Bio 130 Module 4: Reproduction
Finish Module 3
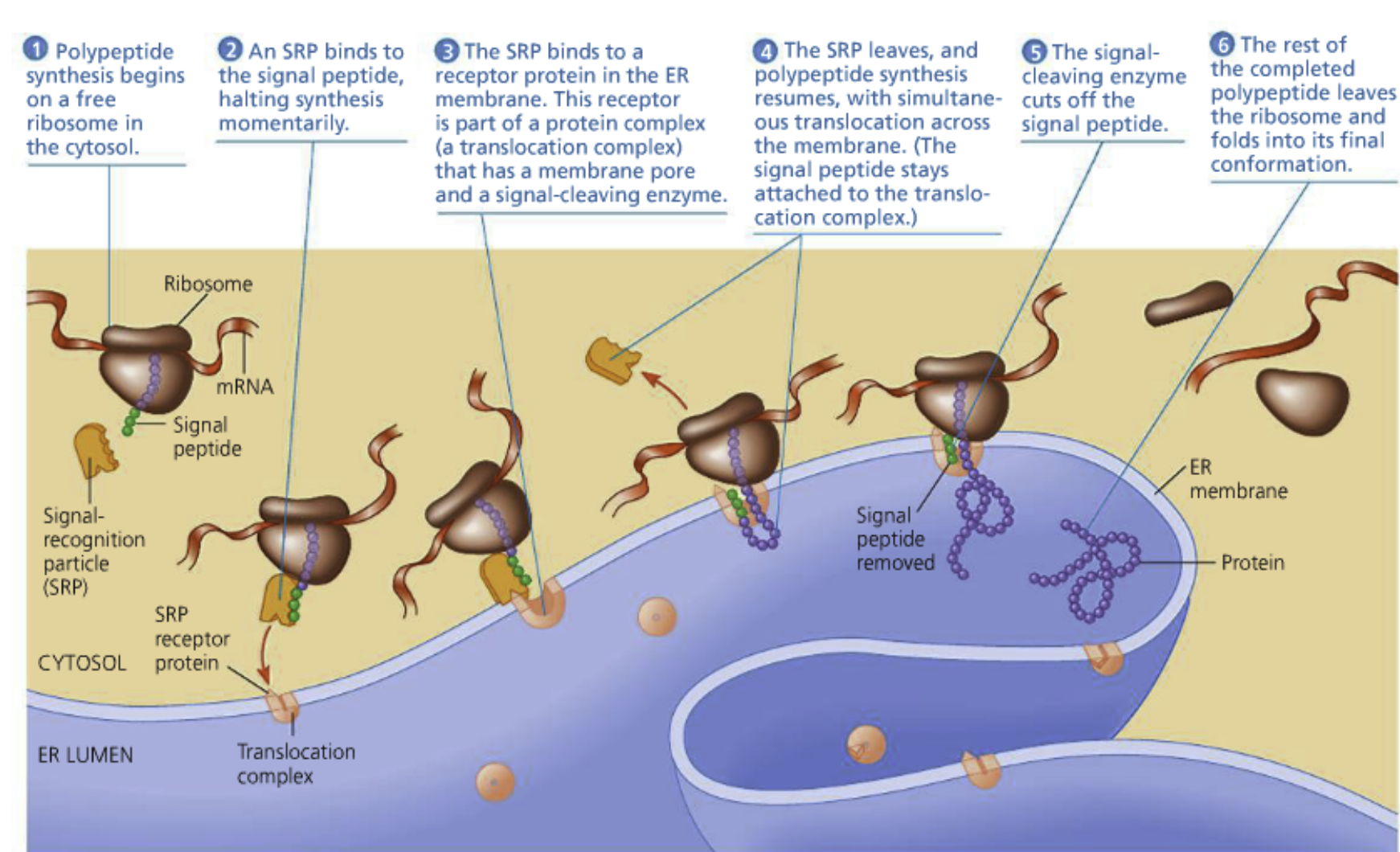
A ribosome performs a cycle of tRNA binding, peptide bond formation, and ejection for each codon of the transcript
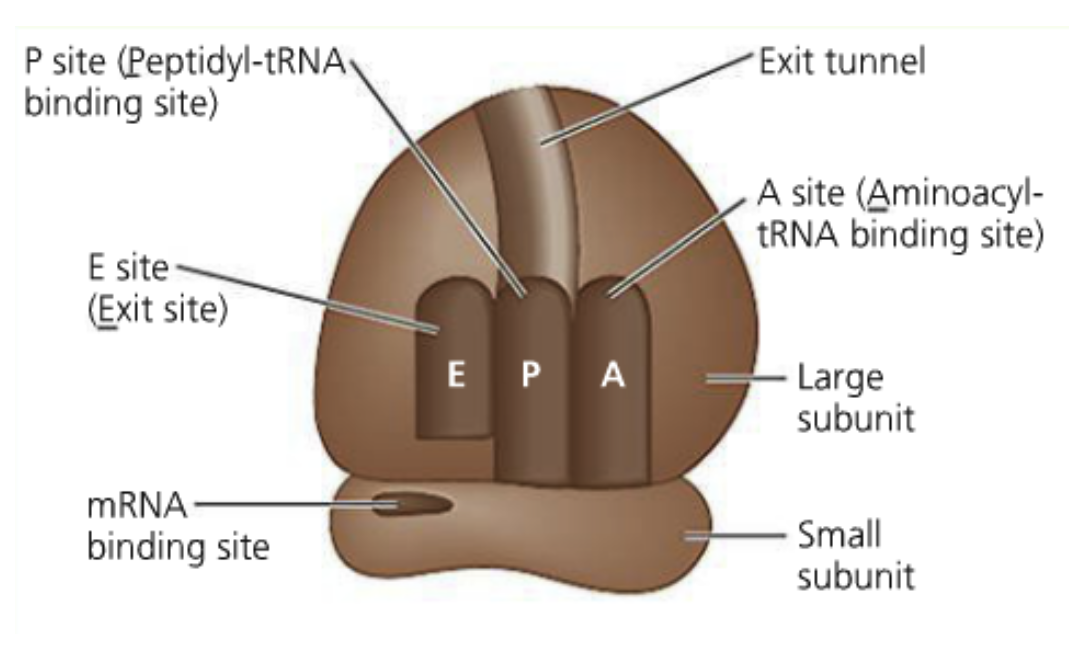
A ribosome has 4 binding sites:
All three types of RNA are represented (mRNA is the transcript we’re reading, tRNA is the adaptor between language of codons and amino acids, rRNA is the ribosome itself (and subunit for ribozyme))
mRNA binding site (The smallsubunit is what binds to it)
The aminoacyl-tRNA site (A site- think of it as the acceptor site)
Codon-anti codon interaction initially occurs here
The peptidyl-tRNA site (P site)
The growing polypeptide is held by tRNA here
The exit site (E site)
tRNAs without amino acids are ejected here
Translation always begins with methionine (AUG)
Ribosome formation begins when the small subunit binds to tRNA charged with methionine
The small subnit-initiator tRNA complex binds to the 5’ cap of the transcript
It will begin scanning in the 5’ to 3’ direction, looking for the first AUG
In bacteria (no cap) a 5’ sequence called the Shine-Dalgarno sequence plays the role of the 5’ cap
When the subunit locates the first AUG, the initiator tRNA is matched to the start codon
The large subunit will bind to the complex and the initiator tRNA is positioned in the P site
At this point we are ready for translation and move into the elongation phase
Peptide elongation cycle:
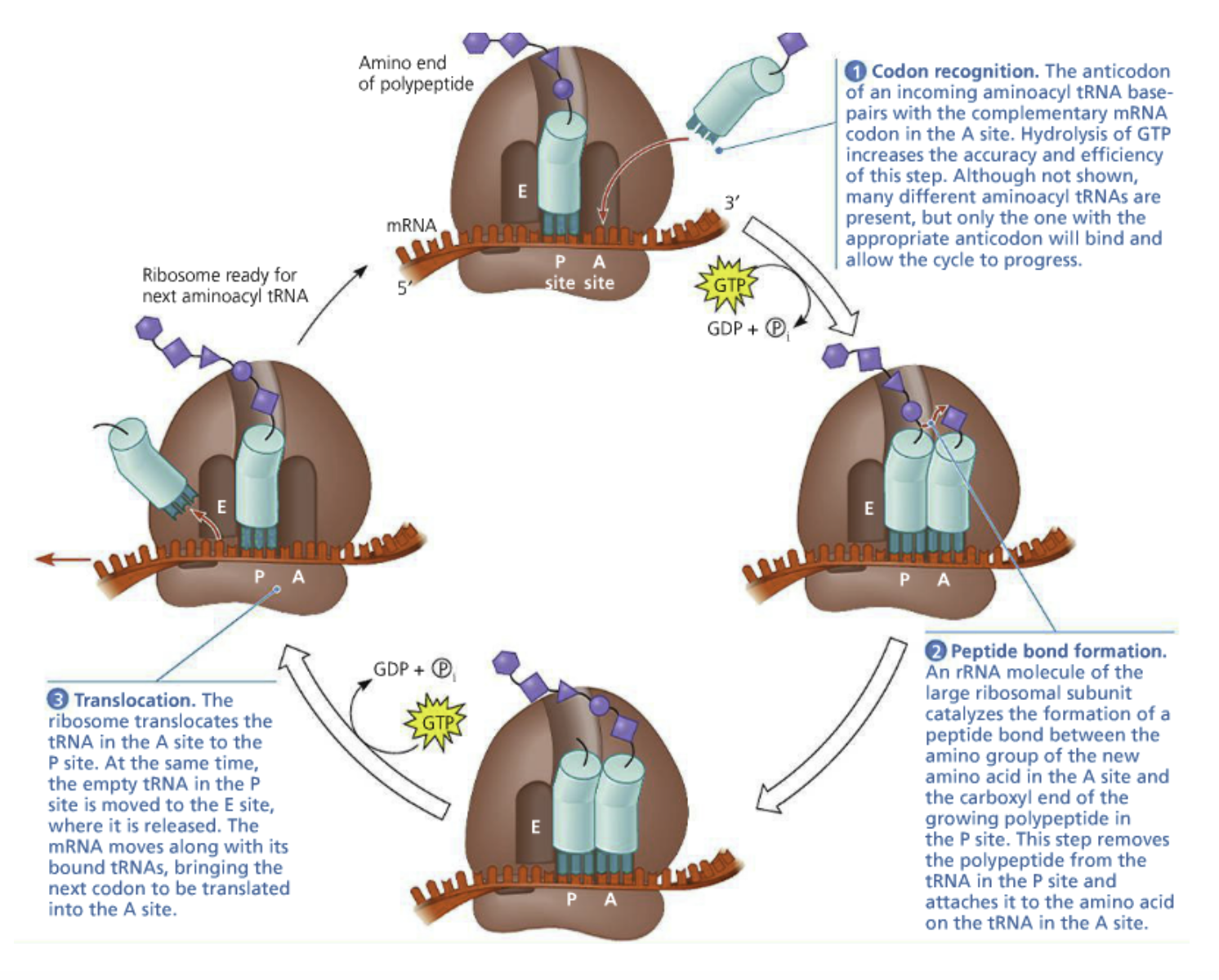
1. Codon recognition: The anticodon of an incoming aminoacyl tRNA base pairs with complementary mRNA codon in the A site (via hydrogen bonds)
GTP hydrolysis increases accuracy
Many aminoacyl tRNAs are present, only one with the appropriate anticodon will bind
2. Peptide bond formation: An rRNA molecule of the large ribosomal subunit catalyzes the formation of a peptide bond between the amino group of the new amino acid in the A site and carboxyl end of the growing polypeptide chain in the P site
Polypeptide is removed from the tRNA in the P site and attached to the amino acid on the tRNA in the A site
Translocation: The ribosome translocates the tRNA in the A site to the P site
The empty tRNA in the P site is moved to the E site and released
The mRNA moves with its bound tRNAs and the next codon is brought to be translated in the A site
You have to move it one codon over
Peptide bond formation: The large ribosomal subunit is a ribozyme and catalyzes the peptidyl transferase reaction (reaction explained below)
The amino group of a tRNA (in the acceptor site) forms an amide (peptide) bond with the C-terminal end (meaning a carbon is attacked) of the growing polypeptide
Therefore, polypeptides grow in the N-terminal (N terminus is made first so it’s always going to have methianine) to C-terminal direction (willl always have tRNAs still on the ribosome)
Termination: A stop codon occupies the A site so there are no tRNA molecules that can recognize it (stick long enough to bond)
A release factor (a protein) moves into the A site, causing a water molecule to be added (hydrolyzing) to the C-terminus of the peptide
This truly makes the C-terminus
This frees the peptide and it floats into the cytoplasm
The two ribosomal subunits dissociate
Mimics the tRNA (L shape)
Termination factors are examples of mimicry
Proteins have a 3D structure very similar to tRNA, allowing them to effectively bind to the A site
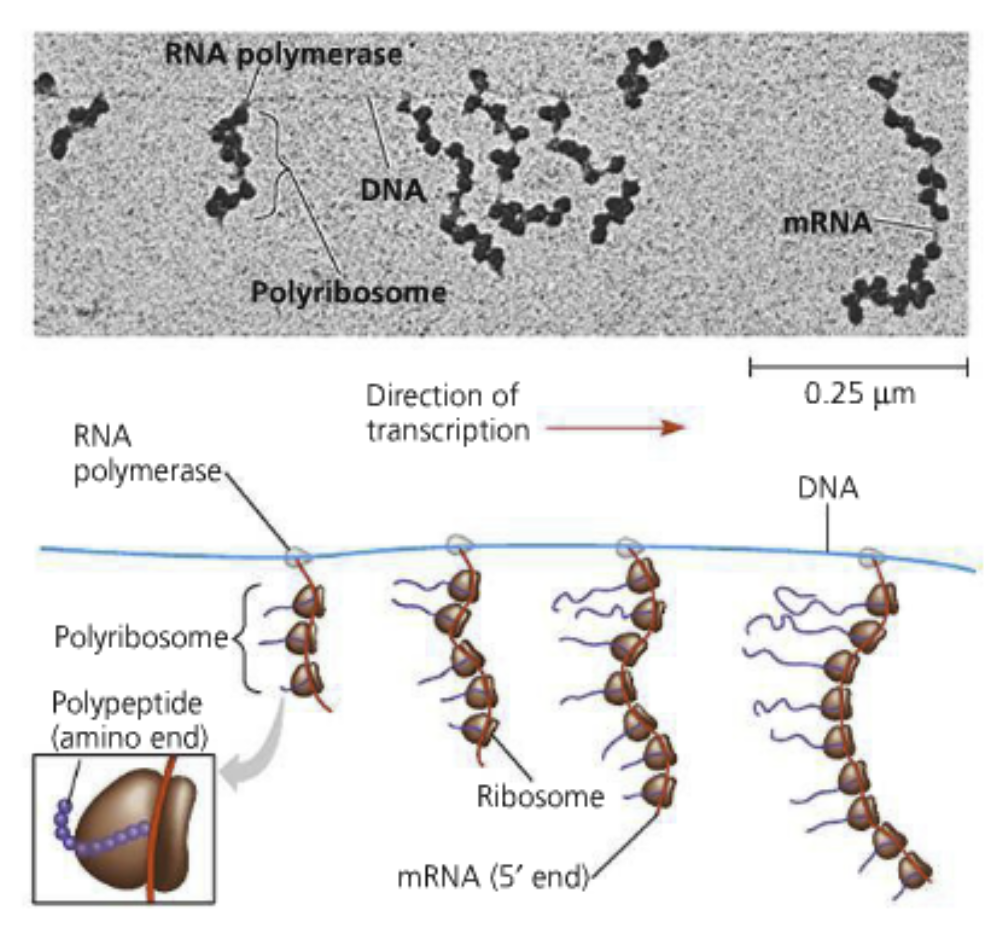
Polyribosomes: When multiple ribosomes simultaneously translate an mRNA
Individual ribosomes form at the start codon (5’ end!), move along the transcript to make the polypeptide, and detach from one another
This is effecient to amplify the message and produce a lot of protein quickly
Bacteria can couple transcription and translation because they don’t have a nucleus or introns
A polypeptide can be made will the gene is being transcribed
Polyribosomes can form on the transcripts
Many proteins fold as they come off the ribosome (the N-terminal may be folded before the C-terminal is complete
Other proteins require chaperone proteins (Hsp70) to fold properly (need to be prevented from folding too early)
The Cell Cycle and Cancer
Remember Virchow and Schwann created cell theory (Virchow did the all cells come from preexisting cells)
In embryos, cells grow/divide rapidly to provide new cells for the developing embryo
More is better!
Zygote: The two gametes (sperm and egg) coming together
When adult tissues are formed, cell division becomes regulated
Cells only divide if new cells are needed
Cell division and cell death are balanced
Ex.: The liver (parenchyma) regrows when damaged
Stem cells: In bone marrow and produce red and white blood cells
They are designed to divide continuously
Neurons: In the nervous system. Very unlikely to divide
We call them post-mitotic
Cancer: Unregulated cell division in adult tissue
Leads to tumors
Tumors parallel reverting back to embryonic form. Divide without limitations
Metastasis: Tumors grow independent of original tissue, colonizing new tissues
The Cell Cycle
Divided into four phases: gap phase 1 (G1), synthesis (S: DNA synthesis) phase, gap phase 2 (G2) and mitosis (M) phase
Lasts about 24 hours
The length of the cell cycle gets longer as you go from embryonic to adult phase
Interphase: G1 + S + G2
A non-dividing cell
In this stage, the nucleus is typically 30nm and 300nm
The histones are in two different shapes (heterochromatin and euchromatin)
The chromosomes are disorganized (and partially decondensed (30nm and 300nm)) in the nucleus
G1 phase: The cell grows, takes in information from environment if it should divide
This cell has just come out of division
Will only transition to DNA synthesis if conditions are favorable
Generally lasts for 5-6 hours
Has the machinery needed to begin replication, it just won’t do it until S phase
G0 phase: Resting stage within G1 if the cell never gets the signal to proceed
Most cells in the body are in this stage
Under certain conditions, some cells can reenter the cycle
S phase: DNA replication
There are certain mechanisms ensuring DNA replications only occurs once
Generally lasts 10-12 hours
Associate nucleotides with them (the 6th question from quiz 9)
At this point we are committed and can’t go back!
G2 phase: After the entire genome has been replicated
This is the second resting phase
The cell continues to grow, making sure it has enough volume, all of its organs, etc.
The cell prepares for cell division
Generally lasts 4-6 hours
M phase: The duplicated genomes are separated, each goes to a daughter cell
Generally lasts 1 hour
This phase includes mitosis and cytokinesis
Mitosis: Making the second nucleus
Cytokinesis: The cleavage (division) between the two cells
The chromatin switch to the mitotic chromosome
Checkpoints: G1-S transition, G2-M transition (quality control), within M phase (anaphase transition)
Ensure outside conditions are favorable and internal processes have been completed
G1 checkpoint: “The restriction point”. If the cell passes this point, it will commit to completing the cell cycle
This is the most important checkpoint
Regulation of the cell cycle
Insight on these mechanisms first came from cell fusion experiments
When cells in G1 and S phase were merged, the G1 nucleus began replicating its DNA (the G1 cell is pulled into DNA synthesis)
When cells in G1 and M phase were fused, G1 began mitosis (the G1 cell is pulled into mitosis)
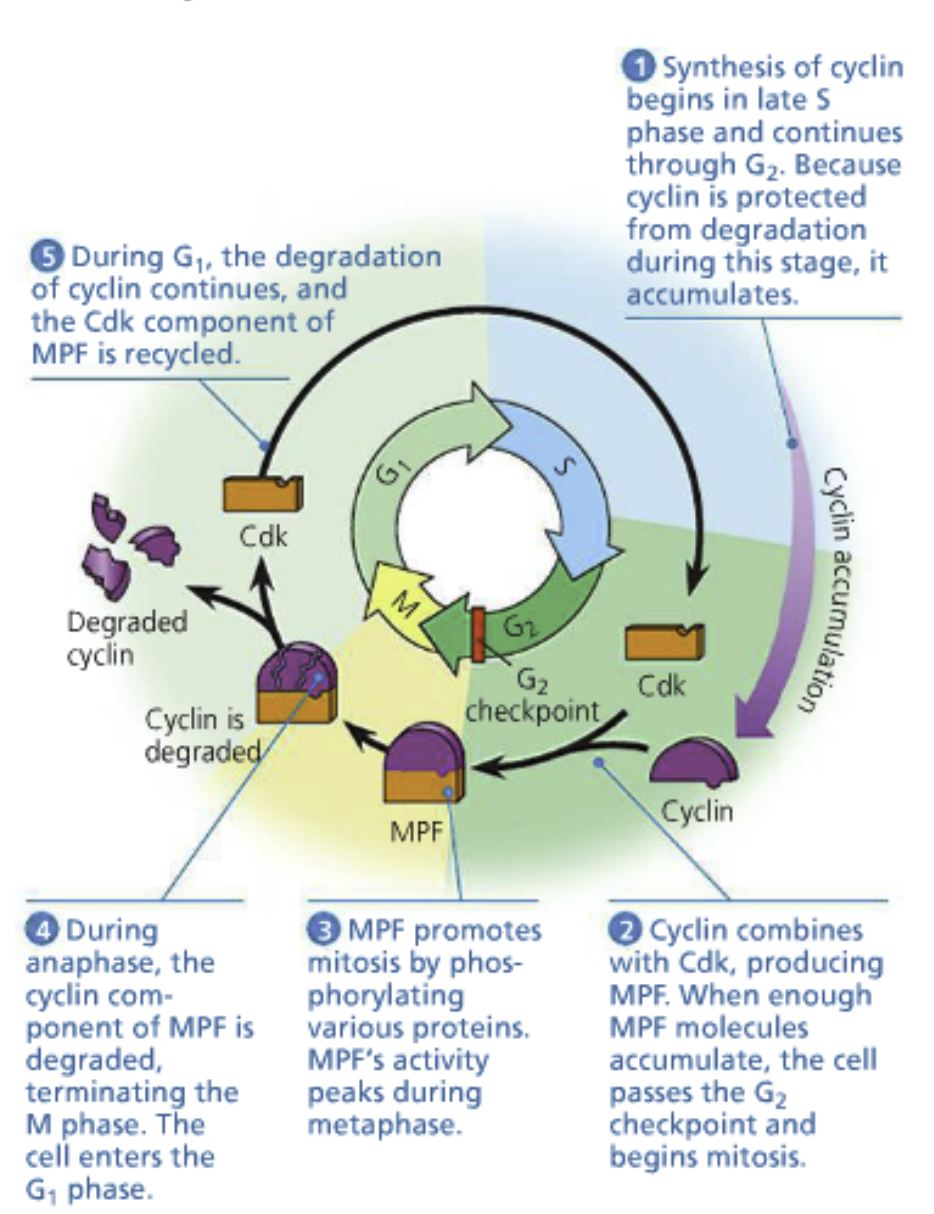
Cyclins: The chemical signal causing the results listed above. Cytoplasmic factors in fusion experiments. Their expression peaks at certain phases of the cell cycle
Cyclins bind to/activate cyclin-dependent kinases (Cdk)
Kinase: An enzyme that phosphorylates proteins→ activating/deactivating them
G1/S cyclins commit the cell to S phase at the end of G1. (Not the same as S cyclins)
G1 cyclins help the cell progress through the G1 checkpoint, not present in all cells
Cyclin-Cdk complex: (The chemical signal causing the results listed) above) Orchestrates events of the next phase of the cell cycle by phosphorylation of specific target proteins
Cdks driving the cell cycle are present through the cell cycle but are only active with their specific cyclin
Proteolytic destruction: Proteins being broken down. The mechanism for controlling cyclin concentration. Occurs to the S-cyclin in S-phase
Mediated by ubiquitinoylation and proteosomal degradations
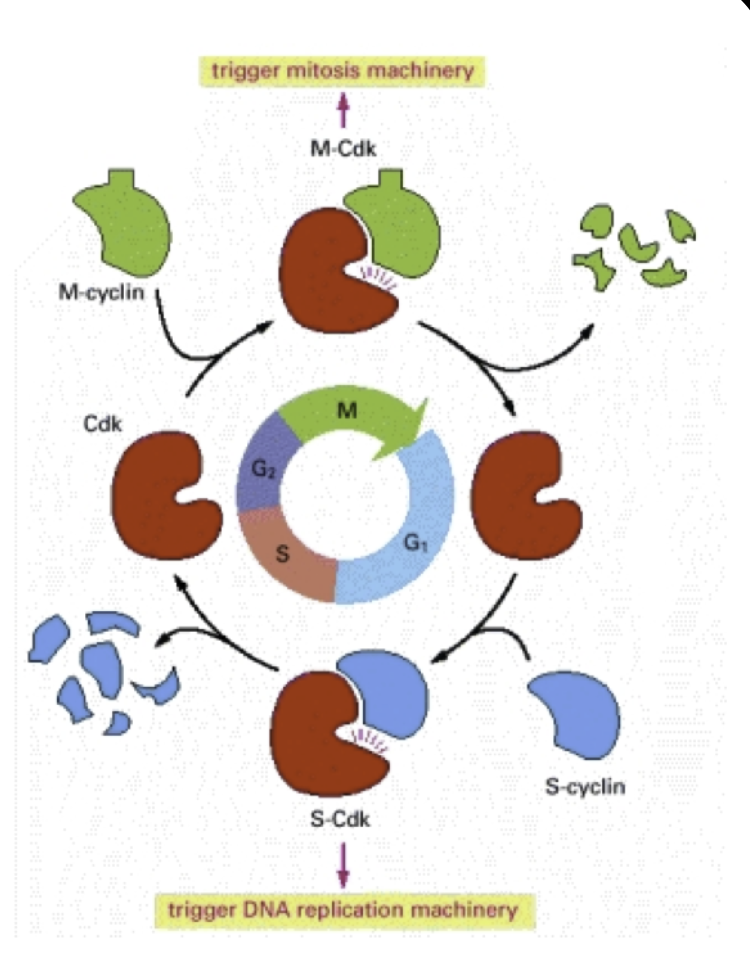
G2 checkpoint: M-cyclins establish M-Cdk complex (the maturation-promoting factor (MPF))
M-cyclins begin accumulating at the end of the S phase, continue through the G2 phase
M-Cdk will trigger M phase after it has reached a certain abundance
Triggered by a sharp increase in MPF activity
In M-phase, M-cyclin degrades and MPF activity stops. The daughter cell passes into G1 phase
Setting up for mitosis:
M-Cdk phosphorylates the nuclear lamina, breaking it down
Happens by breaking down the lamins (intermediate filaments)
M-Cdk activates condensin molecules to condense the chromatin into chromosomes
M-Cdk phosphorylates microtuble-associated proteins, directing spindle formation
Cancer
Mostly occurs in the G1 → S phase
Neoplastic transformation: A cell evolving into a cancer cell
Growth factor: Signals telling a cell to commit to divison
Works through tyrosine-kinase receptors (remember the dimerization thing)
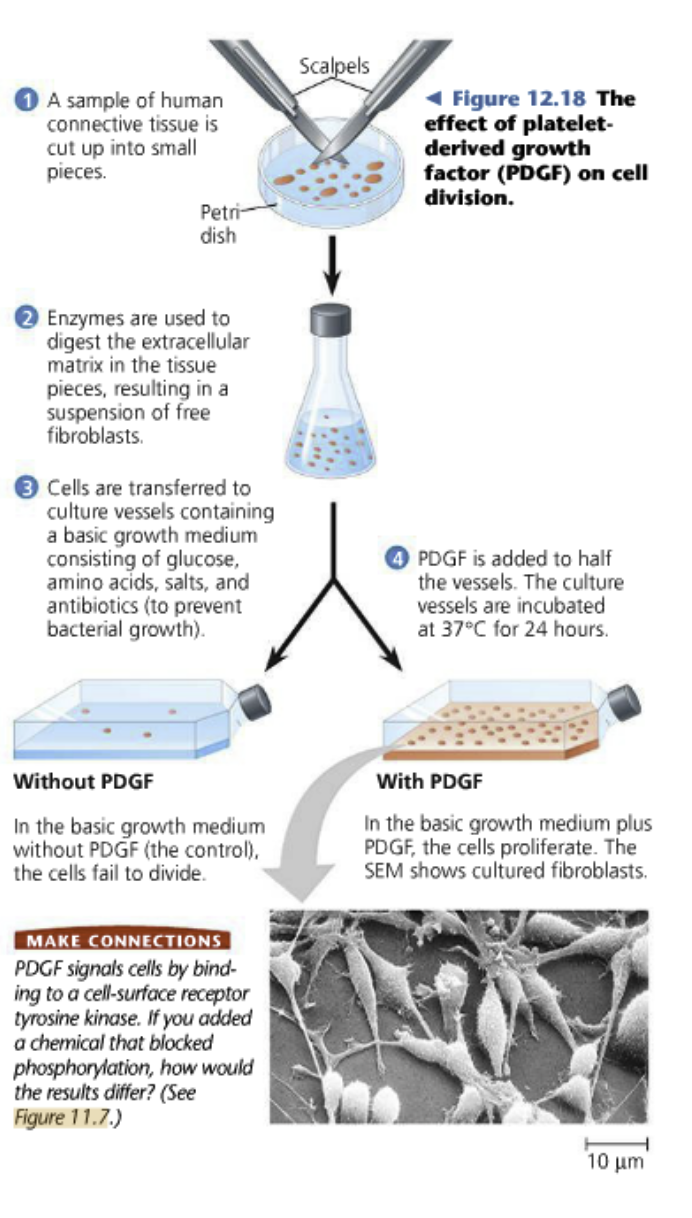
Ex.: Platelet-derived growth factor: Induces a signal transduction cascade in fibroblasts allowing it to pass the G1 checkpoint
Platelets in the blood stimulate PDGF when an injury occurs
Put the cells in an isotonic solution with glucose in the sample of human connective tissues cut up in small pieces
The cells in the experiment don’t grow on top of each other, they spread out in a single layer becaue it will make it harder for them to become cancerous?
Positive growth signal!
Anchorage dependence: Most mammalian cells must attach to a substrate to grow and divide
This keeps the cells from growing independently of others
The cell is told it’s okay to divide because it’s attached to something
Positive growth signal!
Density-dependent inhibition: When cells stop dividing because a cell culture (tissue) has reached a certain density
This occurs by external signals by contact with neighboring cells through cell adhesion
Cells feeling each other know they have reached the density limit, stops them from growing (cancerous cells don’t care)
Prevents passage past the G1 checkpoint
Negative growth signal
Cancer cells lose their anchorage-dependent and density-dependent inhibition
Therefore they can establish tumors and become very large
They will grow on top of each other (ignoring monolayer) or without anchorage
Oncogenes: A code for proteins that sends inappropriate positive growth signals
Ras: A G-protein sending growth factor signals from the membrane to the nucleus
If it becomes hyperactive, the growth factor signal becomes independent of the growth factor
Abnormal activation of Cdks are sent
Proto-oncogenes: Normal genes involved with positive growth signals
Can be converted to oncogenes through control by an inappropriate promoter, gene amplification, and point mutations leading to over-expression/hyperactivity
Tumor suppressor genes: Code for genes that send negative growth signals
p53 binds to damaged regions of DNA, inhibits progression through the cell cycle until the damage has been repaired
The cell is more likely to divide inappropriately when p53 becomes inactive
Tumor suppressor genes underlie density-dependent inhibition
It is unlikely a single mutation will transform a cell into a cancerous cell
Mutations add up: More likely for cancer when the cell expresses oncogenes and loses tumor suppressor gene expression
Dysregulation: Uncontrolled cell division
Angiogenesis: When tumors get large enough they need their own blood supply and take over the circulatory system
The middle is being starved of oxygen because the tumor cells grow on top of each other
You need oxygen for aerobic metabolism. They turn to anaerobic metabolism
The inside of tumors become glycolytic, run glycolysis at a very fast pace
Metastasis: When a tumor has access to the vasculature (blood vessels in an organ), it can escape its home tissue and colonize others
DNA Replication
When cells divide, they need to replicate their genome for the daughter cell
To replicate:
There needs to be a well-regulated system initiating the replication process
Full copies of the genome must be made
The new copy needs to be identical (or at least very close) (mutations can lead to tumors)
Semi-conservative replication
Once Watson and Crick figured out there was a double helix, they soon found out there was a copying system to DNA
Replication: Begins when the double helix is opened and the strands are separated
A complementary daughter strand is synthesized for each of the parental strands
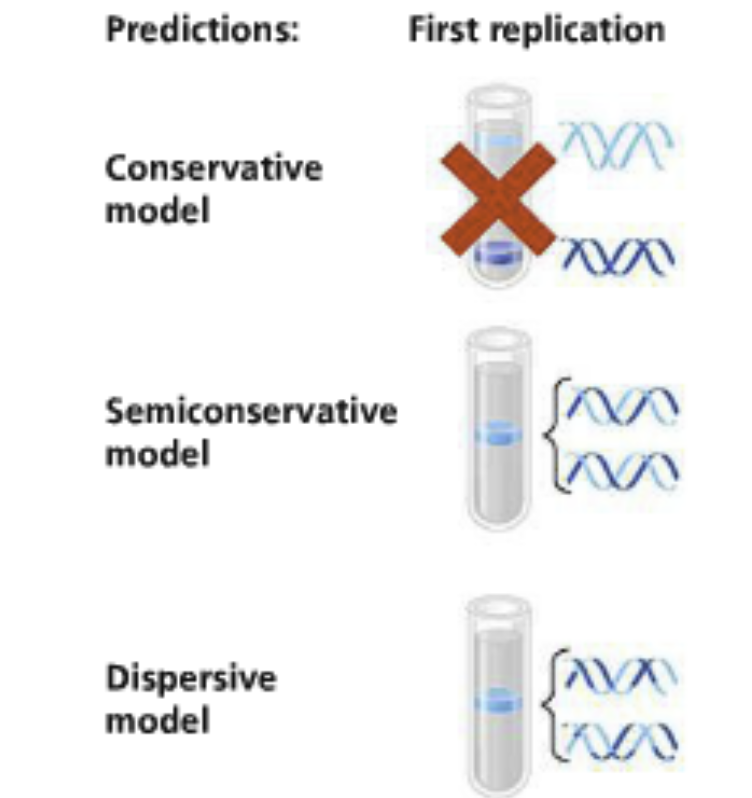
Three possible methods tested by Meselson and Stahl:
Conservative: The two parental strands direct the processing of the daughter strands but after reunite and the the two daughter strands reunite as double helixes
Semi-conservative: The parental strands separate and direct the process for daughter strands and stay related by “hybrid” double helices with their daughter strands
Dispersive: Each strand is a mixture of parental and daughter strands
Unlikely because it requires strand breaks
The parental strand will become more dispersed (diluted) as replication processes keep occuring
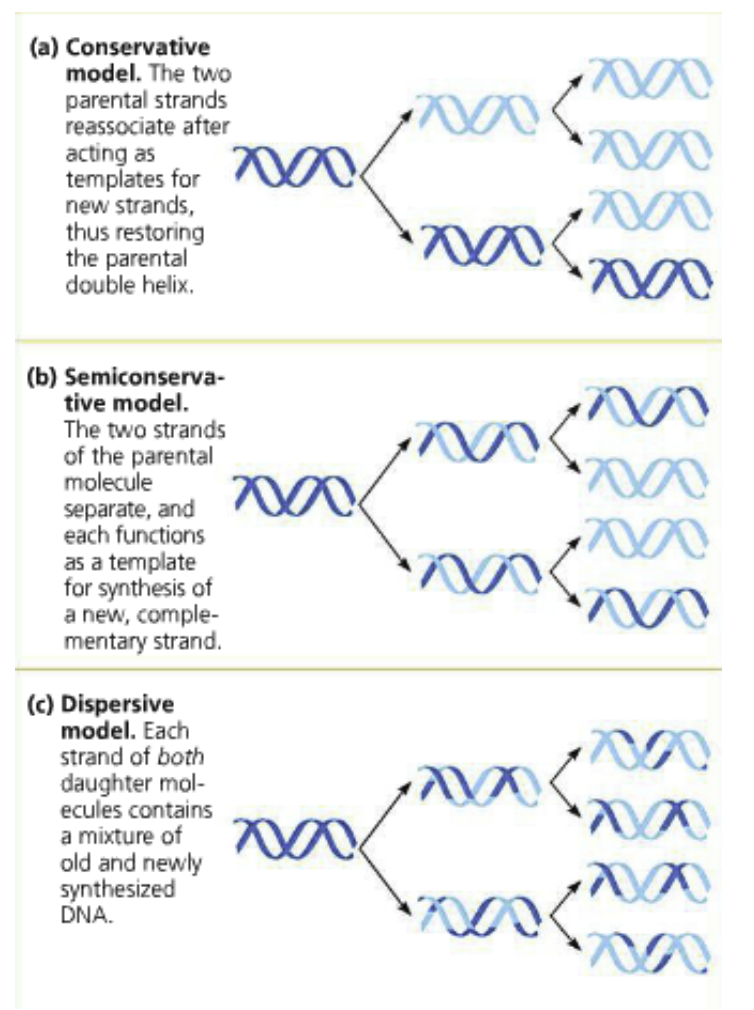
Meselson and Stahl: Performed an experiment to discriminate between the three models of DNA replication
The most elegant experiment in all of biology
The experiment: E. Coli was grown in media containing light (14N) or heavy (15N) nitrogen isotopes
Bacteria was culture in a medium with a heavy isotope then transferred to a medium with a lighter isotope
Results: The DNA sample was centrifuged after the first replication (saw a more dense band) then after the second replication they saw a less dense band too
These were radio isotopes
DNA from each sample was centrifuged. They could distinguish between heavy and light DNA
Bacteria from the 15N was transferred to 14N media and allowed to replicate DNA
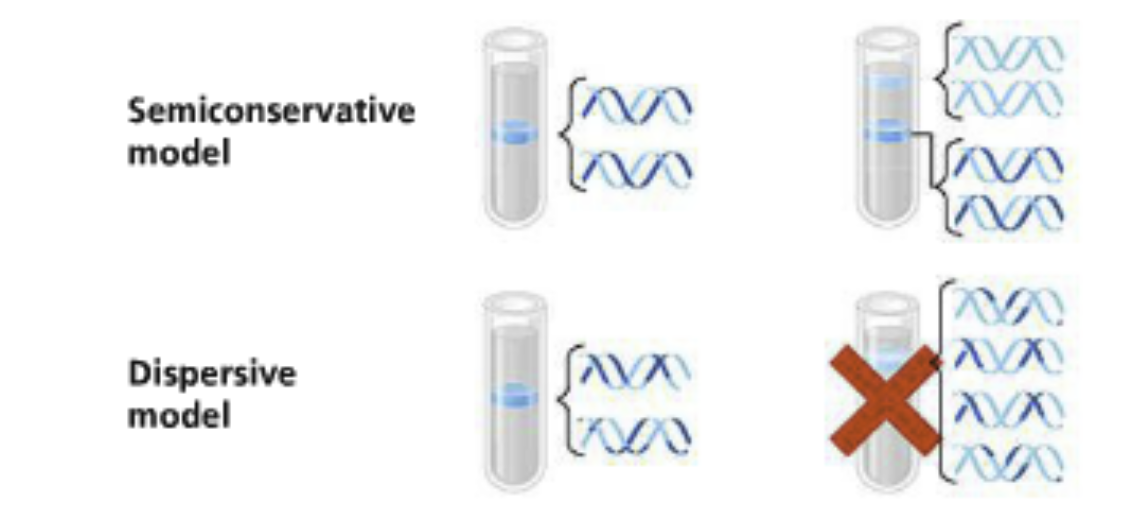
The first replication: A single band of intermediate weight (the heavy 15N) was observed
This excludes the conservative model because it predicted two separate bands
The second replication: Two bands were seen: One of intermediate density and one that was light
Excludes the dispersive model which predicted a single band would be lighter than the intermediate band (the original heavy DNA is distributed among new strands
Conclusion: The semi-conservative replication
DNA replication in prokaryotes
The cell cycle is different (less complex) than for eukaryotes
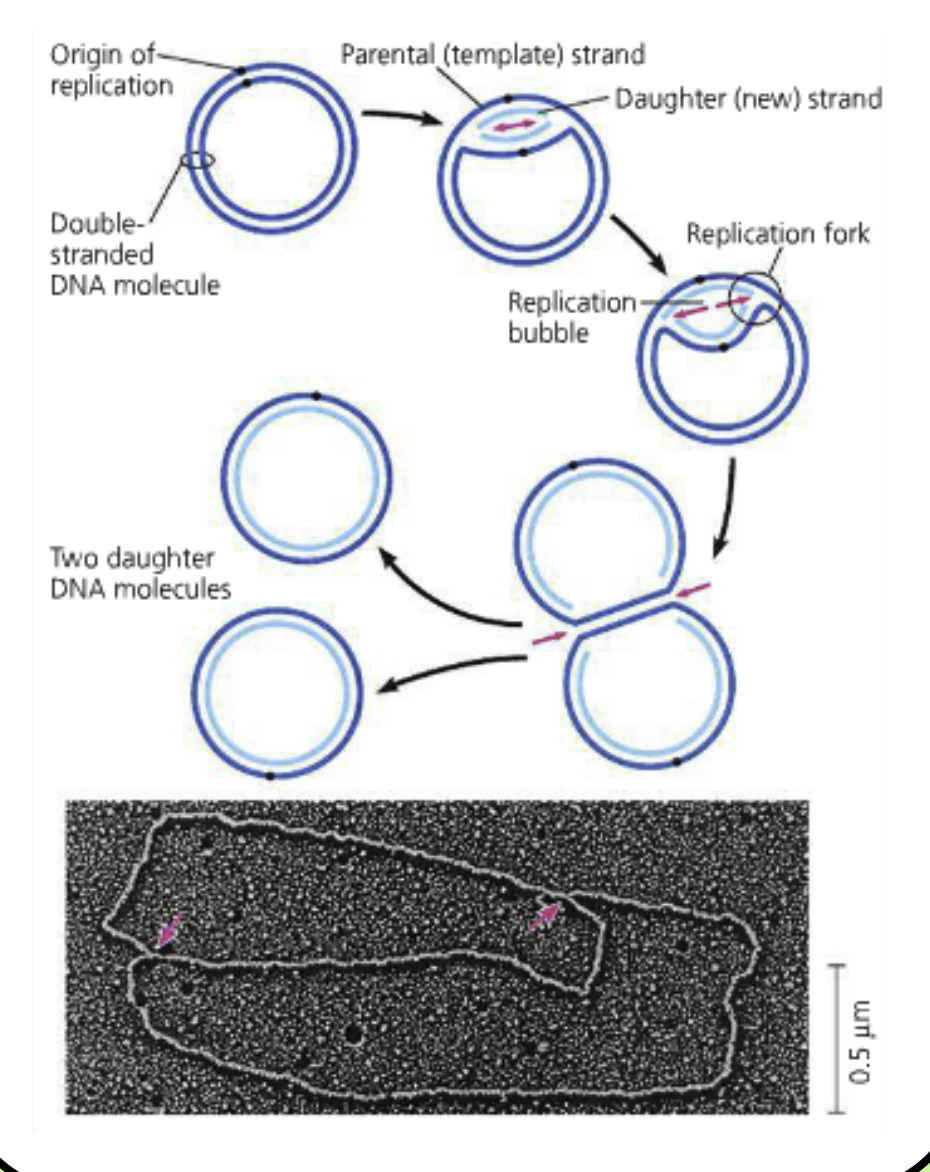
Replication begins at an origation
One replication bubble makes two replication forks
Replication fork: A region of DNA where the parental strands have been separated to replication machinery can access each strand
Replication moves in both directions until two circular chromosomes have been made
Theta replication: Baterial replication. Called this because it looks like the Greek letter theta
The daughter strands remain attached to their parental strands because this is semi-conservative
Supporting proteins: Prepare DNA for synthesis of the complementary strand
Helicase, topoisomerase, single-strand binding proteins
Helicase: Separate parental strands (Opens up the replication bubble, separates strands at replicationn fork- a molecular “bulldozer”)
Large, ring-like proteins encircling a single strand of DNA
It’s ripping open hydrogen bonds utilizing ATP
Double-strand to single-strand
Topoisomerase: When the parental DNA strands are separated, it causes over-winding in front of the replication fork
Is in front of the helicase (still in the double-strand region)
Over-winding is an issue for helicases trying to push the replication fork ahead
Topoisomerase relieves tension in DNA molecules by allowing them to freely rotate.
Topoisomerase makes a little cut (nick) in the backbone of the sugar-phosphate strand, creating a 3’ end that can freely rotate
One side is nicked, the other side can rotate
When the tension is released, topoisomerase unbinds and the backbone is ligated back together
Can use for genetic cloning
Single-stranded DNA binding proteins (SSBs) bind to the single strands made by the helicase
DNA and RNA don’t like to be single-stranded. Will fold up in itself
This binding has several functions (including straightening the DNA strand and preventing the formation of secondary structures that might impede the polymerase)
LOOK AT THE PICTURE!
Synthesizing proteins: Synthesize complementary DNA or RNA strands
DNA polymerase, DNA primase
DNA polymerase: The enzyme that reads the parental (template) strand and adds the complementary nucleotides
We will only talk about DNA polymerase III (there are 3)
DNA polymerases are shaped like hands (palm, finger, thumb)
Adding nucleotides: DNA polymerase cannot add nucleotides without a free 3’ end- DNA MUST grow in the 5’ to 3’ direction
Finds the 5’ triphosphate
Lose two phosphates because it’s an exergonic reaction
Coupled reaction- makes it difficult to reverse reaction. Taking one thing, making two things (entropy)
LOOK AT THE PICTURE!
The polymerase begins by checking on the deoxyribonucleoside triphosphate, making sure it can base pair with its partner
If the deoxyribonucleoside triphosphate can bind, it is hydrolyzed and produces a bound nucleotide residue and a molecule of pyrophosphate
Deoxyribonucleoside triphosphates are the substrate and energy source of the reaction to form the phosphodiester bond
Pyrophosphates: Hydrolyzed by pyrophosphatases: removes a product which prevents the reverse reaction and increases entropy
DNA primase: Makes the short (10 nucleotide) complementary RNA primer allowing DNA polymerase to load onto the strand and begin adding nucleotides
DNA polymerase needs this because it makes the 3’ end
RNA polymerase is De Nova (from nothing), DNA polymerase isn’t
Three steps of replication: When the replication fork is open, DNA primase binds and synthesizes the primer (DNA primase then unbinds)
DNA polymerase binds and adds complementary deoxyribonucleotides using the RNA primer as a starting point
The RNA primer is erased later on- replaced by DNA
DNA primase (basically and RNA polymerase) makes RNA de novo
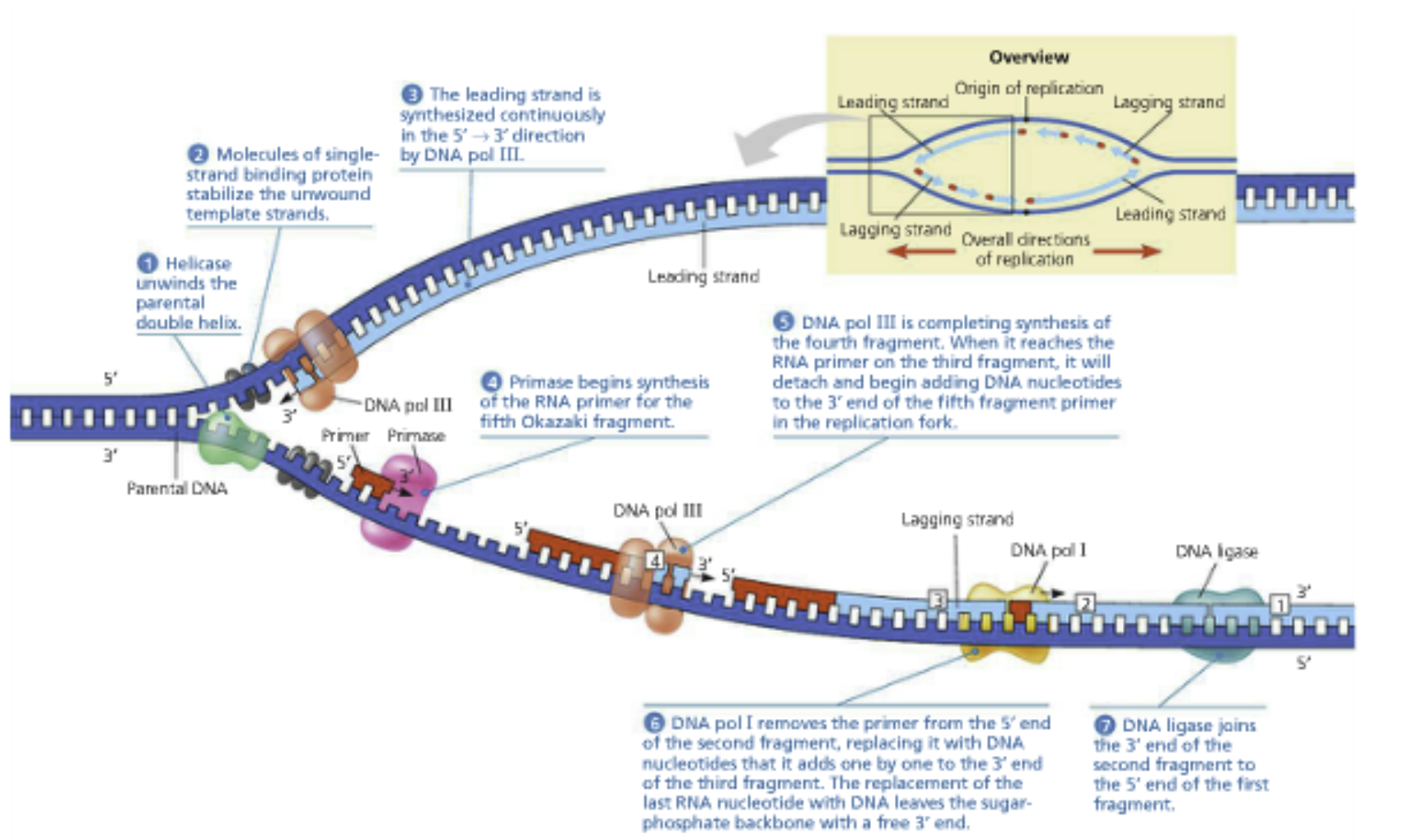
The replication fork: Helicase moves forward, unravels the double helix. Topoisomerase breaks apart supercoils. SSBs bind and prevent hairpins as the single DNA strands become available. DNA primase adds short complementary RNA primer for DNA polymerase to elongate
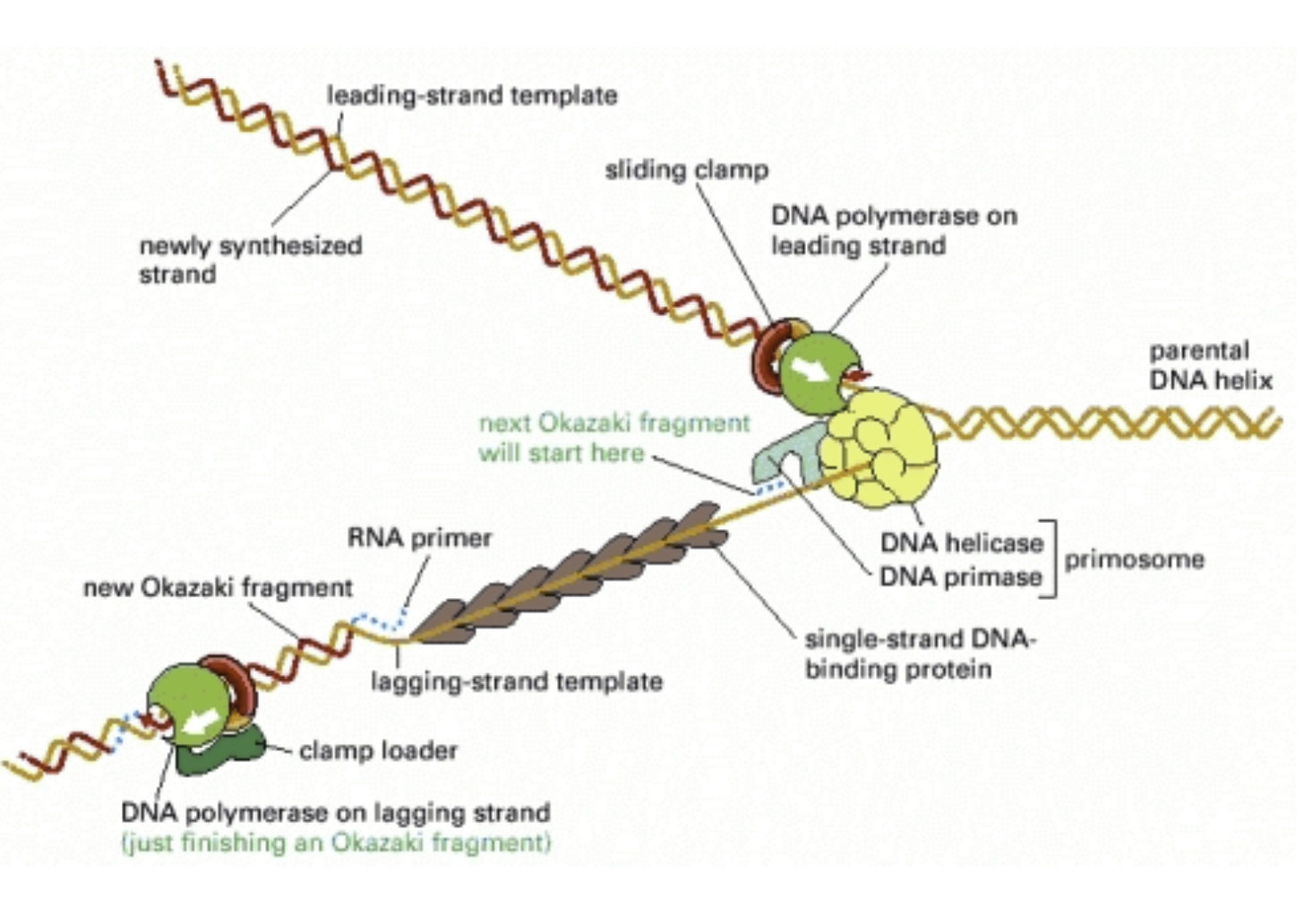
Leading strand: Primed only once and replicated continuously. Begins with a unidirectional polymerase
Primer lies down, creates free 3’ end
The parental strand
Polymerase extends the primer in the 5’ to 3’ direction, reads this parental strand in the 3’ to 5’ direction
In E. Coli, grows continuously at 500bp/sec (10x faster than RNA polymerase (remember 50bp/sec)
Polymerase moves in the same direction as the helicase
Lagging strand: Needs to be backstitched using multiple primers
Replicated in Okazaki fragments: 1-2 kbp segments
Therefore, RNA primers are 1000-2000 base pairs apart
Polymerase moves in the opposite direction as the helicase
A fragment of DNA between the primers
A second Okazaki fragment will be made upstream (towards the fork) and extended to the first primer
DNA Polymerase I will erase the RNA primer and fill it with the correct deoxyribonucleotides
Because we won’t have RNA primer in the finished product
DNA ligase (an enzyme) seals the breaks in the DNA backbone
There’s only one phosphate on the 5’ end so the polymerase gets confused, needs ligase
Ligation- to tie
Antiparallel to the leading strand
The polymerase moves away from the replication fork
An RNA primer is made near the replication fork and DNA polymerase elongates it in the 5’ to 3’ direction
The polymerase will bump into another primer and fall off the template strand
Proteins of the replication fork work efficiently as a replication machine
DNA is linked to helicase to form a primosome which lays down RNA primers as the helices unwind the double helix
The lagging strand is looped so the whole complex moves in the same direction (like a sewing machine)
Couple polymerases and something with helosomes?
DNA replication in eukaryotes
Challenges in replication:
Eukaryotic genes are spread across many linear chromosomes
DNA is packaged into nucleosomes
Histones
Each chromosome ends with telomeres
Eukaryotes have several origins of replication (multiple replication bubbles) on each chromosome
Several replication forks are active at once, helping replicate the chromosome quicker
Replication is under strict control in eukaryotes and only occurs during the S phase
Nucleosomes are an obstacle for the replication fork but it passes through parental nucleosomes without displacing them from DNA
A burst of histone translation accommodates the DNA that must be packaged
A bunch of histones are made in S phase
Behind the replication fork, old and new histones are incorporated into the helices
Telomeres: Short, repeating sequences at the ends of eukaryotic chromosomes that hold many specialize proteins
Play a role both in DNA replication and cell aging
When the RNA primer at the end of the chromosome in the leading strand is erased, it can’t be replaced and there’s no upstream 3’
In the lagging strand, the last bit is laid with a RNA primer
This is a problem because the leading strand has the RNA primer removed. As we do a second round of replication, we don’t have enough because the leading strand is shorter than the lagging strand
Senescence: Dying and aging
Therefore, the chromosome is shortened each time the chromosome is replicated
Eventually this will affect the genes
Therefore, cells have a finite number of replication cycles
Telomerase: An enzyme that extends the 3’ end with a repeating sequence so it can become long enough for another RNA primer
Otherwise, evolution could not occur because we would lose a bit of the chromosome each time. Allows you to get a little end to fit the last Okazaki sequence
This is in the germ line
Germ cells (which produce the gametes) utilize this
Cancer cells can turn on telomerase and divide as much as they want (making telomerase an oncogene)
Mitosis
The G2 checkpoint
G2 Checkpoint: Regulates the progression from G2 to M phase
When the cell commits to M phase, it will divide its genetic material between two nuclei and separate into two daughter cells
Entryway requires an M-cyclin which binds to its Cdk and establishes the M-Cdk Complex (MPF)
There will be a buildup at cyclin which will hit its critical point and begin getting ready for mitosis
M-Cdk: Sets the stage for mitosis
Breaks down the nuclear lamina by phosphorylating the lamin molecules, making them and the nuclear membrane disintegrate
Nuclear lamina: Made of an intermediate filament called lamin, a cytoskeletal structure, directly beneath the inner membrane
The nucleus has two membranes → Outer and inner
The golgi membranes and ER disperse
Activates condensin molecules that condense the chromatin into chromosomes
From the 300nm to 700nm (1400nm with the sister chromatids)
Chromatin condensation → distribution of genetic material between cells
M-Cdk activates condensin (a dimer holding coils of DNA) complexes
Sister chromatids are held by a protein called cohesin
V-shaped, looks a lot like condensin
Cohesin is cleaved by an enzyme during mitosis, leading to rapid dissociation of sister chromatids
Phosphorylates the microtubule-associated proteins, directing (inducing) the formation of the mitotic spindle
Microtubules: Hollow cytoskeletal filaments that are 25nm in diameter, composed of α and β-tubulin
Polarized molecules (+ and - end), organized within the centrosome
Grow by adding tubulin dimers to the + end
In an interphase cell, microtubules are shaped in a radial pattern
Centrosomes: Organize microtubules, contain two centrioles at right angles connected by a protein-rich matrix
Each centrosome forms a pole of the spindle apparatus
Centrosome cycle: At the end of G1, the two centrioles separate. At the beginning of S phase, a daughter centriole forms at the base of the mother centriole. At the beginning of M phase, each centrosome forms an aster (star-shaped structure formed around a centrosome) of microtubules befor disintegrating the nuclear membrane
Mitosis
M phase: Each phase within M phase is characterized by the activity of the chromosomes, spindle apparatus, and nucleus
6 phases of M phase: 5 in mitosis, 1 is cytokinesis
Mitosis: Making a new nucleus
Ccomplete when a new nucleus has formed at the end of telophase
Cytokinesis is required after mitosis to separate the two daughter cells
Interphase: The cell prepares for mitosis by replicating its DNA and centrosomes
The nucleus (and nuclear lamina) is intact and the chromatin inside has not organized into mitotic chromosomes yet
They’re still S chromatin
Connected through cohesin (sister chromatins)
M-Cdk activity hasn’t reached critical mass yet
Prophase: Condensation of the interphase chromatin into mitotic chromosomes
A more subtle change is that the centrosomes begin migrating to either side of the nucleus in anticipation of forming the mitotic spindle
The nuclear membrane (and lamina) remains intact
Centrosomes are beginning to move apart from each other
Prometaphase: The nuclear membrane disintegrates after chromosome condensation and the migration of the centrosomes
Still under direction of M-Cdk
Critical because the microtubules coming from the centrosomes can now attach to the chromsosomes
Other organelles of the endomembrane system such as the ER and Golgi apparatus become fragmented
Centrosomes are on opposite sides of the nucleus
They are now the poles of the spindle body
The nuclear membrane needs to dissolve because sister chromatins of chromosomes need to dissolve
Need to gain access to them- need to physically reach out and touch them
The poles will physically reach in and grab the chromosomes- couldn’t happen if the nuclear membrane didn’t dissolve
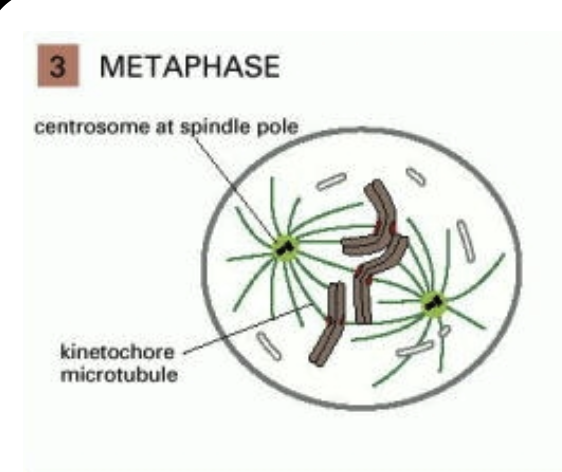
Kinetochore: A protein plaque that forms on the centromere of each chromosome
The plus ends of microtubules emanating from the spindle apparatus bind very stably to the kinetochore
These microtubules are called kinetochore microtubules
The chromosomes are being pulled in two directions at once
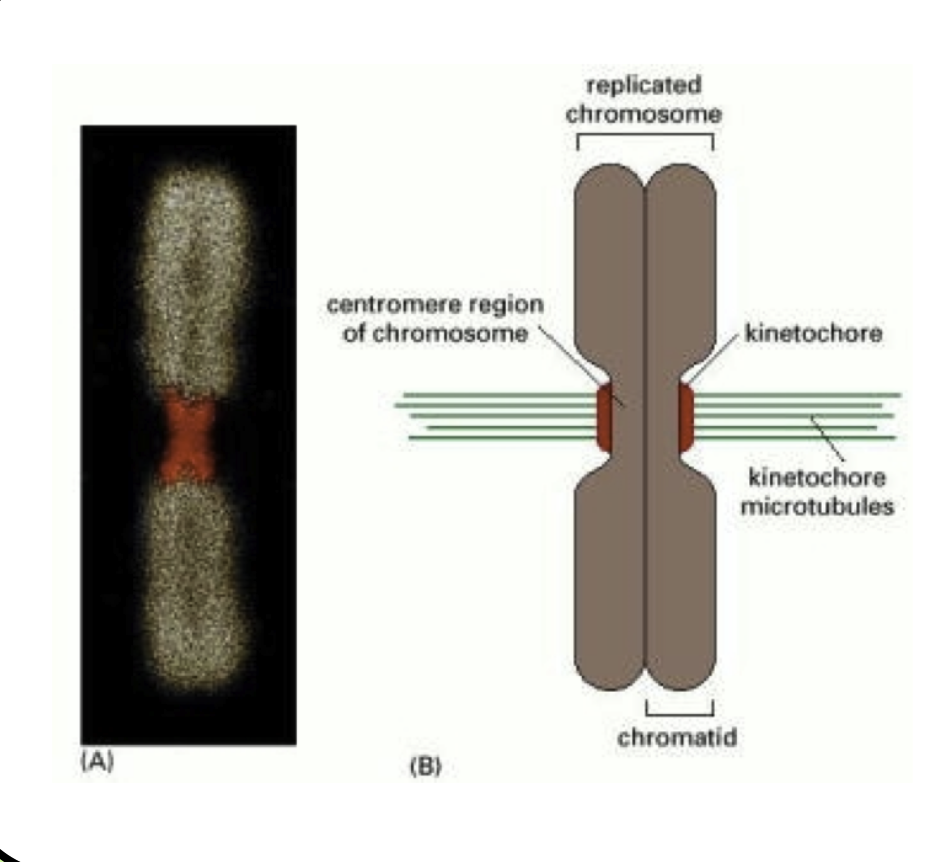
Metaphase: Once the kinetochore microtubules have attached, they push and pull each chromosome until all chromosomes are lined up in a row
This is a tug-of-war of the chromosomes
This is called the metaphase plate: it lies halfway between the poles of the spindle apparatus
Orientation of the plate determines the later plane of division
Three microtubules present:
Kinetochore microtubules physically bind to the chromsome through the kinetochore
Overlap microtubules from the other pole and are connected to each other by motor proteins (kinesins)
Astral microtubules radiate out from the poles of the spindle apparatus in an aster (star) pattern
Spindle attachment checkpoint: (Within M phase) Unattached kinetochores delay the transition from metaphase to anaphase
If the cell progressed into anaphase before all the chromosomes were attached to the spindle, genetic chaos would follow
What would happen if anaphase proceeded before all the kinetochores (holding chromosomes) were attached?
If only one side was attached, one daughter nucleus would get two copies of a chromosome. If neither were attached, gene isn’t given
Aneuploidy: Major displacement of a chromosome if one or both sides aren’t attached
Kinetochores will signal that they’re not attached
Metaphase to anaphase transition: Anaphase promoting complex (APC) is a proteolytic complex driving progression through the spindle attachment checkpoint
APC cleves an inhibitory protein called securin, which allows separase (a protease) to cleave the cohesion complex holding sister chromatids together
Securin was keeping separase from being activated
Separase allows the chromatids to separate by destroying the cohesin keeping the chromatids together
APC degrades M-cyclin, eliminating MPF (M-Cdk) activity
Anaphase: The cell partitions its genetic material
The sister chromatids begin their migration toward the spindle poles now that they are separated from each other
This requires coordination between microtubules and motor proteins
Anaphase A: The retraction of kinetochore microtubules which pull the chromosomes toward each pole (think of it like reeling in a fish
Anaphase B: The poles separate (occurs by two mechanisms)
Motor proteins push the overlap microtubules apart, helps to separate the poles
Motor proteins on the astral microtubules connect with the actin cytoskeleton, helping pull the poles apart
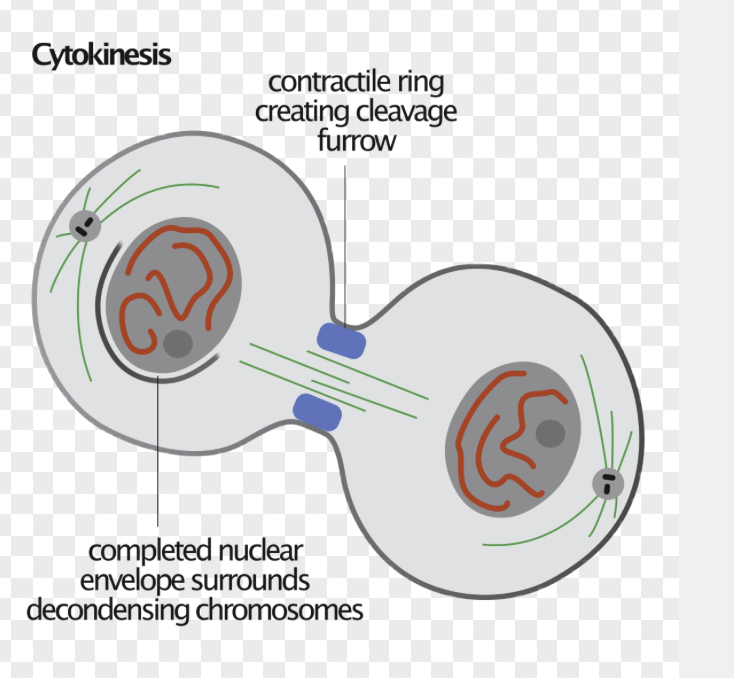
Telophase: Begins when daughter chromosomes complete migration and reach the poles. Marks the end of mitosis (not M phase- cytokinesis!)
Then, a nuclear envelope forms around each pole (M-cyclin has been degraded by this point)
A contractile ring begins to form on the inner surface of the plasma membrane that will partition the cell
See the effects of m-CDK start to be reversed- start decondensing
Cytokinesis
Breaking the cell in two
Form a contractile ring that condenses and gets smaller and smaller as the two new cells are pulled apart
Myosin and actin form the contractile ring
Remnants of the overlap microtubles that separated the poles in Anaphase B remain between the nuclei but they aren’t connected to the poles
Midbody: A tiny bridge separating the two daughter cells at the end of cytokinesis
A mother centriole from a daughter cell separates from the daughter centriole, migrates into the midbody, and stays there for a while before returning to the daughter cell
No one knows why this occurs
When division is complete, the cells separate
The midbody structure leaves a mark on the inside of the plasma membrane
When the cells have finished dividing, they reenter the cell cycle in G1
The activity of M-Cdk is lessened by M-cyclin degradation by the APC complex. It remains low by continued degradation of M-cyclin during G1
Syncytium: A multi-nucleated cell because cytokinesis did not occur after mitosis
In drosophila (fruit flies), 13 nuclear divisions occur without cytokinesis generating 6000 nuclei in one cell
The nuclei migrate to the periphery of the cell and a massive round of cytokinesis occurs through cellularization
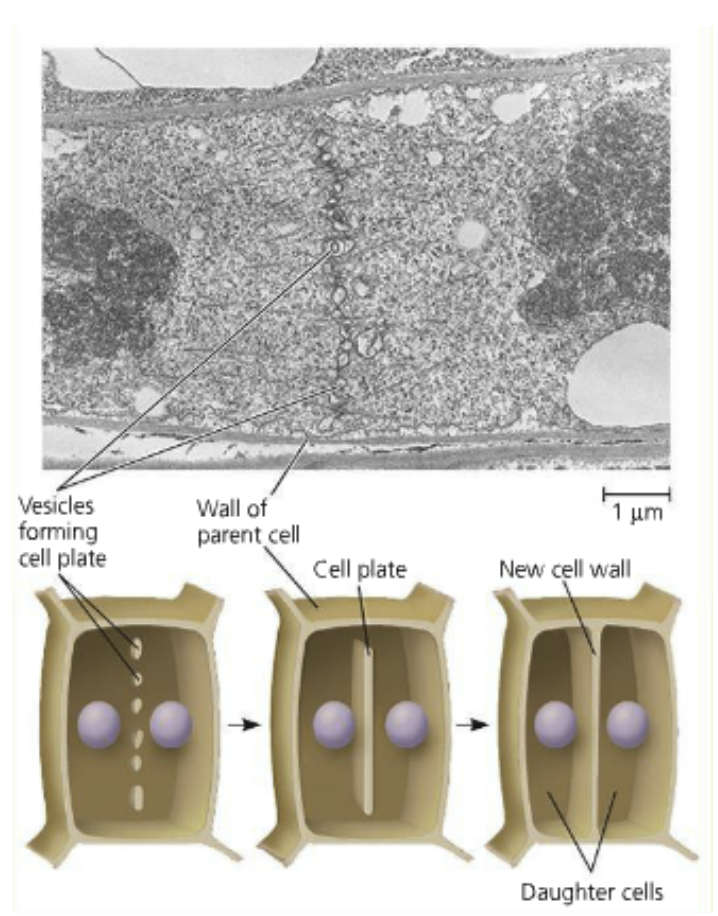
The cell wall of plants is a challenge for cytokinesis
Cell plate: A structure the two daughter cells create which walls themselves off
The cell plate is enveloped by a plasma membrane and grows until it separates the two daughter cells
Cellulose microfibrils are added to the matrix of the cell plate
Mutation
Natural selection: Individuals with inherited traits become evolutionarily more fit because of those traits
Only works if there’s a genetically diverse population to select from
Mutation: Changes to the genetic code- comes from the Latin word for “change”
Can be beneficial, detrimetal, inconsequential, lethal
Any phenotypic outcome of the mutation is inherited by the mutated cell’s lineage
Can create new alleles which will create a new genotype which will create a new phenotype
Replication mistakes, DNA damage, repair
Mutations occur at fixed rates in most organisms
The mutation rate in E. Coli is 1 in 109 and its genome is 4.6 million base pairs so it is likely to replicate its entire genome without mistakes
This states you’d have to go through 217 processes of replication before a mutation (not accurate)
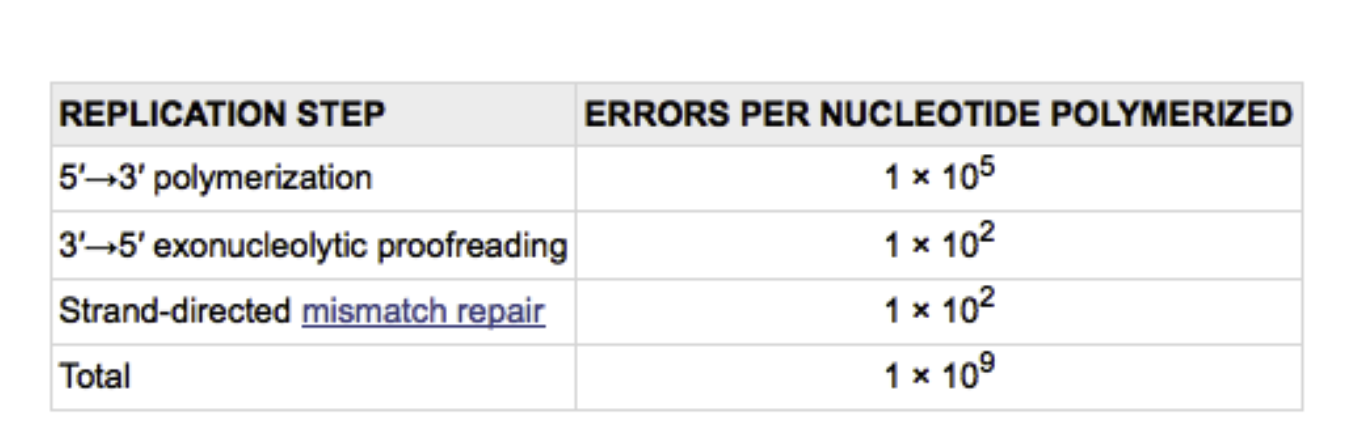
DNA polymerase III makes 1 error in 100,000 (105 base pairs or 46 mutations per E. Coli genome replication
Every once in a blue moon, an incorrect base looks and feels like the correct base to the polymerase
Tautomers: The equilibrium pair of ketones and enols (the evil twin of ketones- can base pair differently than keto form. Has a hydrogen bonded to the ketone’s oxygen (makes an alcohol))
Tautomeric shifts occur very rarely in the ketone groups of nitrogenous bases
About 1 in 100,000 base pairs the DNA polymerase grabs will be in enol form
How incorrect nucleotides end up in DNA
DNA polymerase III can proofread and edit its own work using an editing site within a subunit of the enzyme
If the enol form of a tautomer is added, it shifts it back to keto form
DNA polymerase III can’t add another nucleotide to an unpaired 3’ end so it gets rid of the base and tries again
Will send the enol to the “palm region”, send it out
3’ to 5’ exonucleolytic activity: Taking a step back and removing a mispaired base
E.g.: Adding a cytosine when a thymine was needed because of a tautomeric shift (then there is an unpaired 3’ end so DNA polymerase can’t elongate)
After absolving the mispaired base, it will add the correct one and continue the strand
The keto/enol shift is unstable and flickers back and forth
DNA polymerase III proofreading brings error down to 1 in 107
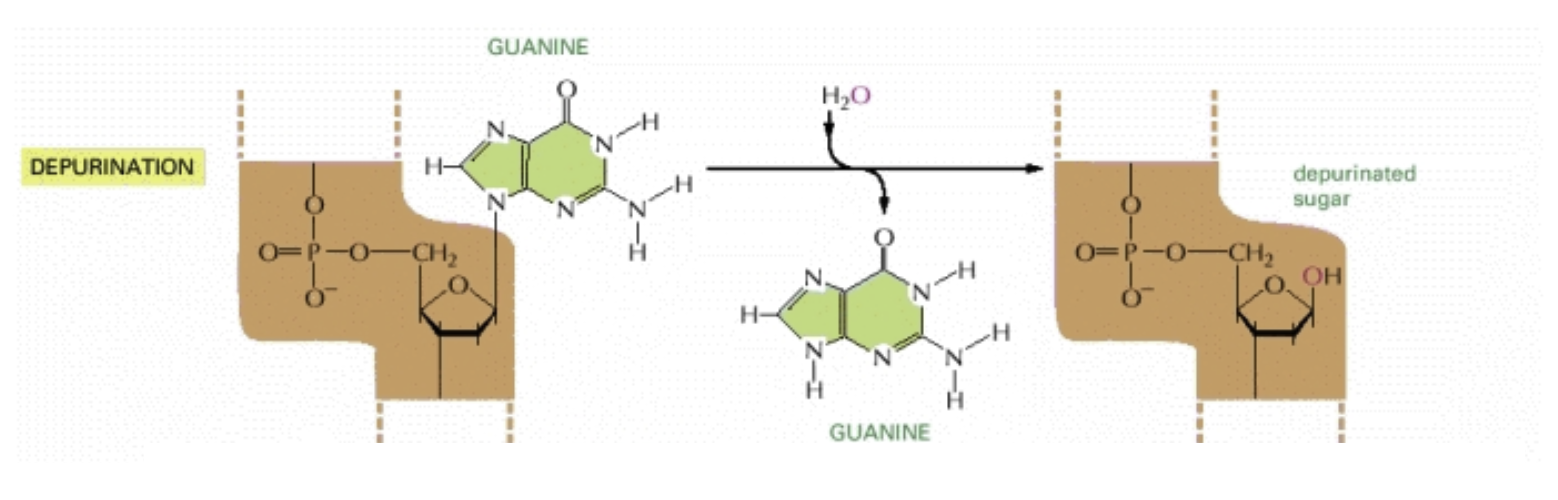
Depurination: The most common form of spontaneous damage
The glycosidic bond between a purine (adenine and guanine) to the nucleotide breaks and the nucleotide is left without an “identity”
A human cell loses 5000 purines per day to this
When left uncorrected, a base pair is deleted in one of the daugher strands
This is called a frameshift mutation
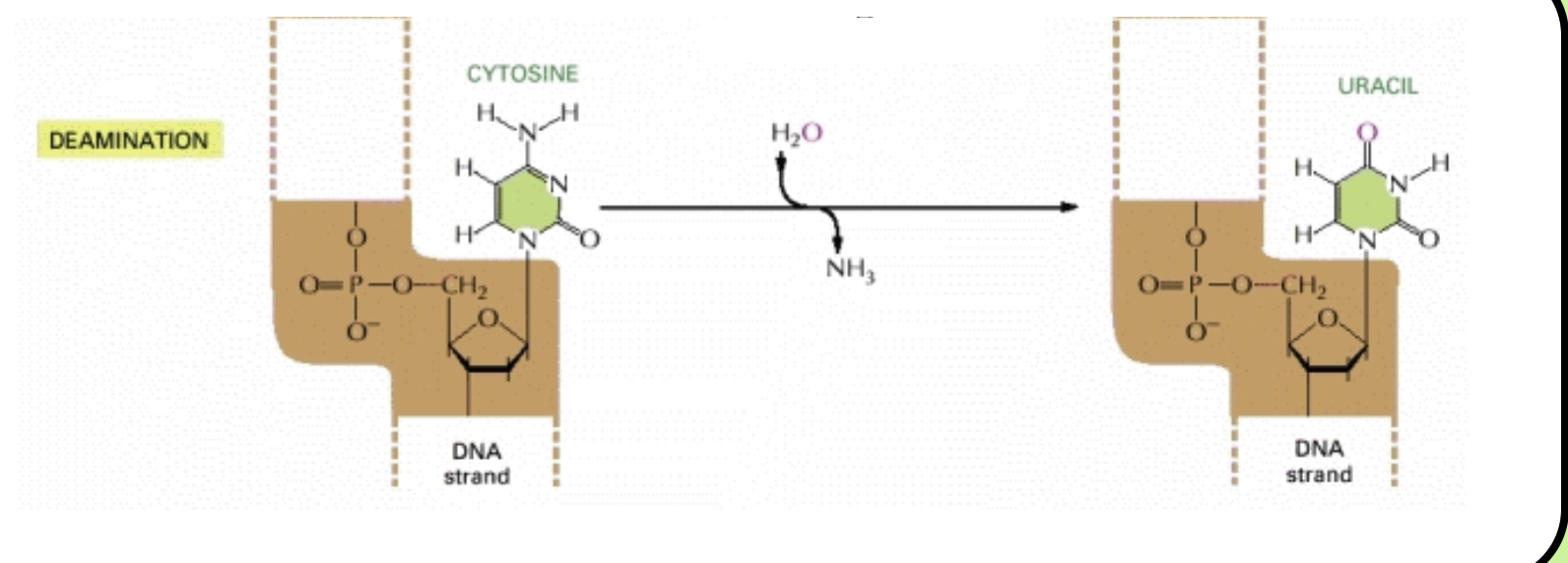
Deamination: Also spontaneous damage
Cytosine loses its amino group and becomes uracil
This occurs in the genome of a human cell 100 times a day
When left uncorrected, deamination causes a guanine residue to be replaced with an adenine residue in one of the daugher DNA strands
This mutation is called a transition
Double-stranded DNA has a built-in “back-up” copy of the genetic code (the complementary strand)
Strand-directed repair mechanisms: Use the complementary strand to repair an inappropriate or damaged base pair
Reduces the mutation rate 100-fold and brings the final mutation rate to 1 in 109 nucleotides
Base excision repair: The repair mechanism for single-nucleotide damage to the genome
The damaged base is removed, the sugar-phosphate bone is broken, the site is filled with the correct base
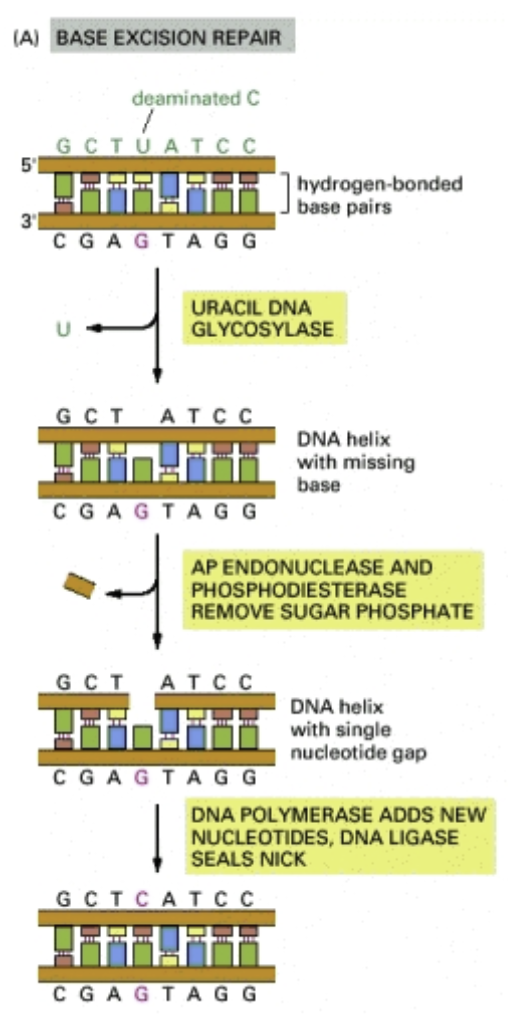
Mutations
Transition: Purine-for-purine or pyrimidine-for-pyrmidine mutation
Less disruptive and more frequent in DNA than transversions (more benign)
Transversion: Purine-for-pyrymidine or pyrimidine-for-purine
Tends to have more consequences
Substitutions: Silent mutations: Due to redundance of the genetic code, many nucleic acid substitutions don’t change the protein’s primary structure
Ex: GGC and GGU (transition of G to A in template causes this but both GGC and GGU code for glycine)
Substitution: Missense mutation: When a nucleic acid substitution changes the protein’s primary structure
Effects can vary from benign to disastrous depending on the amino acid’s chemical nature and location in the protein
Substitution: Nonsense mutation: A mutation changing the codon into a stop codon- bad!
Lead to truncated (shortened) proteins: Cannot perform their normal function
Frameshift-to-nonsense mutation: Insertions and deletions changing the reading frame causing a stop codon immediately after the start codon
Frameshift mutations can produce long string of gibberish, truncations, or lead to protein aggregation (harming the cell)
The most likely way to get a random stop codon (will lead to truncation)
When a full codon is inserted or deleted, this is rare but benign
It doesn’t change the reading frame
Mutations at the phenotype level:
Not always exclusive! Can have more than one
Lethal mutation: Interferes with a critical gene, causes death
Conditional mutation: The phenotype associated with mutation can only be seen in certain conditions
Loss-of-function regulation: (Typically recessive) Results in a decrease in the gene’s ability to function normally
Going back to the pea plants (white flowers, purple flowers, loss of pigment would cause albinism)
Typically recessive because the dominant gene would make up for the loss
Null mutation: Results in the complete loss of gene functioning
Gain-of-function mutation: (Typically dominant) Results in a gene product with new properties
Mutagens
Mutagens are agents (substance) that increase the mutation rate above natural levels
Three forms: Physical, chemical, viral
Physical mutagens
Chemical mutagens
Typically involve radiation which breaks DNA strands or photochemically modifies bases
Chemical agents that induce mutations by interacting with DNA
Physical mutagens: Ultraviolet light induces a photochemical reaction between neighboring pyrimidines forming pyrimidine dimers (often thymine dimers)
DNA polymerases tend to get stuck on pyrimidine dimers and replication stops (they’ll jump over the dimer)
The polymerase will often insert an incorrect nucleotide (mutation)
Pyrimidine dimers are the most common cause of melanoma
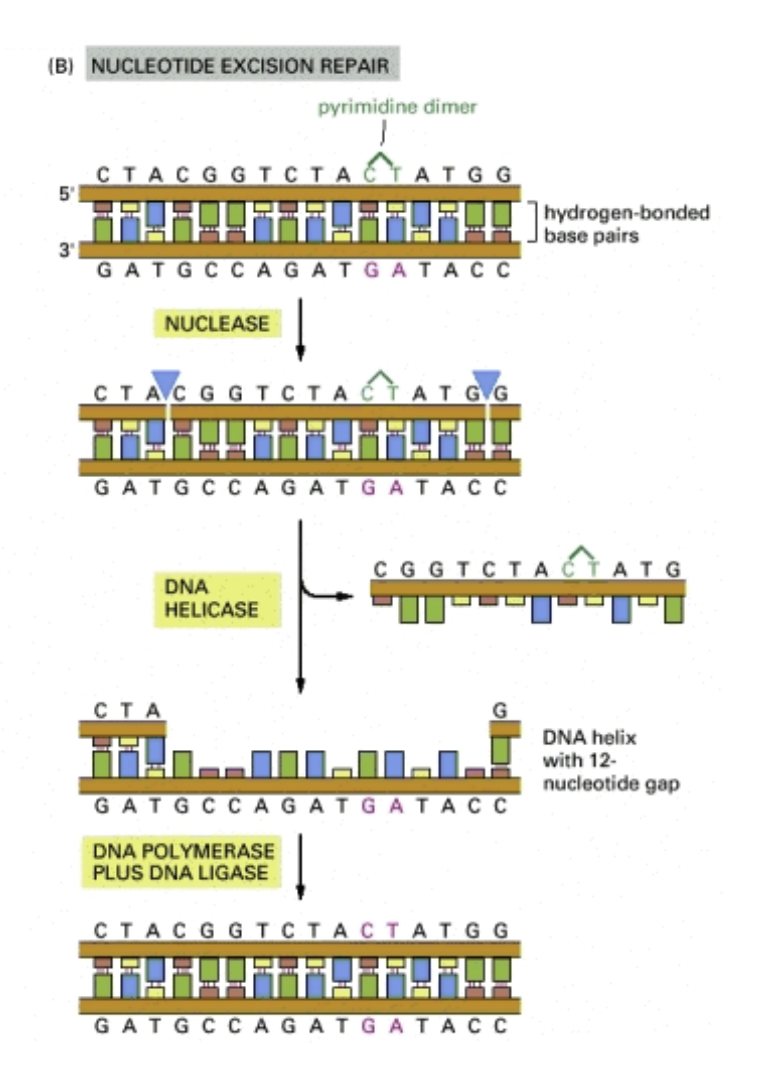
Nucleotide excision repair: Pyrimidine dimers are bulky lesions. They are corrected by nucleotide excision repair
1. Enzymes scan the DNA looking for distortions of the double helix
2. The sugar-phosphate backbon is broken on each side of the lesion and a patch of surrounding nucleotides is removed by helicase
3. The gap is filled in by DNA polymerase with complementary nucleotides
Xeroderma pigmentosum (XP): A condition which comes from defects in the nucleotide excision repair mechanism
Pyrimidine dimers from sunlight UV cause cancer and disfiguring
Chemical mutagens: Some bind directly to nucleotide bases (ex: aflatoxin)
Aflatoxin B1 is produce by a fungus that grows on grains and peanuts in tropical environments
It’s not a problem by itself bus is metabolized in the liver into a very mutagenic epoxide with the bases of DNA
Intercalating agents: Rigid planar aromatic molecules that slip between adjacent base pairs
Cause slight bulges in the double helix leading to frameshift mutations
Will cause spacing between nucleotides to change
The DNA polymerase will get confused
What causes the frameshift mutation
Ex: Ethidium bromide: used to visualize DNA in agarose gels
The Ames test: Quantifying mutagenicity- A potential mutagen is mixed with a homogenized liver extract and a culture of histidine-dependent bacteria
The gene responsible for making histidine is nonfunctioinal (the bacteria requires supplemental histidine)
A substance is mutagenic if it converts the nonfunctional histidine gene into a functional gene
Gametogenesis and fertilization
Mitosis conserves the genetic code
Mutations are unlikely (but are the only ways we’ve learned since now to make new alleles)
Asexual reproduction: An organism reproduces by making identical copies of itself
An organism that reproduces asexually can only generate new alleles through mutation
Limits the population’s genetic diversity
Sex pilus: Two asexually reproducing bacteria can exchange DNA and diversify the complement of genes they posses
Gametes: Containing a single copy of the genome
In sexual reproductions, two gametes come together
This is a zygote (diploid!)
Gametes are haploid, zygotes are diploid
Ploidy: The number of copies of the genome found in a cell
Haploid: Single copy of the genome (Reduction in ploidy is brought by mitosis)
Diploid: Two copies of the genome (Increase in ploidy is brought by fertilization)
Bananas are 3n, wheat is 6n, strawberries are 8n,
Meiosis
The human genome has two sets of 23 chromosomes (46 in total)
Somatic cells: Diploid (2n),
ONLY DIVIDES THROUGH MITOSIS
Germ cells: Involved in forming gametes
The germ line has a complex lineage of germ cells leading to the formation of gametes
THE ONLY CELLS THAT UNDERGO MEIOSIS
Animals are diploid organisms that produce haploid gametes
Other forms of life have different processes for sexual reproduction
Meisosis and fertilization are common to lifecycles for each organism
Homologous chromosomes: The collective maternal and paternal set of chromosomes
Each diploid cell has both a maternal and paternal set
Homologous chromosomes are similar but not identical
You have chromosome I from Mom and chromosome I from Dad. Inherit one copy of each of these chromosomes from each of these parents
The pair from Mom and Dad are homologous. Similar, but not identical
Similar: Same chromosome (and genes in the same order (remember gene is general)
Different: Different alleles
Ploidy reduction: (Diploid-to-haploid) is done by meiosis
Meiosis: (a cell divides twice to produce four haploid genes
Meiosis has two divisions: Reductive and equatorial
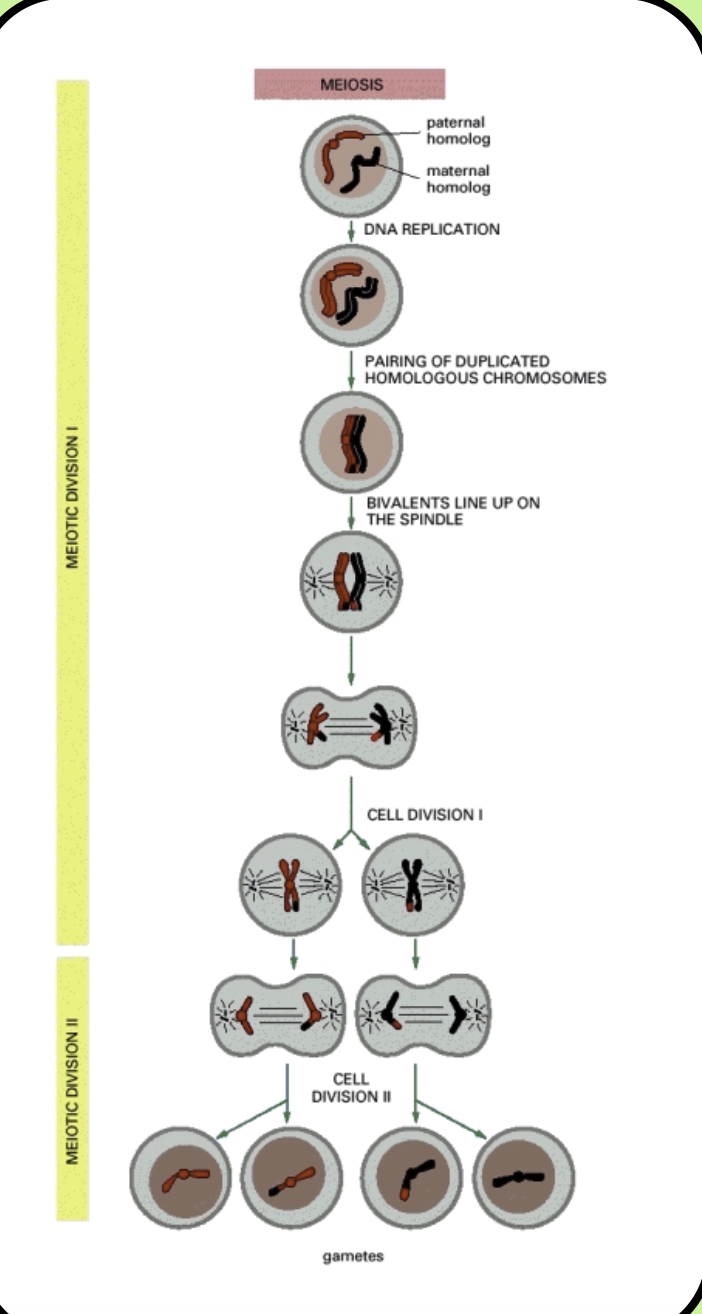
Reductive division: (Meiosis I). Homologous chromosomes find and bind to each other, then are separated and the diploid-to-haploid transition takes place
This occurs in prophase. The pair of duplicated homologous chromosomes pair to form a structure called a bivalent
The chromosomes within the bivalent exchange genetic material- synapsis
Then the bivalent line up at the metaphase plate and are separated by the spindle apparatus
Even though we have two sister chromatids, they are identical copies so there is ONLY one set of alleles. There is ONLY one copy of the genome
Equatorial division: (Meiosis II): The sister chromatids are distributed among four haploid daughter cells (gametes)
The same as mitosis but the sister chromatids of each chromosome are separated
We have formed the spindle
Remember Mendel! The law of segregation states we only inherit Mom’s alleles or Dad’s alleles for a gene
Random segregation occurs during Meiosis I
The law of independent assortment states genes are inherited independently of each other
This is because genes are located on different chromosomes
Mitosis
Meiosis
One division
Two divisions
No synapsis
Synapsis
Produces two diploid cells
Produces four haploid cells
Produces identical cells for growth and tissue prepare
Produces gametes
Gametogenesis
Gametogenesis: The production of gametes in males and females
Sperm: A motile cell that carries paternal genes
The head has an acrosomal vesicle and haploid nucleus
The acrosomal vesicle has enzymes to bury down when it finds the egg
The midpiece is loaded with mitochondria to power the flagella
The flagella allows for motility
9 + 2 (axoneme)
Testes: Sperm production occurs within the seminerferous tubules in an inward-outward production
Moving past the core, you see more mature cells
Males will continue to produce sperm throughout their life (unlike females)
Leydig cells produce testosterone
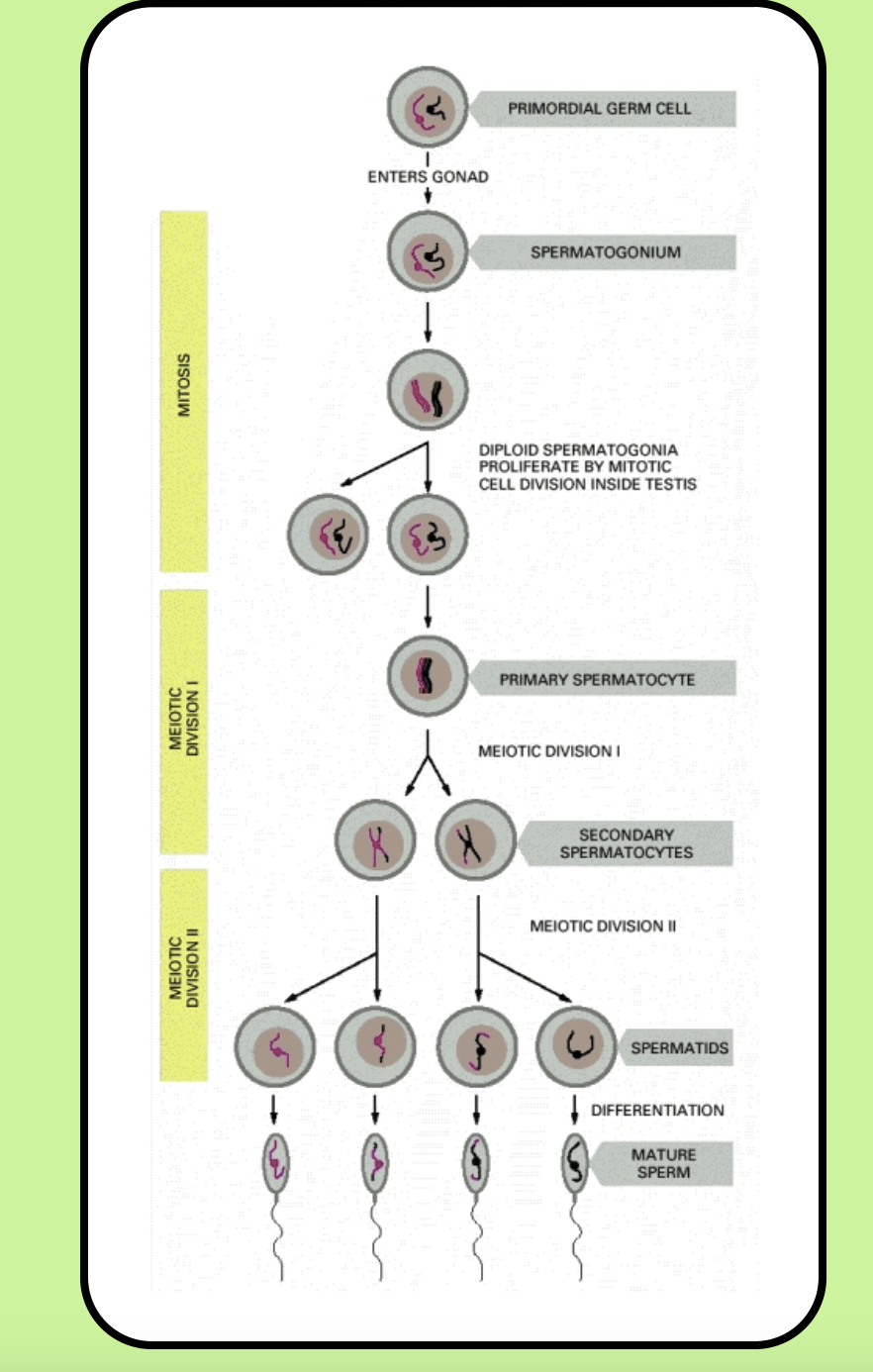
Spermatogenesis: Sperm continually divide by mitosis
Start with a stem cell, primordial germ cell
Some of the daughter cells become primary spermatocytes
Primary spermatocytes pass through meiosis I, resulting in two secondary haploid sperm
Secondary sperm pass through meiosis II- four mature sperm
The mature sperm pass into the seminerferous tubules
Most of a sperm’s maturation occurs after it has become haploid
Cytoplasmic bridges: Cytokinesis after meiosis I and II is incomplete so cytoplasmic bridges between the two spermatids remains
Necessary because survival requires an X chromosome and only half of the sperm have them (the other half have a Y)
The cytoplasmic bridge allows both sperm to be attached to the X for as long as possible
Eggs: (Ova). Mammalian babies have access to their mother’s nutrients as they develop
The egg has a haploid pronucleus and organelles in the cytoplasm
The female reproductive system has ovaries, Fallopian tubes, uterus, corvix, and vagina
Mature oocytes (eggs) are produced in the ovaries one at a time then released into the Fallopian tubes as they wait for fertilization
A fertilized egg will implant into the wall of the uterus and develop into a fetus
Oogenesis: Before puberty. Begins with the mitotic division of the oogonia inside the ovary to make primary oocatytes
Primary oocatytes: Begin in meiosis I, are arrested in this stage and wait for sexual maturity
This happens before birth and all the eggs you’ll ever have are there before you’re born
After puberty, primary oocytes are individually selected for further maturation
They complete meisosis I and divide asymmetrically to produce a polar body (later degenerates) and secondary oocytes
Assymetric for greater chance of survival in one egg, it requires so many nutrients
The secondary oocyte is arrested during metaphase of meiosis II and is released upon ovulation, waiting to be fertilized
Meiosis II is completed after fertilizaiton, producing a second polar body
Fertilization
Fertilization: The formation of a diploid nucleus from the pronuclei from each gamete
Zona pellucida: A glycoprotein coat that covers mammalian eggs
When the sperm binds to its proteins, the hydrolytic enzymes in its acrosomal vesicle digest the zona pellucida
By penetrating the egg’s coat, the sperm can reach the plasma membrane
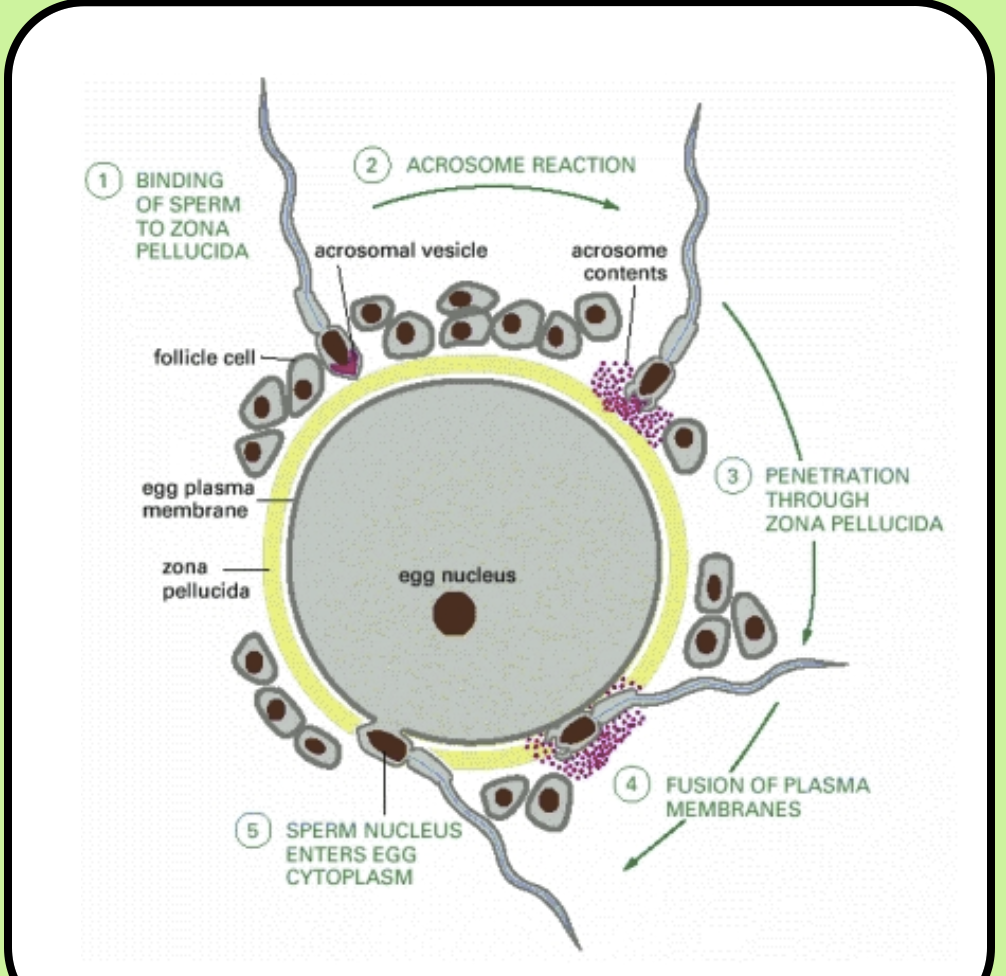
Polyspermy: A cell that has too many chromosomes and isn’t viable because two or more sperm fuse with the egg
Cortical granules: Vesicles loaded with hydrolytic enzymes that are positioned around the egg’s periphery
When the first sperm enters the egg, a wave of calcium crosses the entire cell leading to the fusion of the cortical granules with the plasma membrane
Cortical reaction: Hydrolytic enzymes destroy the zona pellucida, preventing additional sperm from binding
After the sperm’s haploid nucleus has entered, the two pronuclei are separate until the first mitotic division
The sperm provides a centriole that joins with the egg’s centriole- a centrosome!
This centrosome replicates
When the DNA of each pronucleus is replicated, the nuclear envelope breaks down and the first (of many) mitotic divisons begins- fertilization is complete!
Once ferilization is complete, the cell undergoes a series of divisions that turn one cell into a whole organism
Recombination
General recombination- Recombining genes to mix things up genetically
Produces new combinations of alleles that could be useful for an organism in a particular environment
Enhances genetic diversity of gametes, accelerates adaption through natural selection
Recombination
Morgan’s lab:
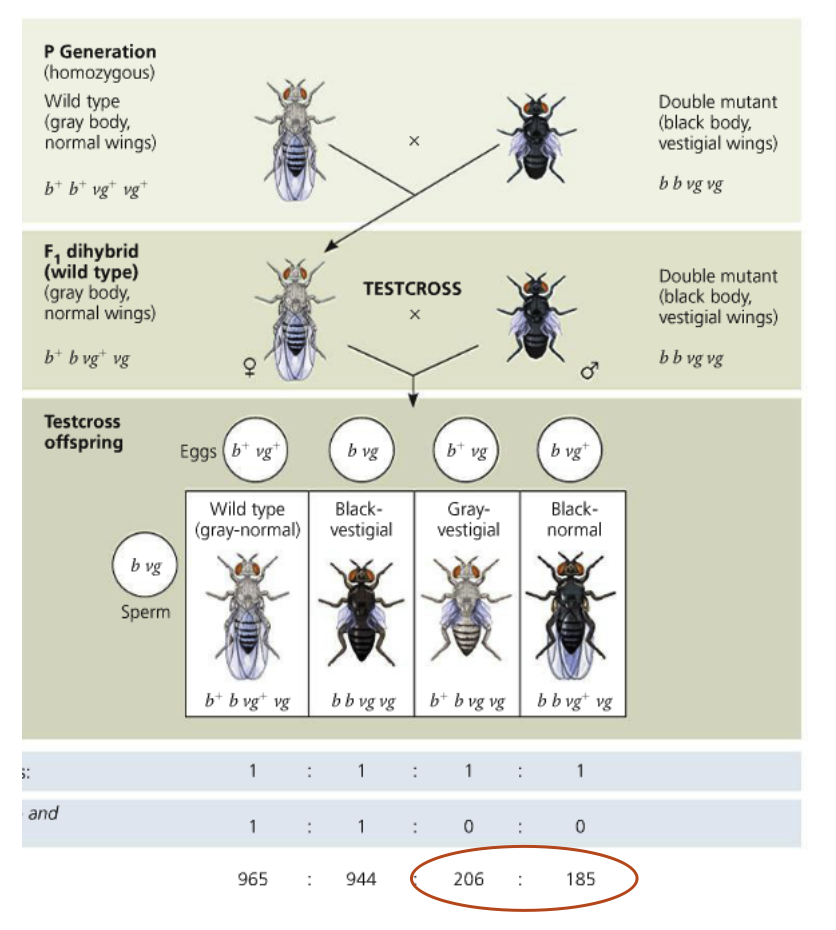
Morgan worked with flies that had different body color and wing size
Wildtype flies have gray bodies and normal sized wings, mutant flies have black bodies and vestigal wings (both mutant alleles were recessive)
Genetic linkage: These genes were located on the same chromosome so they were inherited together
Morgan crossed his flies so he would produce a dihybrid F1 generation
When F1 flies were crossed with mutant flies, the majority of the offspring had the parental phenotype (remember genetic linkage)
Some flies showed a non-parental phenotype, not consistent with genetic linkage (suggests genes had been recombined
50% of the flies should have been grey with normal wings, 50% should have been black with vestigial wings
Follow the chromosomes: The F1 dihybrid can make b+vg or b vg gametes
The mutant fly can only make b vg gametes
The only way to explain this is to assume parts of the chromosome have been exchanged
Now the F1 dihybrid fly can make four possible gametes
The majority of the offspring still have parental phenotypes
MISSED A SLIDE!
Crossing over occurs while DNA is held in a special structure called the synaptonemal complex
The two DNA strands are bound to a ladder-like protein structure called the central element and two lateral elements
In the center of the structure sits a protein complex called the recomination module