Exam 2- Overview
Ch 10- Membranes
1. Be able to recognize phospholipids (ie, phosphatidylserine).
Major lipids in cell membrane: Phosphoglycerides, Sphingolipids, and Sterols
2. Understand the fluidity of membrane and the factors affect it (Lipid phase
transition: be able to interpret Differential scanning calorimetry data).
Lipid Phase Transition: This refers to the temperature at which the membrane transitions from a gel-like state to a fluid state.
Differential Scanning Calorimetry (DSC): DSC measures the heat flow associated with phase transitions. A peak in the DSC curve indicates the melting temperature (Tm), where the membrane becomes more fluid.
Factors: Fatty acid saturation (unsaturated increases fluidity), cholesterol content (buffer), and temperature all affect membrane fluidity
At higher temps: increase fluidity
Lower temps: decreased fluiditySaturated: straight structure allows tighter packing, so less permeable
Unsaturated: Kinks prevent tight packing so increased permeability
3. Understand the role of Cholesterol in membrane fluidity.
High Temperatures: Cholesterol stabilizes the membrane, reducing excessive fluidity.
Low Temperatures: Cholesterol prevents tight packing of phospholipids, maintaining some fluidity.
Buffer: Overall, cholesterol acts as a fluidity buffer, ensuring the membrane remains functional across different temperatures.
4. Be able to describe the asymmetric distribution of lipids on the biological bilayers
Inner vs. Outer Leaflet: Phospholipids like phosphatidylserine and phosphatidylethanolamine are predominantly found in the inner leaflet (negatively charged) , while phosphatidylcholine and sphingomyelin (neutral phospholipids) are found in the outer leaflet.
Lipid rafts: cholesterol-rich microdomains in cell membranes that organize and facilitate efficient signal transduction and protein interactions
Functional Significance: This asymmetry is crucial for various cellular processes, including membrane curvature, vesicle formation, and cell signaling.
1. Understand the Hydropathy plot.
Definition: A hydropathy plot is a graphical representation that displays the hydrophobic (water-repelling) and hydrophilic (water-attracting) regions of a protein sequence.
Purpose: It helps predict transmembrane segments of proteins based on the hydrophobicity of amino acid residues.
Interpretation: Peaks above a certain threshold indicate hydrophobic regions likely to be within the lipid bilayer, while valleys below the threshold indicate hydrophilic regions likely to be outside the membrane
2. State different types of membrane proteins and what non-covalent interaction
drives their incorporation into the lipid bilayer.Integral (Transmembrane) Proteins:
Hydrophobic Interactions: These proteins span the lipid bilayer and their hydrophobic regions interact with the hydrophobic core of the membrane. This helps anchor the protein within the membrane.
Example: Ion channels, receptors.
Peripheral Membrane Proteins:
Electrostatic Interactions: These proteins are attached to the surface of the membrane or to integral proteins through electrostatic interactions (attraction between opposite charges).
Hydrogen Bonds: They can also interact through hydrogen bonds with the polar head groups of lipids or with other membrane proteins.
Example: Cytochrome c, certain enzymes.
Lipid-Anchored Proteins:
Covalent Attachment: These proteins are covalently attached to lipids within the membrane, such as fatty acids, prenyl groups, or glycosylphosphatidylinositol (GPI) anchors.
Stabilization: The hydrophobic lipid moiety helps anchor the protein in the membrane through hydrophobic interactions.
Example: G-proteins, prion proteins
3. Understand the basic rationale behind isolating functional membrane proteins
using detergent.Purpose: Detergents are used to solubilize membrane proteins while maintaining their functional integrity.
Mechanism: Detergents mimic the lipid environment by surrounding the hydrophobic regions of the membrane protein, preventing aggregation and maintaining their native conformation.
Preservation: By preventing denaturation, detergents ensure that the proteins retain their function during purification
4. Understand a way to measure membrane protein mobilityFluorescence Recovery After Photobleaching (FRAP): A method where a specific region of the membrane is bleached with a laser, and the recovery of fluorescence is monitored over time. The rate of fluorescence recovery indicates the mobility of membrane proteins.
Single Particle Tracking (SPT): Tracks the movement of individual membrane proteins labeled with fluorescent tags, providing detailed information on their diffusion and mobility within the membrane.
Understand “Electrochemical gradient” and “Membrane potential”
Electrochemical Gradient:
Definition: An electrochemical gradient is the combined effect of both the concentration gradient of ions and the electrical gradient across a membrane.
Components: It consists of two parts:
Chemical Gradient: The difference in the concentration of ions across the membrane.
Electrical Gradient: The difference in charge (voltage) across the membrane.
Significance: The electrochemical gradient drives the movement of ions across the membrane, influencing various cellular processes, including transport and signaling.
Membrane Potential:
Definition: Membrane potential is the voltage difference across a cell's membrane, resulting from the unequal distribution of ions.
Measurement: It is typically measured in millivolts (mV) and is established by the activity of ion channels and pumps.
- State different types of membrane transport and the proteins that are involved.Passive Transport:
Simple Diffusion: Movement of molecules from high to low concentration without energy input.
Proteins Involved: Not typically involved.
Facilitated Diffusion: Movement of molecules through membrane proteins from high to low concentration.
Proteins Involved: Carrier proteins, channel proteins (e.g., aquaporins for water transport).
Active Transport:
Primary Active Transport: Movement of molecules against their concentration gradient using energy from ATP.
Proteins Involved: Pumps (e.g., Na+/K+ ATPase).
Secondary Active Transport: Uses the energy from the electrochemical gradient of one molecule to drive the transport of another molecule against its gradient.
Proteins Involved: Symporters and antiporters (e.g., SGLT for glucose transport).
- What is “Resting membrane potential” and how to measure it?Definition: The resting membrane potential is the voltage difference across the cell membrane when the cell is at rest (typically around -70mV in neurons).
Maintenance: It is maintained by the Na+/K+ pump, which pumps three Na+ ions out of the cell and two K+ ions into the cell, creating a negative charge inside the cell relative to the outside
Measured by the inner leaflet. In animal cells should always be negative
- Describe how the plasma membrane potential is generated.
Ion Distribution: The difference in ion concentrations across the membrane, mainly Na+, K+, and Cl-, contributes to the membrane potential.
Ion Channels: Selective permeability of the membrane to different ions through ion channels (e.g., K+ leak channels).
Na+/K+ Pump: Actively transports Na+ out and K+ into the cell, contributing to the negative charge inside the cell.
Overall Effect: The combination of ion distribution, selective permeability, and active transport mechanisms generates and maintains the plasma membrane potential
1. What are the differences between “Transporter” and “Channel”?
Transporter: A protein that facilitates the movement of ions or molecules across the cell membrane through a conformational change, often requiring energy (e.g., ATP). AGAINST THE GRADIENT
Channel: A protein that forms a pore in the membrane, allowing ions to flow through passively and rapidly, driven by concentration gradients without the need for energy. FACILITATED DIFFUSION
2. Be able to describe how Na+-Glucose symporter utilizes the electrochemical gradient of
Na+ for active transport of Glucose.
This symporter allows glucose to be transported into the cell against its concentration gradient by coupling its movement to the flow of sodium ions down their electrochemical gradient, which is maintained by the sodium-potassium pump. Binding of NA and glucose to transporter is cooperative (one facilitates the other). They have opposite gradients. Na concentration is high outside cell, so it moves inward. Glucose is high inside cell but is brought into cell anyway using Na gradient.
3. What are the three (or four) components that enables the transcellular “active” transport
of Glucose?
Asymmetric distribution of transporters in epithelial cells including:
Sodium-potassium pump (Na+/K+ ATPase) - Maintains the sodium gradient by actively transporting Na out and K into the cell.
Sodium-glucose symporter (SGLT) - Facilitates the simultaneous transport of Na and glucose into the cell.
Glucose transporter (GLUT) - Allows glucose to exit the cell into the bloodstream once it is inside.
4. Be able to describe the steps for “Action potential” generation and propagation.
Action potential generation begins with a depolarization phase where sodium channels open, allowing Na+ to rush into the neuron, causing the membrane potential to become more positive.
This is followed by the repolarization phase, where sodium channels close and potassium channels open, allowing K+ to exit the cell, restoring the negative membrane potential.
Finally, the hyperpolarization phase occurs as the membrane potential temporarily becomes more negative than the resting potential before returning to equilibrium.
5. How does “excitatory” and “inhibitory” neurotransmitter work in terms of membrane
potential?
Excitatory neurotransmitters cause
depolarization of the postsynaptic neuron
(ie, Acetylcholine, glutamate)
Inhibitory neurotransmitters cause the
postsynaptic cell to hyperpolarize (ie,
GABA, Glycine)
6. Be able to describe how “channel-rhodopsin” can generate action potential upon light
illumination.
Channelrhodopsin is a light-sensitive ion channel that, when exposed to specific wavelengths of light, opens to allow the influx of cations such as sodium (Na+) into the neuron. This influx leads to a depolarization of the membrane potential, which can trigger an action potential if the threshold is reached, thereby enabling precise control of neuronal activity through optogenetics.
1. What is Chemiosmotic mechanism and how it is used in bacteria?
The chemiosmotic mechanism refers to the process by which ATP is produced using the energy derived from the electrochemical gradient of protons (H+) across a membrane. In bacteria, this mechanism is utilized during cellular respiration and photosynthesis, where protons are pumped across the cell membrane, creating a proton motive force. This force drives ATP synthase to convert ADP and inorganic phosphate into ATP as protons flow back into the cell, providing the energy necessary for various cellular functions.
2. Understand how mitochondria generates and utilizes the electrochemical gradient
of H+ for ATP synthesis.
- Remember how mitochondrial membrane system is organized: The outer
membrane envelops the inner boundary
membrane. The inner membrane is highly
folded into tubular or lamellar cristae, which
crisscross the matrix. Note that the
inner membrane is compartmentalized into
the inner boundary membrane and the
crista membrane. There are three distinct
spaces: the inner membrane space, the
crista space, and the matrix.
- Remember what the electron transport system does
The electron transport system facilitates the transfer of electrons through a series of protein complexes, ultimately leading to the pumping of protons (H+) from the matrix into the inner membrane space, thereby establishing the electrochemical gradient necessary for ATP synthesis. NADH Transfers Its Electrons to Oxygen Through Three Large Enzyme Complexes (Complex I, II,and III) Embedded in the Inner Membrane of mitochondria
- Remember the direction that H+ gradient (or pH)
Protons are pumped from the matrix into the inner membrane space, creating a higher concentration of H+ ions in the inner membrane space compared to the matrix, which is crucial for ATP production during oxidative phosphorylation.
3. Remember that mitochondrial is constructed from Nuclear gene and Mitochondrial
gene
- Be able to describe the function and organization of nucleus and nuclear
envelop.
Nucleus function: Contains genome, contains sites for transcription and ribosome assembly
Nucleus: Center is the nucleolus (site of ribosome assembly), surrounded by chromatin (DNA and proteins), Then the nuclear lamina (fibrous protein meshwork), then the inner nuclear membrane and outer nuclear membrane (inner and outer make up the nuclear envelope).
Envelope: There are channels in the envelope (nuclear pores containing NPCs). The endoplasmic reticulum extends from the outer membrane. Ribosomes are bound to systolic surfaces of ER and nuclear outer membrane. The outside of the ER is the ER membrane and the inside is the ER lumen. There is a perinuclear space in between the inner and outer nuclear membrane of the nucleus.
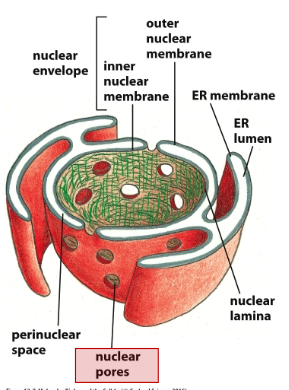
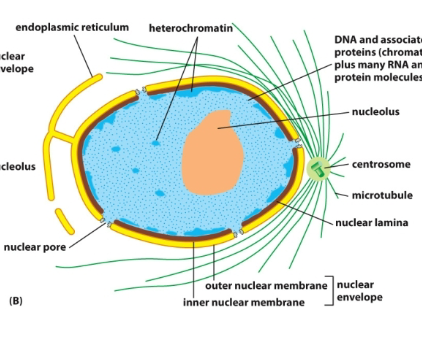
- Understand how the NPC controls the selective passage of cargos in and
out of nucleus (FG-repeat meshwork through the pore).
Gate transport: Nuclear pores contain nuclear pore complexes (NPCs), which forms a permeable barrier for large macromolecules. Filled with disordered regions (contains a lot of phenylalanine and glycine (FG-repeats)) to block passive diffusion of large macromolecules. Nuclear localization signals (NLSs) direct nuclear proteins to nucleus. Then Importins/Exportins (nuclear import receptors) bind to NLSs , facilitating the transport of proteins into the nucleus by recognizing and interacting with the FG-repeat meshwork of the NPC (weakens mesh to be able to move in and out AND determine cargo exchange rate).
-Understand the role and mechanism of Ran-GTPase (or Ran) in nuclear import
as well as export through the GDP-bound and GTP-bound conformation (ie;
cargo, importin/exportin, Ran-GAP, Ran-GEF)
Ran GTPase-activating protein (Ran- GAP) is located in the cytosol. Ran guanine nucleotide exchange factor (Ran-GEF) binds to chromatin and is therefore located in the nucleus. Antagonistic for import because importins can bind to RAN GTP or to cargo, not both. The cooperative relationship between exportins and RAN-GTP, is that it can bind to both at the same time. Ran-GTPase cycles between GTP-bound (active) and GDP-bound (inactive) states to regulate nuclear import and export. During nuclear import, cargo with NLS binds to importin and translocates through the nuclear pore complex. Inside the nucleus, Ran-GTP binds to the cargo-importin complex, causing a conformational change in importin that releases the cargo. For nuclear export, cargo with NES binds to exportin and Ran-GTP in the nucleus. The complex moves through the nuclear pore, and in the cytoplasm, Ran-GAP hydrolyzes Ran-GTP to Ran-GDP, causing a conformational change that releases the cargo. Ran-GDP and exportin are then transported back to the nucleus, where Ran-GEF catalyzes the exchange of GDP for GTP on Ran, reactivating it for another cycle
- Be able to state the functions of the ER (Smooth and Rough ER).
Smooth: processing or storing non-protein molecules (drug detox, Ca storage, and lipid bilayer synthesis)
Drug detox: Monooxygenase enzymes (cytochrome p-450 family) add hydroxyl groups to hydrophobic drugs to increase solubility (hydroxylation) so easier to export)
Ca storage: Lumen has lots of Ca binding proteins. Ca ions are pumped into ER by calcium ATPases and released when needed (like muscle contraction or intracellular signaling). Process happens in sarcoplasmic reticulum for example.
Lipid bilayer synth: ER is primary source of membrane lipids because fatty acids are made in cytoplasm and incorporated into ER membrane on cytoplasmic side, then transferred to ER lumen by phospholipid translocators (flippases). Then membrane proteins are made in cytosol and then exchange requires protein translocation across bilayer.
Rough ER: production and processing of membrane secreted proteins.
- What are the ER targeting signal and a signal receptor and how
they work?
ER signal sequence: a short (nonpolar, hydrophobic) peptide sequence that directs the protein to the ER. It opens aqueous channel in translocator during translation so that co-translational transport across the membrane can occur, allowing newly synthesized proteins to enter the lumen of the ER directly as they are being made. In transmembrane proteins, ER signal sequence remains in bilayer as a helix to anchor it (single pass). In multipass transmembrane proteins, there is a combo of start transfer (internal ER signal sequence) and stop transfer signals
Signal recognition particle (SNP): a ribonucleoprotein complex that recognizes and binds (hydrophobic pocket filled with methionine residues) to the ER signal sequence, and directs the ER Signal Sequence to a specific receptor in the rough ER membrane while pausing the protein translation on ribosome.
Most Proteins Synthesized in the Rough ER Are Glycosylated by the Addition of a
Common N-Linked Oligosaccharide (on Asn)
- Be able to describe how the ER copes with accumulated unfolded
proteins.
Molecular chaperones assist in the proper folding of these proteins by preventing aggregation and facilitating correct conformations, thus ensuring that proteins reach their functional state before exiting the ER. Also, glucosyl transferase adds a glucose moiety to the oligosaccharide, which serves as a recognition signal for chaperones, ensuring that only properly folded proteins are allowed to exit the ER.
ERAD: Endoplasmic Reticulum-Associated Degradation, a quality control mechanism that targets misfolded proteins to export them from ER to be degraded in cytosol
UPR: Unfolded Protein Response, a cellular stress response that is activated by the accumulation of misfolded proteins in the ER, aiming to restore normal function by halting protein translation, degrading misfolded proteins, and enhancing the production of molecular chaperones. It will trigger cell death if stress is too severe.
IRE1 pathway: This pathway is activated by the accumulation of unfolded proteins in the ER and plays a crucial role in the UPR. IRE1 is a transmembrane protein that, upon activation, splices XBP1 mRNA to produce a potent transcription factor that enhances the expression of genes (chaperones) involved in protein folding and degradation, thereby alleviating ER stress.
- Be able to tell co-translational and post-translational protein
transport.
Co-translational: Happens in the ER as ribosomes synthesize proteins while simultaneously translocating them into the endoplasmic reticulum lumen, allowing for immediate folding and modification.
Post-translational: mitochondrial precursor proteins are imported as unfolded polypeptide chains. They have an amphiphilic alpha helix that acts as a mitochondrial signal sequence.
- What are the functions of peroxisome and how proteins are
transported into peroxisome?
Functions: Detoxification reaction, beta oxidation (long chain fatty acid breakdown), and biosynthesis of unusual molecules (like plasmogens)
Transport: native, folded proteins traverse the lipid bilayer of the peroxisome (posttranslational)