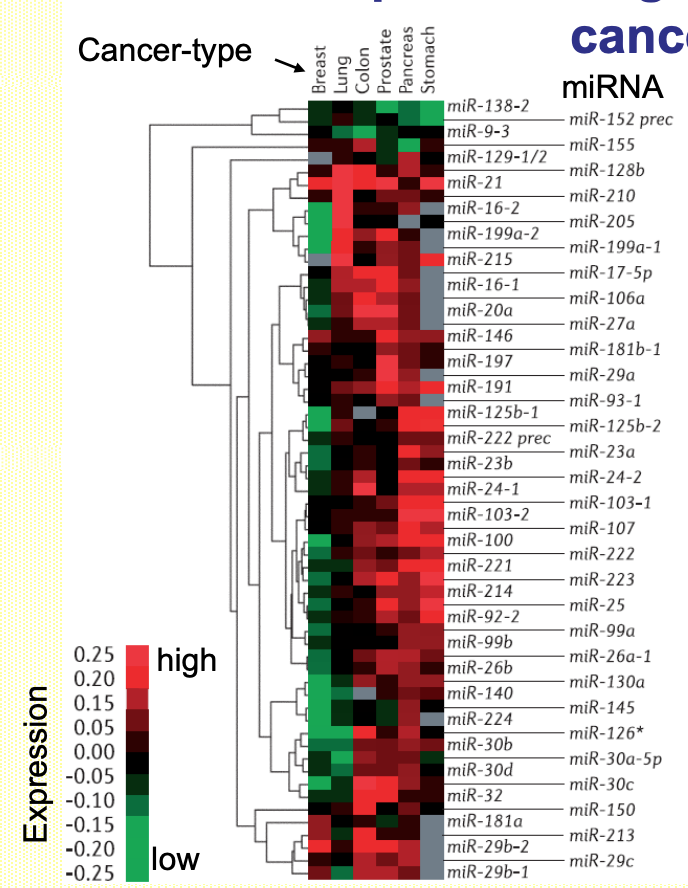
miRNAs expression signatures classify human cancers
Profiling of miRNA expression in diff issues (tumours) enables to establish disease (tumour) specific ‘cancer markers’ for diagnostics
Many cancer-related miRNAs specifically target mRNAs coding for proteins w/ key functions in cell proliferation, migration & immune-response
Heat-map representing expression of miRNAs in 6 solid tumours - expression signature
Long non-coding RNAs (lncRNAs)
- Human: 16,000 lncRNA genes annotated (GENCODE v26) - giving raise to ~30,000 diff transcripts
- Mainly transcribed by Pol II & sharing similarities w/ mRNAs → most lncRNAs contain a 5’ cap & a poly(A) tail at 3’end
- Not translated into proteins but functional mols
- Tissue/cell type specific expression - many of them v. low abundance (1-2 copies/cell)
- Involved in many cellular processes e.g. gene imprinting, cell differentiation & development, antiviral response etc.
- Function of thousands of incRNAs is unknown
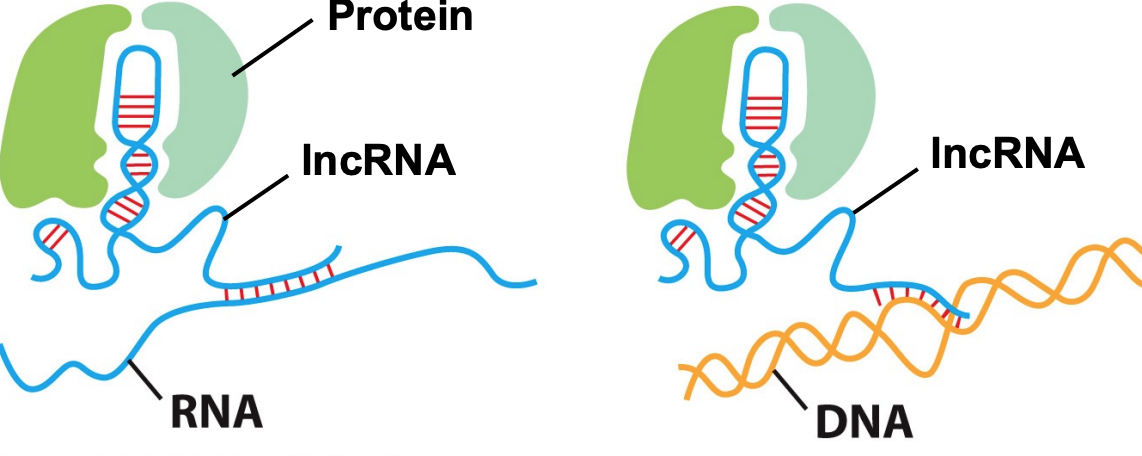
LncRNAs can interact w/ proteins, RNA & DNA to execute regulatory functions in nucleus & cytoplasm
Nuclear lncRNAs control chromatin structure/transcription in cis or trans
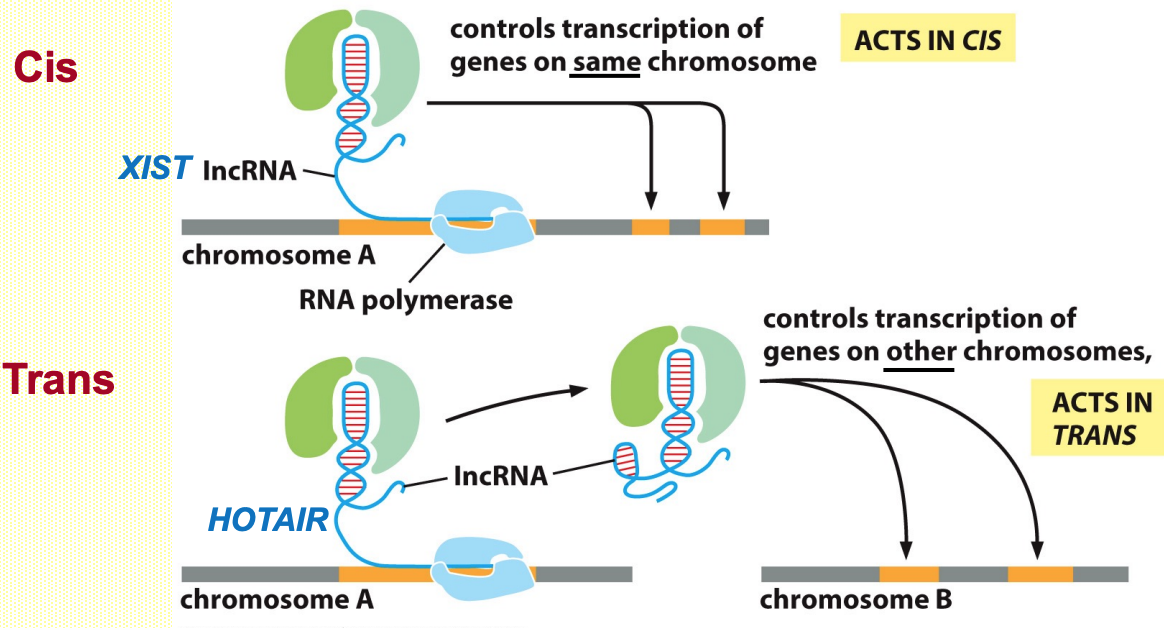
XIST - 1st lncRNA discovered controls mammalian dosage compensation
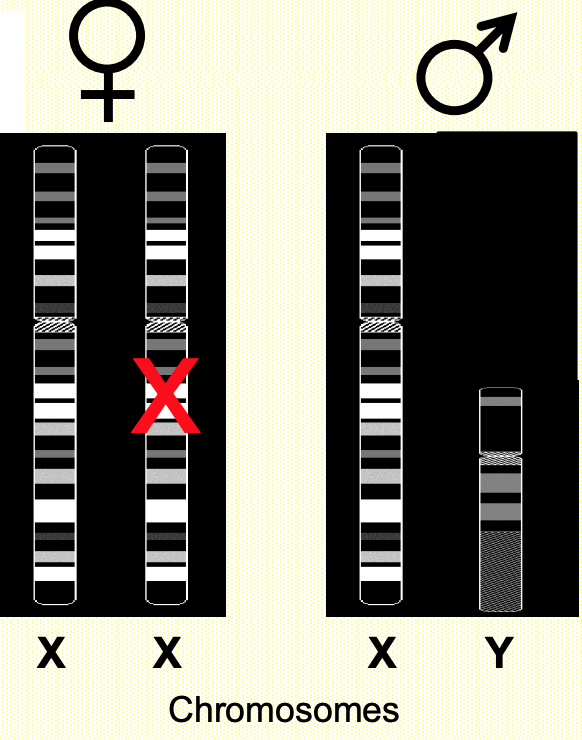
XIST: X inactivation specific transcript
Xist is a large (17kb) cis-acting regulatory lncRNA
XIST associates w/ X-chromosome that is was expressed from (cis regulation)
Initiates histone modifications (methylation, deacetylation) →results in heterochromatin formation
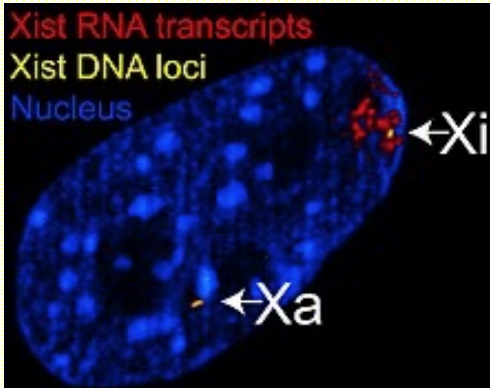
Deletion of Xist gene abolishes X inactivation
Xa: active X chromosome
Xi: inactivated X chromosome
Cytoplasmic lncRNA have diverse functions
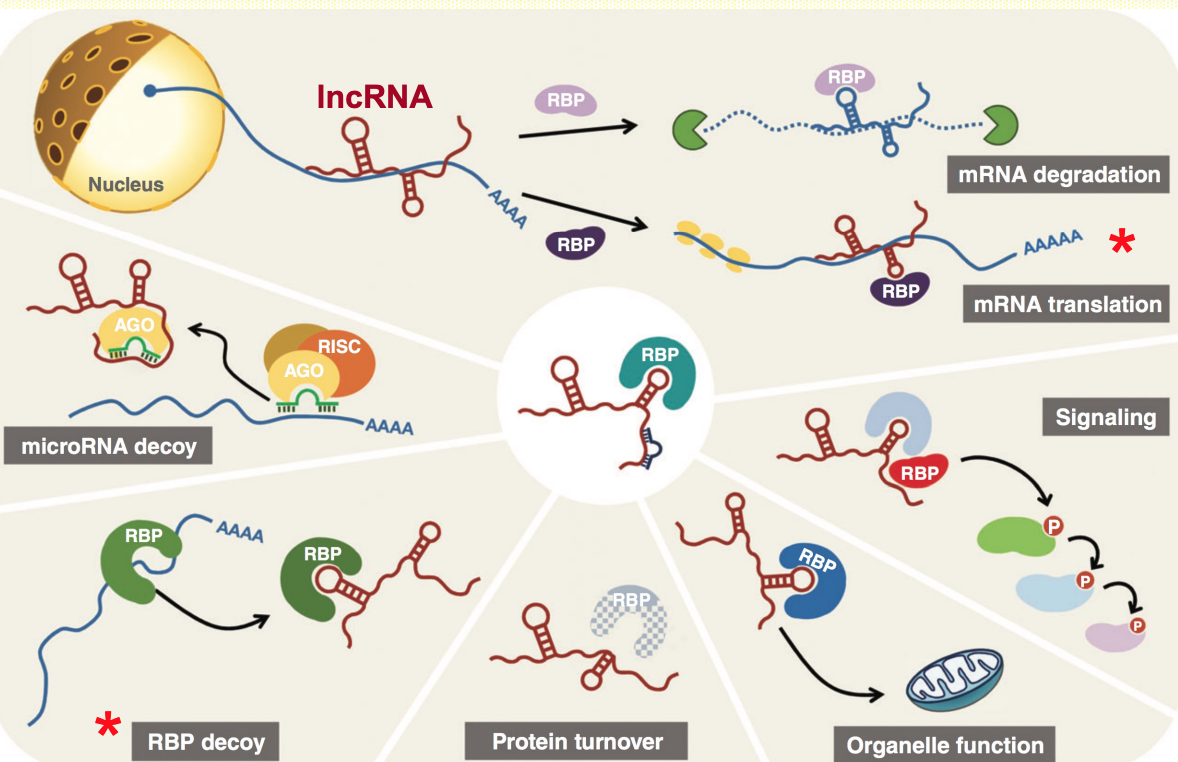
LncRNAs can regulate mRNA stability & translation (examples)
mRNA stability
lncRNA 𝘛𝘐𝘕𝘊𝘙 interacts w/ complementary sequences in a target mRNA & recruits RNA-binding protein (STAU1) → promotes stability of mRNA
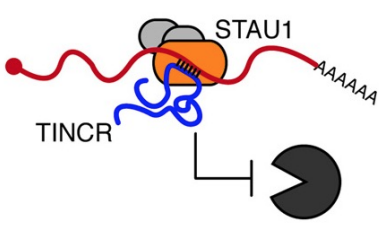
Translational control
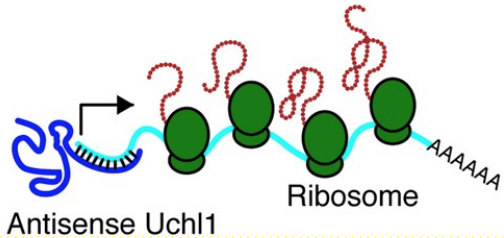
Under stress conditions, lncRNA antisense to Uchl1 moves from nucleus to cytoplasm & binds the end of Uchl1 mRNA to promote cap-independant translation
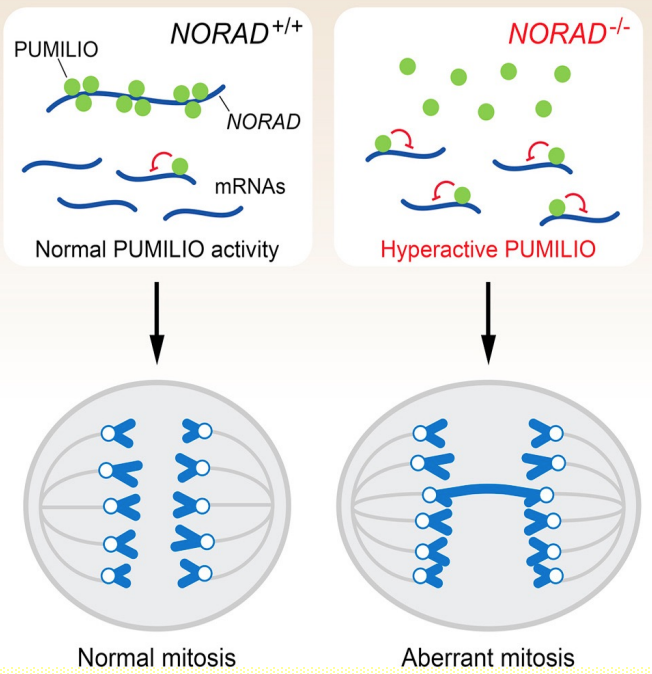
lncRNAs (NORAD) can act as decoy for RNA-binding proteins
- NORAD is lncRNAs activated by DNA damage
- NORAD IS ~5,3 kb polyadenylated transcript predominantly localised in cytoplasm
- NORAD sequesters PUMILO RNA-binding proteins - acting as -ve regulator by limiting their availability to interact w/ mRNA targets
- Involved in control of cell mitosis
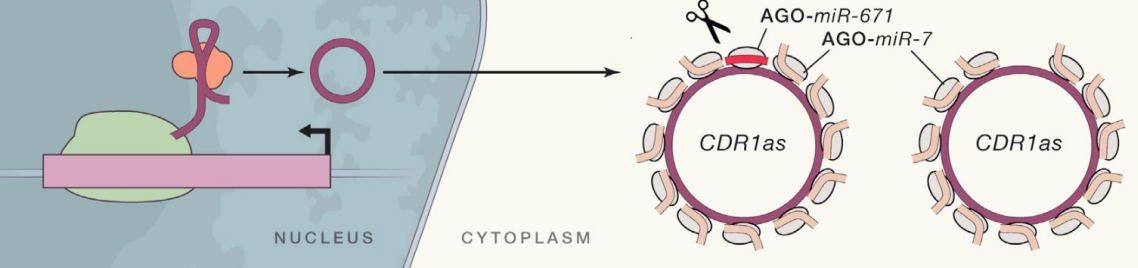
Circular RNAs (circRNAs) can act as decoys for miRNAs/RBPs
- 100s of circular RNAs (e.g. CDR1as) have recently been discovered in eukaryotes
- CDRs are generated via back-splicing mechanisms by joining 5’ & 3’ end of linear RNA molecules originating from protein coding genes
- CircRNAs lack cap/poly(A) tails - highly abundant & stable
- Originally thought to be non-coding, but recent data shows circRNAs can be translated into proteins ( not really non-coding RNA)
SUMMARY
- tRNAs and rRNAs are most abundant ncRNAs w/ fundamental functions in protein synthesis & ribosome architecture, respectively.
- Small nuclear (snRNAs) are required for splicing
- Small nucleolar RNAs (snoRNAs) guide site-specific modification of rRNA
- Regulatory small ncRNAs such as miRNAs/siRNAs control gene expression post-transcriptionally by annealing to sequences in the 3’UTRs of mRNA targets.
- Processing of miRNAs/siRNAs involves Drosha, Dicer & other proteins to finally form the RISC complex that assembles on mRNA target.
- 1000s of lncRNAs (>200 nts) exist in eukaryotes that have nuclear & cytoplasmic functions in gene expression control.
- Circular RNAs (circRNAs) are highly abundant & stable; produced by a back-splicing mechanism in eukaryotes.