Biology Exam Review
Unit 1: Biochemistry
Functional Groups
Hydroxyl (Alcohols)
Carbonyl (Aldehydes or Ketones)
Carboxyl (Carboxylic acid)
Amino (Amines)
Sulfhydryl (Thiols)
Phosphate (Organic Phosphates)
Chemical Bonding
Hydrogen Bonding
Water forms weak bonds with each other (Making it polar)
Due to oxygen being positively charged, it attracts more electrons that it shares with Hydrogen
Electrons spend more time near oxygen
Hydrogen Bonding involves bonds to highly electromagnetive atoms
Covalent bonds: Electrons being shared between two atoms
Ionic Bonds: Positively charged ion bonds with a negatively charged ion
Intramolecular forces: Ionic or covalent
Intermolecular forces
Dipole-Dipole: the positive end of one polar molecule and the negative end of another attracts each other
London Dispersion: Electrons in two adjacent atoms occupy positions that make the atoms form temporary dipoles
Dehydration vs. Hydrolysis
Dehydration: The removal of a water molecule when two molecules join
Hydrolysis: The splitting of a large molecule by adding water
Macromolecules
Carbohydrates(C6H12O6)
Made of Carbon, Hydrogen, and Oxygen
Used for short term energy storage and cellular respiration
Monosaccharides (Fructose, Glucose, Galactose): Monomer containing a carbonyl and multiple hydroxyls
If the carbonyl is at the end, it is an aldose. If not, it is a ketose
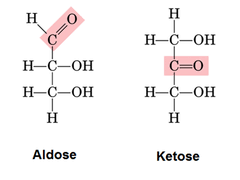
Dissacharides: two monosaccharides formed by a glycosidic linkeage
E.g. Glucose + Glucose = Maltose
Polysaccharide: polymer of many monosaccharides
Starch: Energy store in plants
Glycogen: energy store in the liver and muscles
Cellulose: structural components of plant cell walls
Chitin: used for exoskeleton
Lipids
Made of Carbon, Hydrogen, and Oxygen
Used for long term energy storage, insulation, organ cushoning, and nerve impulses
Glycerol: three carbon skeleton with a hydroxyl attached
Fatty acid: Carboxyl with a long hydrocarbon tail
Triglyceride: three Fatty acid’s and one glycerol joined together by an ester linkage
Saturated fats: has no carbon-carbon double bond, solid at room temperature, found mostly in animals
Unsaturated fats: Contains one or morecarbon-carbon double bond, liquid at room temperature, found mostly in plants
Proteins
Functions
Enzymes (catabolyze metabolic reactions)
Antibodies and bloodclotting
Motion
Component of the cell membrane
Structural support
Transport
Made of the same sets of 20 monomers (Amino acids)
Polymer is polypeptides
Made of a hydrogen, A carboxyl group, an amino group and an R chain
Polypeptides are made through a Peptide Bond via dehydration
Primary Structure: long sequence of amino acids Secondary structure: Amino acids form hydrogen bonds along the backbones
Tertiary Structure: R group to R group bonds (and R group to backbone bonds)
Quaternary Structure: Aggregation of two or more polypeptides
Denature: the unfolding of proteins due to extreme conditions (temperature and pH)
Nucleic Acids (RNA and DNA)
Directs growth and development of organisms
Monomer is nucleotides
Nucleotide components
Pyrimidines: single ringed structures (Cytosine, Thymine, and Uracil)
Purines: double ringed structures (Adenine and Guanine)
Pentose Sugar: deoxyribose (DNA) or ribose (RNA)
Phosphate group
The connection of the sugars and phosphate creates a polynucleotide via phosphodiester linkage
Adenine bonds with Thymine or Uracil, Guanine bonds with Cytosine
Enzymes
Speeds up reactions and acts as a Catalyst (Chemical agent that changes the rate of reactions)
Functions
Lowers the activation energy for a reaction to occur
Provide a favourable space for the breaking/forming of bonds
Promotes the conversion of reactions into products
Enzymes binds to susbtrates at an active site, forming an enzyme-substrate complex
Lock and Key model: outdated hypothesis determining how enzymes are locks that fit into a specific substrate (key)
Induced fit hypothesis: Active site of an enzyme is a pocket where a substrate fits (Determined by the four protein structures)
Factors affecting enzymes
Enzyme and Substrate Concentration
A low substrate concentration creates a faster conversion of products due to the excessive enzyme capacity
Vice-versa will create a slower conversion as enzymes are occupied
Reactions will speed up with there are more enzymes added
Enzyme Inhibitors
Inhibitors competes with subtrates for an active site, making them not converted into products
Allosteric Regulation
Inhibitor binds to an allosteric site and changes the shape of an enzyme
Changes the activity of the enzyme
Temperature and pH
Enzymes work at a specific temperature and pH range. Too much heat can either speed up collision or weaken bonds (or denature)
Cell Transport
Isotonic: equal amounts of water into and out of the cell
Hypotonic: net movement of water into the cell (Causes cells to burst aka. Lysis)
Hypertonic: net movement of water out of the cell (Causes the cells to shrink aka. Plasmolysis)
Active Transport: movement of substances against the concentration gradient (Low concentration to High concentration)
Achieved through a cell membrane protein (E.g. Sodium/Potassium Pump)
Diffusion: movement from a high concentration to a low concentration area
Pinocytosis: cells intake a small drop of ECF
Phagocytosis: cells intake large molecules and organic matter
Endocytosis:
Plasma Membrane
Proteins and Functions
Integral/Intrinsic: provides structural support, recognizes cells through binding to protein sites of other cells, signaling, and transport
Peripheral: connected to other proteins
Glycoproteins: Enters and communicates with the cell
Unit 2: Metabolism
Part 1: Cellular Respiration
Structure of mitochondria
Matrix: where cellular respiration occurs (Pyruvate Oxidation, Krebs Cycle)
Outer and inner member membrane(Electron transport chain)
Intermembrane space
Oxidation vs. Reduction
Oxidation: Atom loses an electron
Reduction: Atom gains an electron
Endothermic: absorption of heat energy
Exothermic: release of heat energy
Endergonic: absorption of energy (Free energy is positive and nonspontaneous)
Exothermic: release of energy (Free energy is negative and spontaneous)
Glycolysis
Breaking down glucose into 2 pyruvate molecules
Net Yield: 2 ATP, 2 NADH
Hexokinase used towards glucose makes Glucose-6-phosphate
Phosphoglucoisomerase used towards G6P makes Fructose-6-phosphate
Phosphofructokinase used towards F6P makes Fructose-1, 6-Bisphosphate
Aldolase is used to make Dihydroxyacetone Phosphate
Isomerases is used to make Glyceraldehyde-3-Phosphate
Triose Phosphate Dehydrogenase is used to make 1, 3-Bisphosphoglycerate
Phosphoglycerokinase is used to make 3-Phosphoglycerate
Phosphoglyceromutase is used to make 2-Phosphoglycerate
Enolase is used to make Phosphoenol pyruvate
Pyruvate Kinase is used to make Pyruvate
Pyruvate Oxidation
Pyruvate turns into acetyl-CoA
Net Yield: 2 NADH
Krebs Cycle
The production of ATP, NADH, and FADH2 with acetyl-CoA
Net Yield: 2 ATP, 6 NADH, 2 FADH2
Citrate synthetase is used to make Citrate
Aconitase is used to make Isocitrate
Isocitrate dehydrogenase is used to make Alpha-ketogluterate
Alpha-Ketogluterate dehydrogenase is used to make Succinyl-CoA
Succinyl-CoA synthetase is used to make Succinate
Succinate dehydrogenase is used to make Fumarate
Fumarase is used to make Malate
Malate Dehydrogenase is used to make Oxaloacetate
Electron Transport Chain
Series of transport proteins and electron carriers forming H2O when oxidized
Net Yield: 2 NADH from glycolysis makes 4-6 ATP
2 NADH from Pyruvate Oxidation makes 6 ATP
6 NADH and 2 FADH2 makes 22 ATP
Total: 32-36 ATP
ETC Proteins/Complexes: NADH Dehydrogenase Complex, Ubiquinone, Cytochrome b-c complex, Cytochrome c, cytochrome oxidase complex, ATP Synthase
NADH and FADH2 passes their electrons to complexes
NADH to NADH Dehydrogenase Complex, FADH2 to Ubiquinone
Final electroon acceptor is Oxygen (2H+ + 2 electrons + ½ O2 = H2O)
ETC is Exergonic, which pumps Hydrogen from matrix to intermembrane space and gets transported through each complex
ATP Synthase: makes ATP from ADP and Pi
Chemiosmosis: using stored energy of an electrochemical gradient and the ATP Synthetase to generate ATP
Fermentation
Generation of some ATP without oxygen
Alcohol: pyruvate is converted to ethanol (2 Pyruvates = 2 acetaldehydes + 2 CO2)
(2 Acetaldehydes + 2 NADH + 2 H+ = 2 Ethanol + 2 NAD+)
Lactic acid: pyruvate is reduce to lactate (2 Pyruvates + 2 NADH = 2 Lactate + 2 NAD)
Part 2: Photosynthesis
Pigments: substances that absorb light (photons)
The colour we see is NOT absorbed, but reflected by the pigment
Examples: Chlorophyll a (P680 and P700)
Light Harvesting Complex: consist of various pigment molecules that are bound to proteins
Noncyclic Electron Flow
Energy transfer through photosystems
Step 1: Photosystem II absorbs a photon of light striking a pigment molecule. It boosts its electrons to a higher energy level. Electron reaches P680 making it excited and transfers to the primary electron acceptor
Step 2: Z proteins splits water to release O2
Step 3: Excited electron passes through an Electron Transport Chain (Plastoquinone, Cytochrome complex, and plastocyanin)
Step 4: Electron falls to a lower energy level for the synthesis of ATP
This is noncyclic phosphorylation
Step 5: photon strikes Photosystem I pigment P700, boosting the electron level
Step 6: Primary Electron Acceptor passes electron down to the second ETC (Ferredoxin and NADP Reductase)
Cyclic Electron Flow
When ferredoxin donates back an electron to plastoquinone (repeats continually by moving through the membrane)
Occurs when a plant has enough NADPH
Energy from light converts to ATP without the oxidation of H2O or reduction of NADP+ to NADPH
Calvin Cycle
ATP and NADPH converts CO2 to sugar
Production of Glyceraldehyde-3-phosphate (G3P)
Calvin cycle must occur three times to produce G3P
6 CO2+Glucose
Carbon Fixation: CO2 attaches to Rubilose Bisphosphate (RuBP) by the catalyzation from rubisco
The product is too unstable and splits in half to make two molecules of 3-phosphoglycerate
Reduction: Each 3-phosphoglycerate recieves a phosphate group to form 1, 3-Bisphosphoglycerate
Product is reduced by NADPH to make G3P
Regeneration: Carbon skeletons of 5 G3P are rearranged to 3 RuBP
C4 Plants
Physically separated Calvin cycle and light reactions, O2 is not exposed by rubisco
CO2 is limited, C4 Cycle feeds CO2 to rubisco
CAM Plants
Opens stomata at night, allowing CO2 to enter and minimize H2O loss
Converts malate which is stored till day
Unit 3: Molecular Genetics
Gene: Functional and physical unit of heredity passed from parent to offspring
Carries a code (information) for making a specific protein
Allele: variant form of a gene
DNA Structure
In a form of a double helix (Found by Rosalind Franklin)
Contains a phosphate group, deoxyribose sugars, and a nitrogenous base
Phosphate is attached to the sugar of one nucleotide, creating a backbone of alternating Phosphate and sugars
Nucleotide consists of purines (Adenine and Guanine), and pyrimidines (Cytosine, Thymine, Uracil)
Complementary Base Pairs: Adenine makes two hydrogen bonds with Thymine, Guanine makes three hydrogen bonds with Cytosine
RNA Structure
Single Strand
Phosphate group, ribose sugar, nitrogenous bases
DNA Replication
Semiconservative process where a daughter cell gets a copy of a genome
Begins at the origin of replication, where short stretches of DNA have a specific sequence that proteins recognize
Replication bubble: made when proteins bind to sites in DNA
Replication fork: Y-shaped region at the end of the Replication Bubble
Enzymes Used (also in order of steps)
Helicase: untwists the double helix model
Single-strand binding proteins: binds to unpaired DNA
Topoisomerase: relieves strand by breaking and rejoining strand
RNA Primer: short stretch of RNA nucleotides made by primase
DNA Polymerase III: adds DNA nucleotides to the RNA primer
DNA Polymerase I: replaces RNA nucleotides with DNA nucleotides
Ligase: forms a bond between DNA fragments to previous fragments (glue)
Okazaki Fragments: discontinuous segments
New DNA Strand Synthesis
DNA Polymerase III adds DNA nucleotides that comes from a Nucleoside Triphosphate
Hydrolysis of pyrophosphate (molecule released when nucleotide is added) becomes two Pi
Anti-parallel Elongation
DNA Polymerase is only able to add nucleotides in the 3’ end
Strand elongates only in the 5’-3’ direction
Leading Strand
DNA Polymerase III adds nucleotides to the 3’ end of the growing strand
Synthesizes a complementary strand by elongating in the 5’-3’ end
Lagging Strand
DNA Polymerase III works away from the replication fork
Strand is discontinuous and occurs in segments (Okazaki fragments)
Proofreading and Repairing DNA
Base pair mistakes occurs at a rate of 1/10,000 base pairs
DNA Polymerase proofreads and removes wrong nucleotides
Nucleoside incision repair: the enzyme nuclease cuts a segment of a damaged strand
DNA Damage: occurs from reactive processes (X-rays, UV Light, Emissions)
Mismatch repair: enzymes fix incorrectly paired nucleotides
Protein Synthesis
Transcription
Turning DNA into RNA in the nucleus
Initiation
RNA Polymerase unwinds DNA and binds at the prometer (a sequence of DNA at the start of the gene)
TATA Box: Section of DNA with a high percentage of Adenine and Thymine. Enables the binding of RNA Polymerase (Two hydrogen bonds needed to open)
Elongation
RNA Polymerase adds nucleotides to the 3’ end of the growing strand
RNA is made using the 3’-5’ strand (the template strand), while the 5’-3’ strand is the coding strand
Termination
RNA Polymerase recognizes a termination sequence (A protein-coding gene that ends transcription)
Can be a string of Adenines that are transcribed to Uracils in RNA
Post-transcriptional Modifications
Newly transcribed RNA is now pre-RNA and is vulnerable to be attacked by RNA digesting enzymes
Poly(a) tail: modification where pre-RBA acquires a chain of 50-250 adenines to the 3’ end by the help of poly-A-polymerase
5’ cap: modifications where pre-RNA acquires 7 Guanines at the 5’ end to prevent itself from hydrolytic enzymes
Exons (Coding regions) and Introns (non-coding regions) are transcribed but makes dysfunctional. Introns are deleted by RNA splicing
Spliceosome: complex formed between pre-mRNA and small nuclear ribonucleproteins that binds to introns and removes them
Translation
Turning mRNA into a polyeptide in the ribosome with tRNA
Codon: 3 base pairs for an amino acid in the 5’-3’ order
tRNA: small RNAs that are 70-90 nucleotides long
Picks up amino acids in the cystol and deposits in the ribosome
Structure: Has regions that base pair with themselves forming four double-helical segments
Contains an anticodon at the tip (3 nucleotide segment that pairs with a codon in mRNA)
Example: mRNA codon: 5’ AAG tRNA codon: 3’ UUC
Wobble Hypothesis: pairing of the anticodon with the first two nucleotides are precise, but the third has flexibility and codes for the same amino acid
Aminoacylation: the addition of an amino acid to tRNA
Catalyzed by 20 aminoacyl-tRNAs that drives its energy to form a peptide bond
Ribosomes: made up of rRNA (ribosomal RNA) and ribosomal proteins
Facilitate the coupling of tRNA anticodons with mRNA codons
Binding sites to tRNA molecules
A-site: aminoacyl site holds the incoming aminoacyl-tRNA that is carrying the next amino acid to the growing polypeptide chain
P-site: Peptidyl site holds the tRNA that is growing the polypeptide chain
E-site: Exit site is where tRNA leaves the ribosome
3 Stages of Transcription
Initiation
Initiator tRNA has the anticodon specific to the codon AUG (start codon). The tRNA carries methionine (now called Met-tRNA)
Met-tRNA forms a complex that binds to the mRNA at the 5’ cap
Large ribosomal subunit binds to the mRNA and met-tRNA is automatically in the P-site
Elongation
Step 1: Hydrogen bonding forms between the mRNA codon under the A-site at the anticodon carrying the amino acid
Step 2: Second tRNA attaches to the codon at the A-site of the ribosome
Methionine is cut off from the P-site tRNA and attached to the tRNA in the A-site
Growing polypeptide chain is attached to the tRNA in the A-site
Step 3: Ribosomes moves to the next codon on the mRNA (tRNA moves from the A-site to the P-site, Steps repeat)
Termination
Ribosome reaches a stop codon (UAA, UAG, UGA) to stop elongation
A protein release factor binds to the A-site to release the newly formed polypeptide
Regulating Gene Expression
Lac Operon: regulates lactose metabolism
Trp Operon: regulates tryptophan production
Both are controlled by negative feedback loops
Lac Operon
Consists of a promoter, an operator(controls transcription), and coding regions for enzymes
Lac repressor is a gene upstream that controls the production of lactose-metabolizing proteins by sensing lactose levels
Lac repressor binds to the operator when lactose is absent, making it active. This stops RNA Polymerase from attaching to the promoter that blocks the production of lactose metabolizing enzymes
Trp Operon
Same structures as Lac Operon (promoter, operator, coding regions)
When tryptophan is absent, the repressor is inactive and allows RNA Polymerase to bind to the promoter, which transcribes the genes to make tryptophan
When tryptophan is present, tryptophan acts as a corepressor and activates the repressor proteins (binds to promoter and stops RNA pol from transcribing genes)
Genetic Mutations
Causes of Mutations
Induced: chemicals and radiation
Spontaneous: Inaccurate DNA Replication
Small scale mutations
Point Mutations
Substitution: one base pair is replaced by another
Insertion: A single base pair is added
Deletion: A single base pair is removed
Inversion: Two adjacent base pairs are swapped
Single Nucleotide Polymorphisms (SNPs): Differences in DNA caused by Point mutations
Missense mutations: changes in base pairs that make a different amino acid
Protein ends up either dysfunctional, function different, or beneficial
Nonsense mutations: base pair change results in a premature stop codon
Protein is incomplete and non-functional
Silent mutations: base pair change that doesn’t alter the function of the gene
DNA codes for the same amino acid, protein is unchanged
Frameshift mutations: insertion or deletion of nucleotides
Affects all amino acids leading to multiple missense or nonsense mutations
Large Scale Mutations
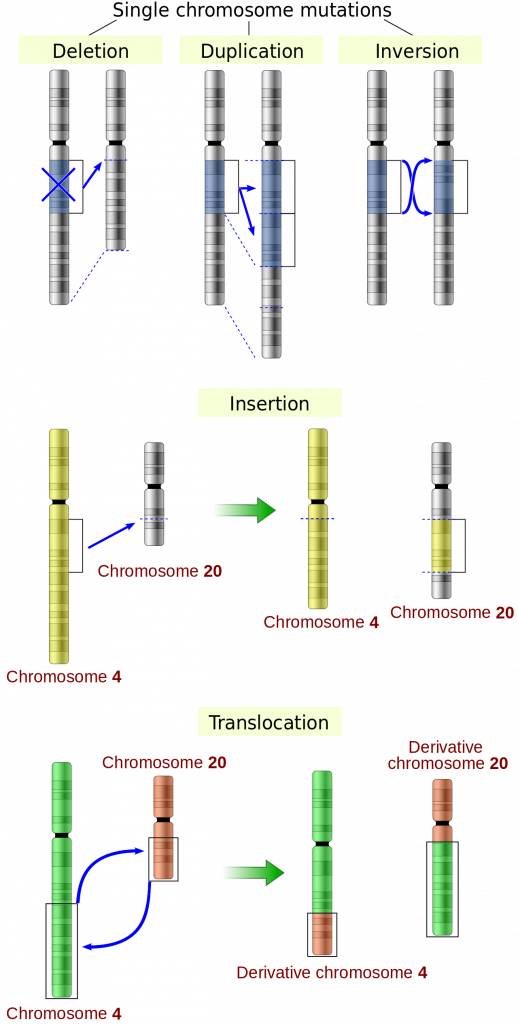
Gene duplication: one or more genes copied to multiple regions of a chromosome
Large scale deletion: Entire coding regions of DNA is removed
Translocation: genes move from one chromosome to another
Inversion: portion of DNA reverses direction within the genome
Trinucleotide repeats: repeating sequences in the genome (E.x CAG CAG CAG)
Unit 4: Homeostasis
The maintenance of a constant environment in the body
Body maintains 1. Temperature 2. Water levels 3. Glucose concentration 4. pH levels
Negative Feedback systems
Homeostatic control system with three components
Receptor: detects a change in the body
Control center: processes the information from the receptor
Effector: creates a response
ADH Feedback mechanism: Increases blood osmotic pressure when there’s an increase of solutes in blood and decrease of water concentration
Osmoreceptors: located in the hypothalamus that detects a change in blood osmotic pressure
Control Center: hypothalamus shrinks due to water moving out of the cell. Sends nerves to the pituitary gland to release ADH in the blood stream
Effector: Kidneys reabsorb water to dilute solute concentration, making more concentrated urine
Nervous System
Nerve signaling
Membrane Potential: electrical gradient across the membrane
Anions (-): Concentrated inside the cell
Proteins, amino acids, sulfate inside, Cl outside
Cations (+): Concentrated outside of the cell
K+ inside, Na+ outside
Resting potential: -70mV
Action potential: All or Nothing depolarization thats triggered if the potential reaches -55mV
Chemical Synapse
Depolarization: the action potential depolarizes the plasma membrane at the synaptic terminal
Ca2+ influx: Voltage gated Ca2+ channels open to allow Ca2+ into the presynaptic terminal
Vesicle fusion: The high Ca2+ concentration causes synaptic vesicles to fuse with the presynaptic membrane
Neurotransmitter release: Vesicles release neurotransmitters into the synaptic cleft
Receptor Binding: Neurotransmitters bind to receptors on ligand-gated ion channels on the postsynaptic membrane
Neuron Structure
Dendrites: ends that receive information and conducts nerve impulses
Cell body: contains the nucleus and other organelles
Axon Hillock: junction between body and axon
Axon: carries nerve impulses away from the cell and towards the neurons
Myelin Sheath: white coat of fatty protein that insulates the neruons to prevent itself from losing charged ions
Node of ranvier: Indentations between sections of myelin sheath
Central Nervous System
Brain and spinal cord
Gray matter: unmyelinated axons, cell bodies, and dendrites
White matter: myelinated axons
Peripheral Nervous System
Consists of nerves that carry information between the organs of the body and the CNS
Somatic Nerves (Voluntary): controls skeletal muscles, bones, and skin
Sensory Somatic Nerves: relays information about the environment to the CNS
Motor Somatic Nerves: Initiates appropriate responses to external stimuli
Automatic Nerves (Involuntary): manages the internal organs, smooth muscle, and cardiac muscle
Sympathetic Nervous System: prepares the body for stress
Parasympathetic Nervous System: restores normal balance
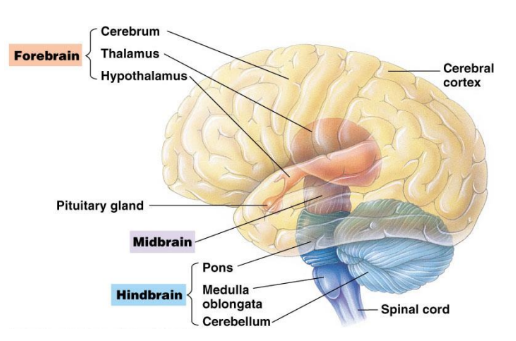
Brain Structure
Brainstem: lower part of the brain containing the Medulla Oblongata, pons, and the midbrain
Maintains homeostasis, coordinates movement, and conducts impulses to and from higher brain centers
Medulla and Pons: Controls automatic nerve functions (Breathing, digestion, swallowing)
Relays information to and from higher brain centers
Cerebellum: coordinates motor activities and perceptual functions
Sends sensory information to the cerebrum via pons
Thalamus: relays sensory information to the cerebrum
Hypothalamus: regulates automatic activity and controls the pituitary gland
Excretory System
Kidney: removes waste, maintains water balance, supplies blood in its renal artery, filters waste
Renal Cortex: outer layer of the kidney
Medulla: inner layer beneath the cortex
Renal pelvis: cavity connecting the kidney to the ureter
Nephron: functional unit of the kidney
Bowman’s Capsule: encircles the glomerulus
Glomerulus: capillaries in the Bowman’s Capsule that makes the first step of filtration
Afferent Arteriole: blood supplying unfiltered blood from the body to the Glomerulus
Efferent Arteriole: blood vessel carrying blood away from the Glomerulus to the rest of the body
Proximal Tubule: connects the Bowmans capsule to the loop of Henle
Peritubular Capillaries: reabsorbs essential ions and minerals from filtered blood
Loop of Henle: U shaped portion connection the proximal tubule to the distal tubule
Distal Tubule: connects the Loop of Henle to the ducts
Urine Formation
Filtration: blood to the Bowman’s Capsule at a fast rate
Bowman’s Capsule cells and the Glomerulus makes a selectively permeable membrane
Pores of the Glomerulus allows blood contents to enter the nephron (But large blood proteins aren’t allowed)
Amino group is removed when proteins enter, creating a by-product of ammonia
Ammonia + CO2 = Urea
Reabsorption: transfer solutes and water to the blood
Begins in the proximal tubule where water, ions, and nutrients are transferred back into the interstitial fluid
Amino acids, glucose, nutrients, and salts are reabsorbed from the filtrate
Microvilli in the tubule increases the surface area for better reabsorption
Water concentration increases in the filtrate by osmosis
Aquaporins (water channels) in the Descending LoH enables more water reabsorption
Peritubular Capillaries allows reabsorbed substances while remaining filtrate thats high in Urea moves to the Loop of Henle
Ascending LoH: absorbs salt by passive diffusion, if concentration decreases it is absorbed by active transport
Secretion: transfer of materials from blood to nephron
Waste from blood and interstitial fluid are removed and secreted into the proximal tubule
H+ ions and detoxified substances are secreted in the nephron and excreted through urine
Distal Tubule secretes K+ and more H+ ions
Kidney secretes excess H+ ions when acidity rises
Endocrine System
Hormone released by negative feedback
Endocrine glands: Secrete hormones (Hormones regulate growth and speed or slow metabolism)
Regular endocrine glands: glands that act exclusively like endocrine glands (e.x. pituitary gland, pineal gland, thyroid gland, adrenal gland)
Tissues and Organs: structures that don’t function like glands, but still secretes hormones (e.x. hypothalamus, pancreas, thymus, ovaries & testes)
Steroid Hormones: lipid based hormones made of cholesterol
Diffuses easily though the cell membrane (since its lipid soluble) and easily binds to the nucleus’ receptor
Water soluble hormones: Amino acid/protein based hormones
Cannot diffuse through the cell membrane, but binds to the cell membrane’s receptor instead to activate cAMP. cAMP activates enzyme cascades
Posterior Pituitary
Part of the nervous system that does NOT produce hormones
Uses ADH and oxytocin (made by the hypothalamus)
Hormone Travel: Hypothalamus produces ADH and Oxytocin in neurosecretory cells
Both hormones move down the axon and gets released into the blood when reaching the endings
Anterior Pituitary
Produces its own hormones
Releases 6 important hormones (TSH, ACTH, PRL, HGH, FSH, LH)
Hormone Travel: Hypothalamus produces releasing or release inhibiting hormones that uses a network of blood vessels (The portal system)
Hormones stimulate the secretion of other hormones which are later released into the blodstream)
Dwarfism: less hGH Gigantism excess HGH in childhood Acromegaly: more HGH in adulthood
 Knowt
Knowt