19.1-19.5
For some genes, the RNA transcript is the final product, but for others it’s protein. mRNAs encode instructions for translation, the process of assembling amino acids into polypeptide.
19.1 The Genetic Code
Summary: -The base sequence of each mRNA molecule dictates the sequence of amino acids in a polypeptide chain.
-When guiding the synthesis of a polypeptide chain, mRNA is read in units of three bases called codons.
-The table of the genetic code indicates which amino acid (or stop signal) is specified by each codon. The code is unambiguous, nonoverlapping, degenerate, and nearly universal.
The relationship between DNA base sequence and linear order of amino acids in protein products is based on rules called gene code.
Beadle + Tatum studied bread mold, Nuerospora crassa, in the 1940’s. They noticed a link between gene mutations and proteins, and used X-rays to generate mutation that were unable to survive on minimal medium, though they could grow on complete medium (supplemented). The mutants had lost the ability to make amino acids and vitamins, surviving only when given supplements. They used a variety of supplemented media to see which vitamins/amino acids they couldn’t make. Mutants grew on minimal medium with metabolic precursors of an amino acid/vitamin, and determine which precursor helped each mutant grow. Conclusion: each mutation disabled a single enzymatic step of a metabolic pathway, and formed one gene–one enzyme hypothesis.
Pauling studied sickle-cell anemia, where red blood cells aren’t round. He analysed hemoglobin using electrophoresis, found that it migrated differently from the normal one.
Ingram used the protease trypsin to cleave hemoglobin into fragments, and examined the peptides. He found just 1 amino acid difference from normal (glutamic acid, negative) and sick (valine, neutral). Changed one gene- one enzyme (since hemoglobin isn’t an enzyme) → one gene–one polypeptide theory.
Yanofsky showed that mutations in bacterial tryptophan synthase gene corresponded to changed amino acids in the polypeptide.
Most eukaryotic genes have noncoding sequences, and coding sequences can be read in many combinations, each coding for a unique polypeptide chains (alternative splicing). Some genes encode functional RNAs. Genes: functional units of DNA that encode one or more polypeptides or functional RNA.
There are 4 DNA bases and 20 amino acids. A doublet code (2 bases = 1 amino acid) is wack since only 16 combos are possible. Triplet code (3 base = 1 amino acid) gives 64 combos, more than enough for 20 amino acids.
1961: Crick + Brenner found evidence of triplet code. Proflavin was used as a mutagen in bacteriophage T4, which induces an indel mutation (insertion, deletion). Some mutations induced appeared to go back to wild type, and when examined, rather than reversing OG mutation, they had a second mutation near the first.
By itself, either the OG mutation or revertant will cause a mutant, but together they cancel out, called the pseudo wild-type.
The gene is written in 3 letters, and the indel causes the rest to be read out of phase, aka shift in reading frame= frameshift mutation.

Crick + Brenner made frameshift mutations and found they can acquire a second mutation that returned the phage to pseudo wild-type. When 3 nucleotides are added/removed, the reading frame doesn’t change = nucleotides are read in 3.
64 combos, but only 20 amino acids = degenerate code, meaning 1 amino acid specified by more than 1 triplet, and also nonoverlapping, reading frame advances 3 nucleotides at a time. Although it’s always nonoverlapping, there are times when a segment of DNA is translated in more than 1 reading frame (ex. some viruses with small genomes have overlapping genes, and some have only slight overlap).
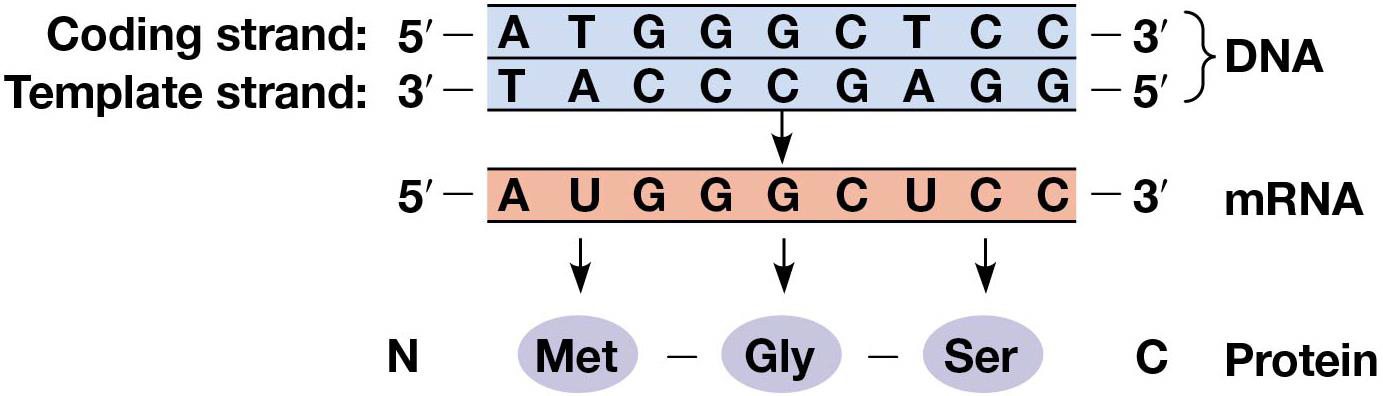
The gene code refers to the order of nucleotides in the mRNA molecules that direct protein synthesis. mRNA is transcribed from DNA similar to how DNA is replicated, but 2 main differences: 1. Only 1 strand of DNA is copied (template strand), other is the coding strand, similair to mRNA sequence 2. Uracil is used instead of Thymine.
Nirenberg and Matthaei created cell-free systems for studying protein synthesis, adding synthetic RNA of known sequence to it. They used polynucleotide phosphorylase to make synthetic RNA of predictable base composition. When 1 ribonucleotide is used to make RNA, the RNA = homopolymer. When poly (U), but not other homopolymers, was added to the cell-free system, a large amount of phenylalanine was incorporated, suggesting that UUU specifies phenylalanine.
RNA triplets (codons) are read by transcriptional machinery, written in 5→3 manner. Further homopolymer experiments showed AAA codes for lysine, and CCC for proline. Copolymers were tested (mix of 2 nucleotides), but it was difficult to figure out which codon was which amino acid.
Khorana used an approach with 1 difference: he made the RNA molecules in an alternating sequence, so only 2 codons (UAUAUAUA= UAU and AUA), so they could narrow the assignment to either tyrosine or isoleucine. Eventually, this showed the assignment of all codons.
All 64 codons are used in translation of mRNA, but 61 specify addition of specific amino acids to the growing chain. One is AUG, which is a start codon. The last 3 (UAA, UAG, UGA) are stop codons. Every codon has only one meaning, unambiguous, and degenerate, many of the amino acids are specific by more than 1 codon. With that in mind, most mutations that cause codon changes ALSO cause changed amino acid. Every organism uses the same genetic code, except mitochondria and a few bacteria. (Ex. AGA is stop for mitochondria mammals, and some organisms have nonstandard amino acids.
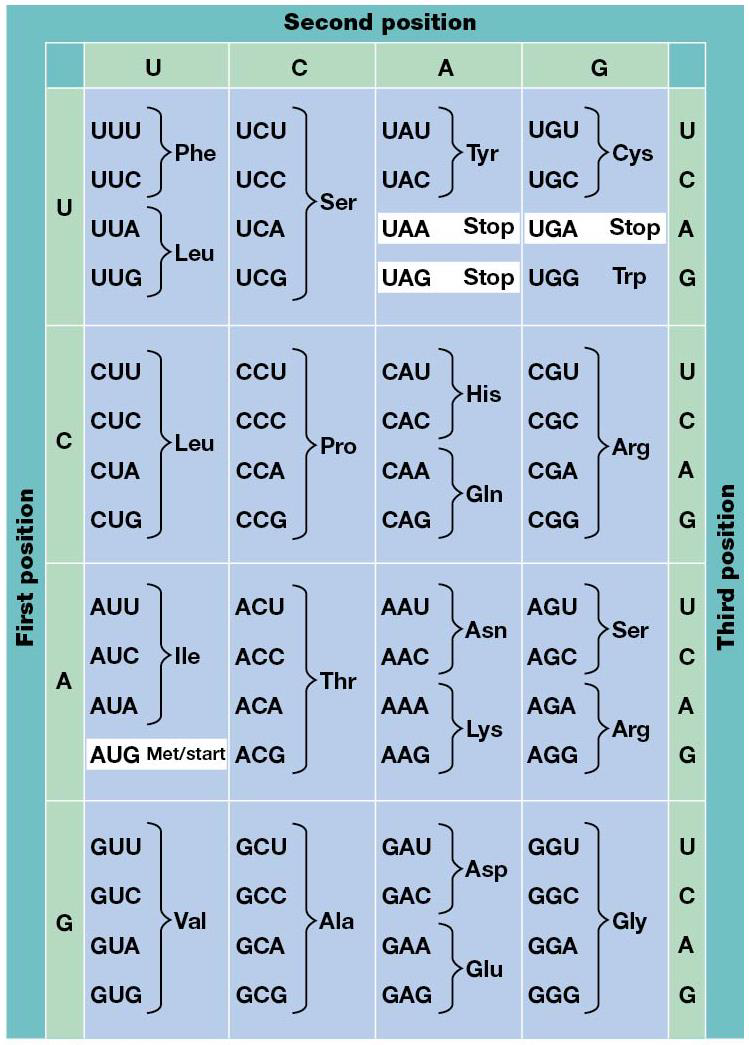
Although more amino acids have more than 1 codon, there’s evidence some are preferred over the other. Codon usage bias is the presence of some synonymous codons at higher frequency in the RNA.
19.2 Translation: The Cast of Characters
Summary: -Translation refers to the synthesis of polypeptide chains on ribosomes using a process that employs mRNA to determine the amino acid sequence.
-Ribosomes have a large and small subunit, each of which is assembled from a large number of proteins and rRNAs.
-The rRNA component of the ribosome helps position the mRNA and catalyzes peptide bond formation; aminoacyl-tRNA synthetases link amino acids to the tRNA molecules that bring amino acids to the ribosome; and various protein factors trigger specific events associated with the translation cycle.
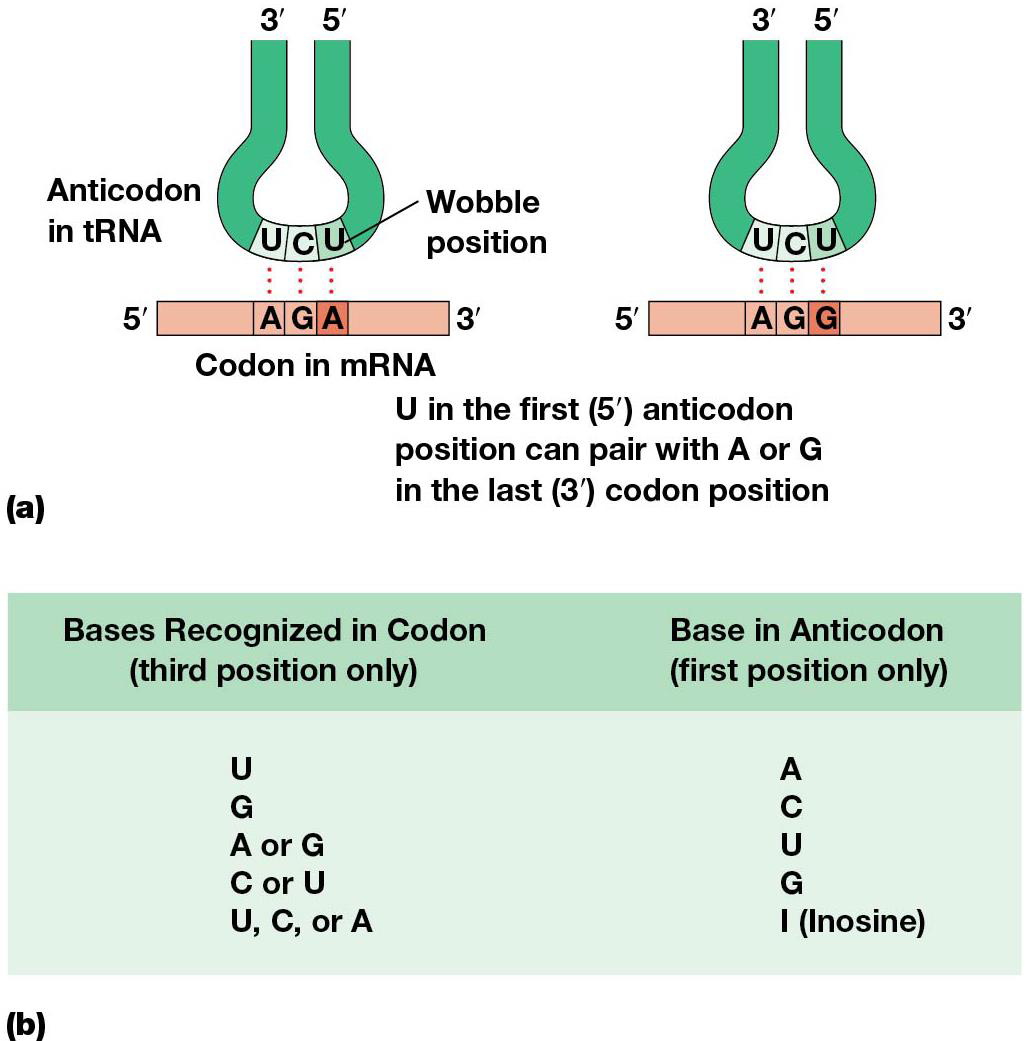
-Wobble allows flexibility in pairing of a tRNA anticodon with more than one codon in the mRNA.
Ribosomes carry out the process of polypeptide synthesis. tRNA align amino acids in the correct order. Aminoacyl-tRNA synthetases attach amino acids to their appropriate tRNA molecules. mRNA encode the amino acid sequence info. Protein factors facilitate some steps of translation.
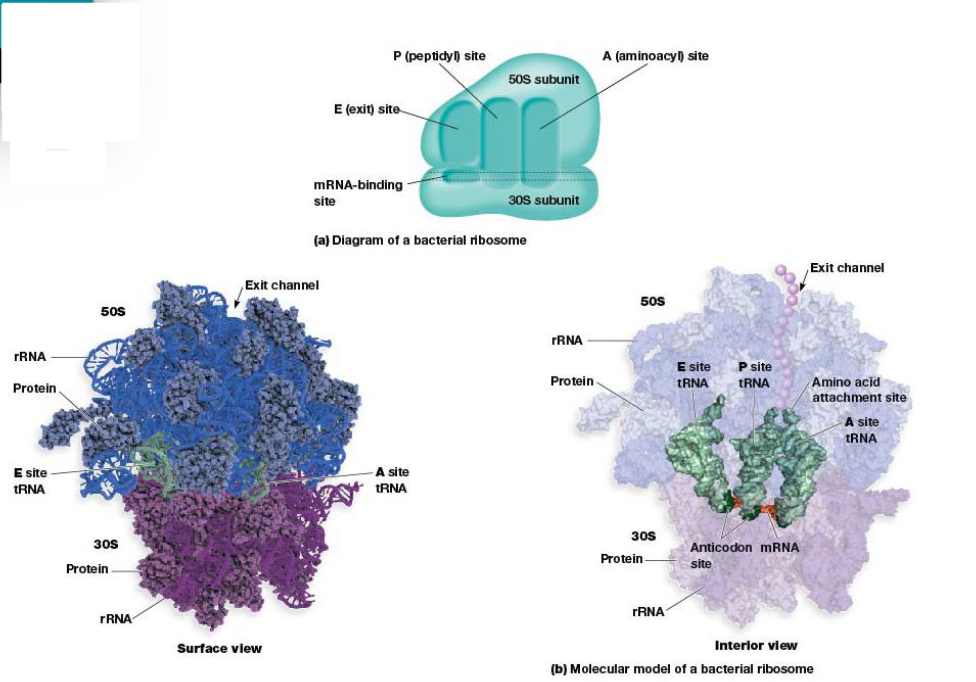
Ribosomes are made of rRNA and protein. Eukaryotes are found free in the cytoplasm and bound to ER + outer nuclear envelope. In prokaryotes, the ribosomes are smaller. They’re made of dissociable units (large and small). Bacterial ones are sensitive to different inhibitors of protein synthesis and are made of less proteins/RNA.
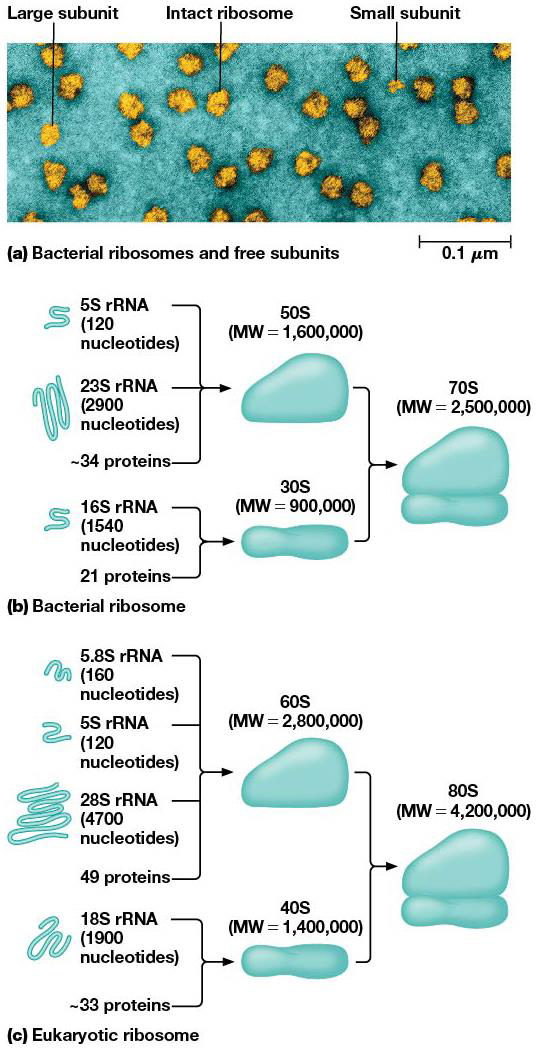
They have a role in making polypeptide synthesis by being a very large enzyme, with rRNA performing many of the functions.
The 4 ribosome sites important for protein synthesis are mRNA-binding sites and the 3 sites for tRNA bind to A, P and E sites.
A (aminoacyl) site binds tRNA with attached amino acid.
P (peptidyl) site tRNA carrying the growing peptide resides.
E (exit) site tRNAs leave ribosomes after discharging their amino acid.
tRNA molecule: adaptor that binds a specific amino acid AND mRNA sequences that specify amino acid. It’s linked to it's amino acid by an ester bond, and named for the amino acids attached to them (tRNAAla). tRNAs attached to the amino acid are called aminoacyl tRNAs , which is ‘charged’, and the amino acid is ‘activated’. Each tRNA recognizes codons in mRNA by complementarity to anticodon in tRNA. Soem tRNAs recognize more than 1 codon. Anticodons permit tRNA molecules to recognize codons in mRNA by complementary base pairing.

mRNA and tRNA line up on ribosome in a way that gives flexibility/wobble in the pairing between the 3rd base of the codon and the corresponding base of the anticodon = wobble hypothesis, which allows for some unexpected base pairing.
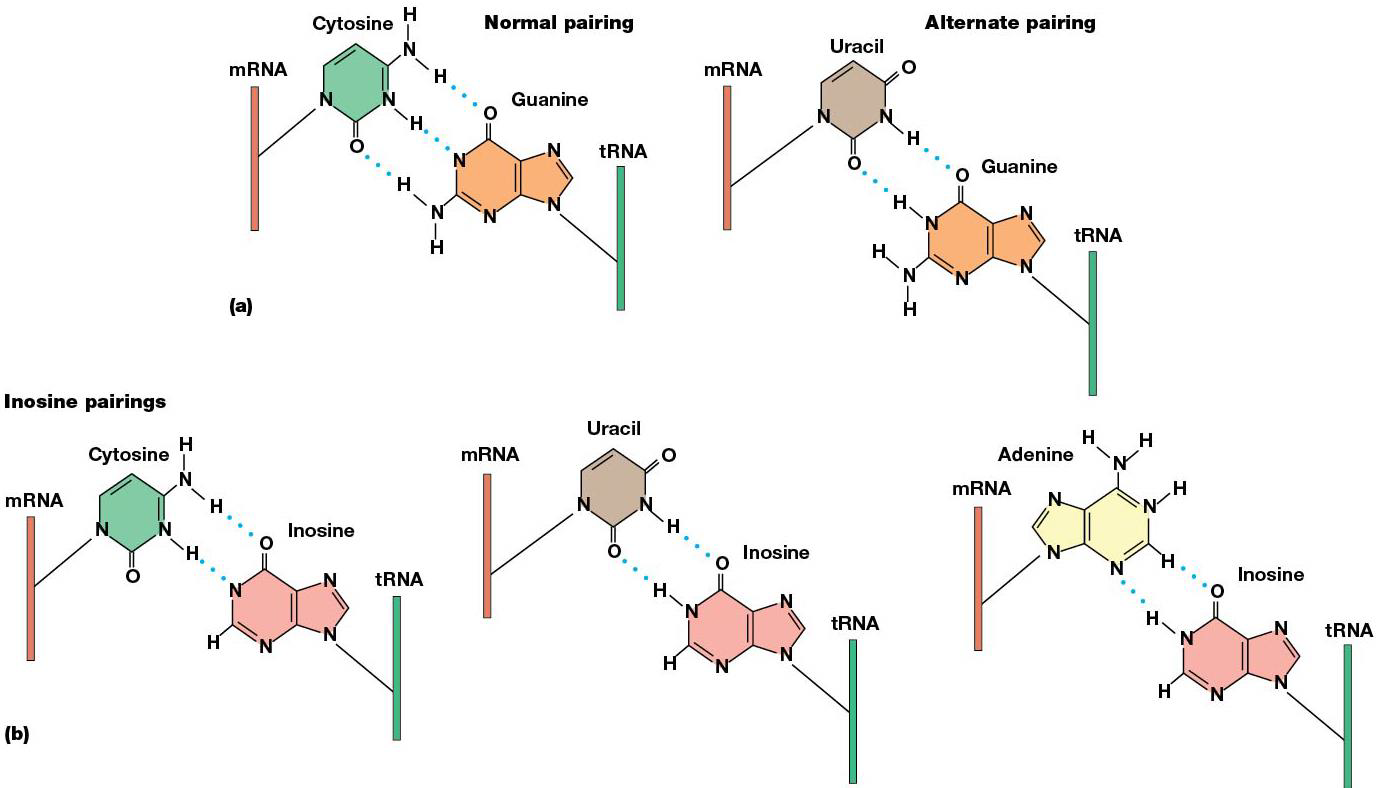
Adenosine → inosine at specific locations in tRNA through RNA editing. Inosine occurs in the wobble position and pairs with U, C or A. (ex. tRNA anticodon 3’-UAI-5’ can recognize AUU, AUC, or AUA, which turns in isoleucine). Because of wobble, less tRNAs are needed for some amino acids than # of codons.
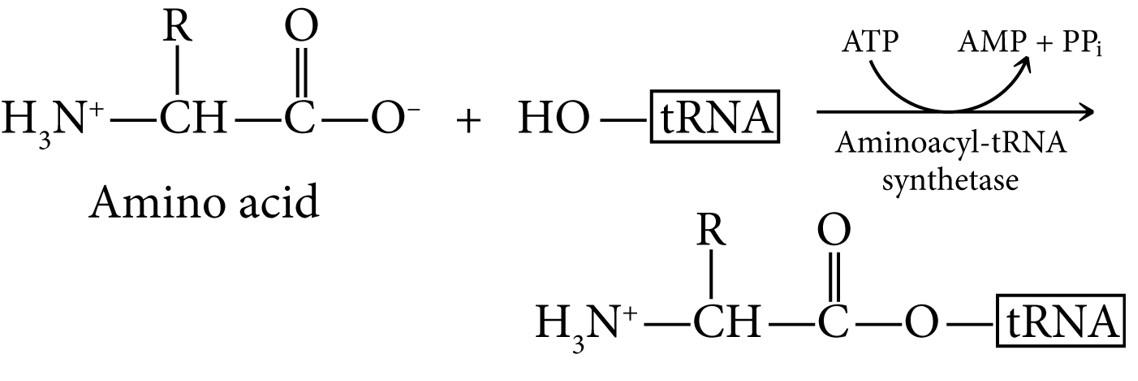
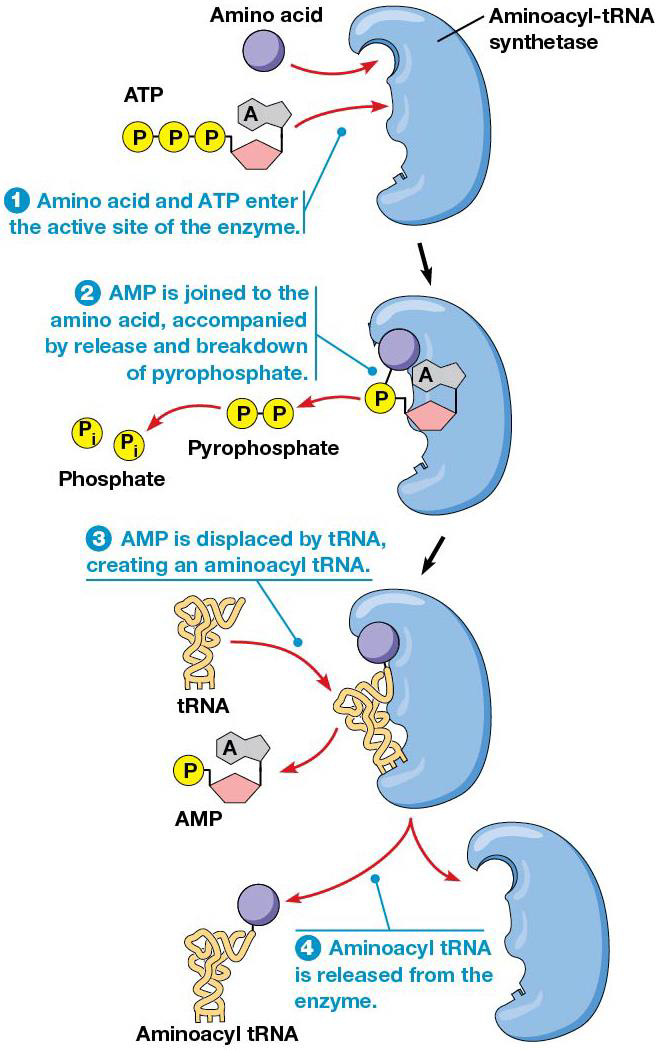
Before a tRNA can bring an amino acid to the ribosome, the amino acid must be attached covalently to the tRNA by aminoacyl-tRNA synthetase. Cells have 20 different ones, one for each amino acidm and cells with nontraditional amino acids have special tRNAs/aminoacyl-tRNA synthetases for them.
They catalyze the attachment of amino acids to tRNAs via ester bond using ATP hydrolysis. Both anticodon and 3’ end of tRNA are needed to specify the correct amino acid. After adding an amino acid (amino acid activation), the synthetases proofread the final product to ensure the correct one was added. It’s the tRNA that then recognizes the right codon in the mRNA.
The codon sequence in mRNA directs the order of amino acids in a polypeptide. mRNA must first be exported from nucleus → cytoplasm. An untranslated sequence at 5’ end of the message is before the start codon, and first to be translated (AUG). Untranslated region at 3’ end of mRNA follows the stop codon (UAG, UAA, UGA). 5’ and 3’ untranslated regions vary in length, and are needed for mRNA function, which also have a 5’ cap and 3’ poly(A) tail.
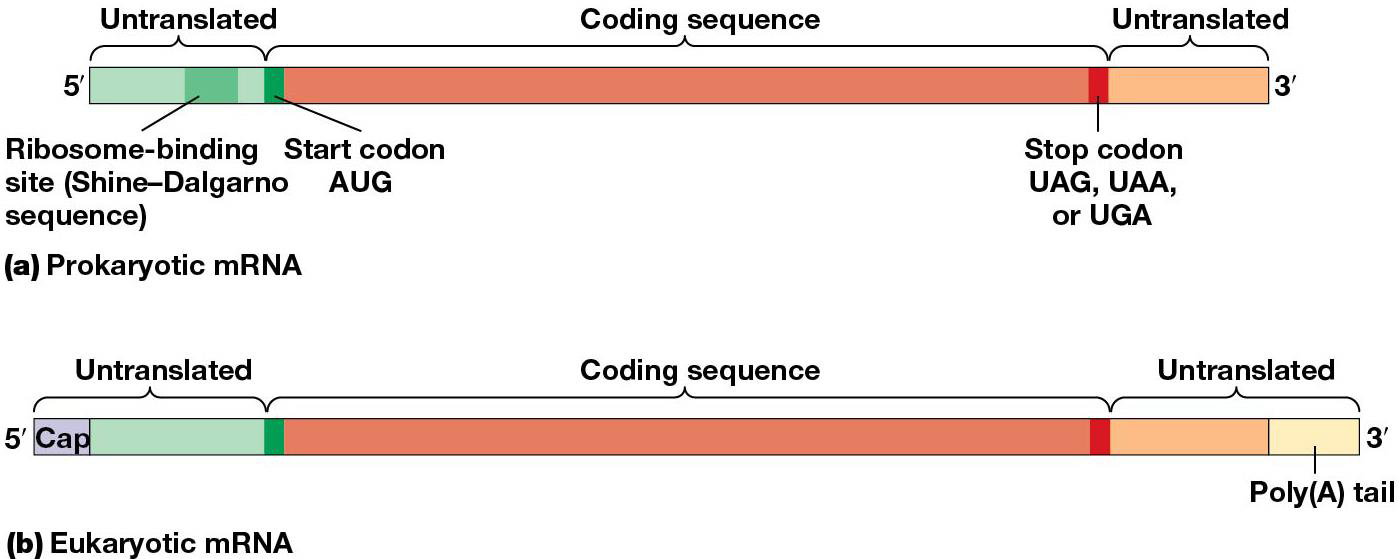
most mRNAs in eukaryotes are monocistronic(encode one polypeptide)
bacteria and archaea are polycistronic (encode several polypeptides with related functions, called operons).
19.3 The Mechanism of Translation
Summary:
-Translation involves initiation, elongation, and termination stages.
-During the initiation stage, initiation factors trigger the assembly of mRNA, ribosomal subunits, and initiator aminoacyl tRNA into an initiation complex. Ribosome binding sequences on the mRNA promote binding of the small ribosomal subunit. In eukaryotes, both the 5' and 3' ends of mRNAs promote translational initiation.
-Chain elongation involves sequential cycles of aminoacyl tRNA binding, peptide bond formation, and translocation, with each cycle driven by the action of elongation factors. The net result is that aminoacyl tRNAs add their amino acids to the growing polypeptide chain in an order specified by the codon sequence in mRNA.
-More than one ribosome can move along a single mRNA at the same time.
-Chain termination occurs when a stop codon in mRNA is recognized by release factors, which cause the mRNA and newly formed polypeptide to be released from the ribosome.
-GTP binding and hydrolysis are required for the action of several initiation, elongation, and release factors.
-Proper folding of newly produced polypeptide chains is assisted by molecular chaperones.
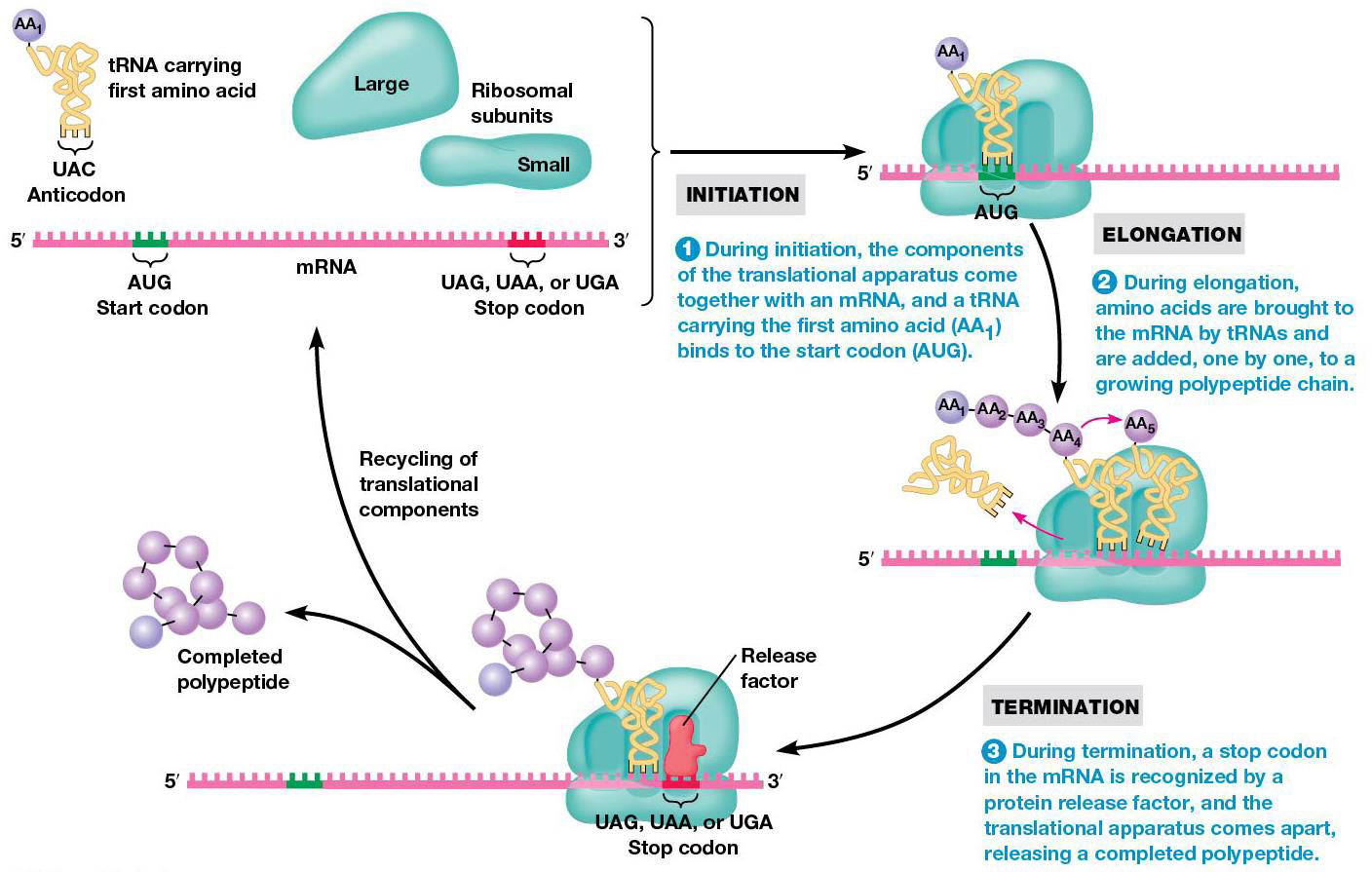
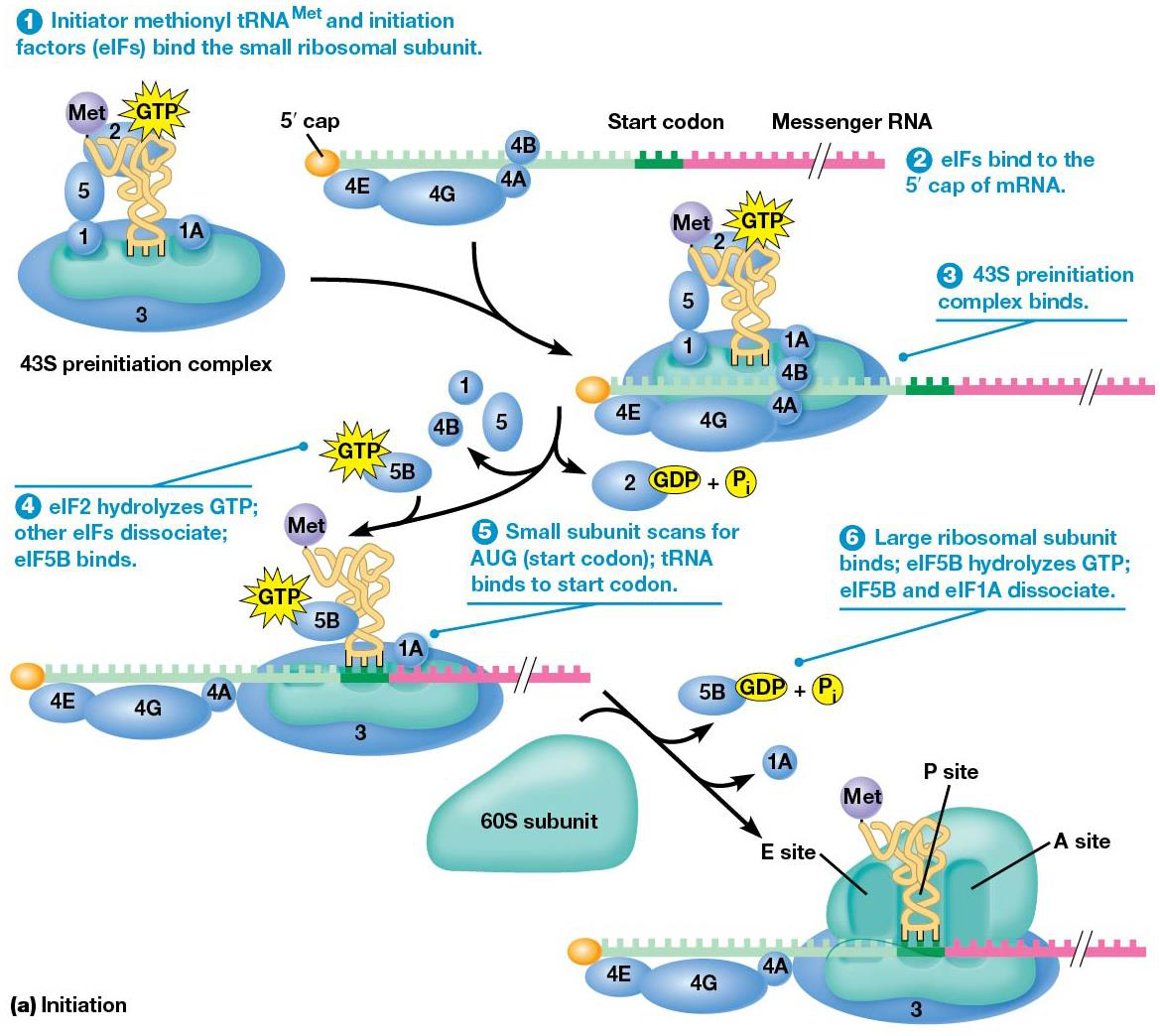
Translation begins at the N-terminus of polypeptide and adds amino acids to the growing chain until C-terminus is reached. mRNA is read 5→3. Translation is divided into 3 stages: 1. initiation mRNA is bound to ribosome, positioned for translation 2. elongation amino acids are joined via peptide bonds 3. termination mRNA and polypeptide are release from ribosome.
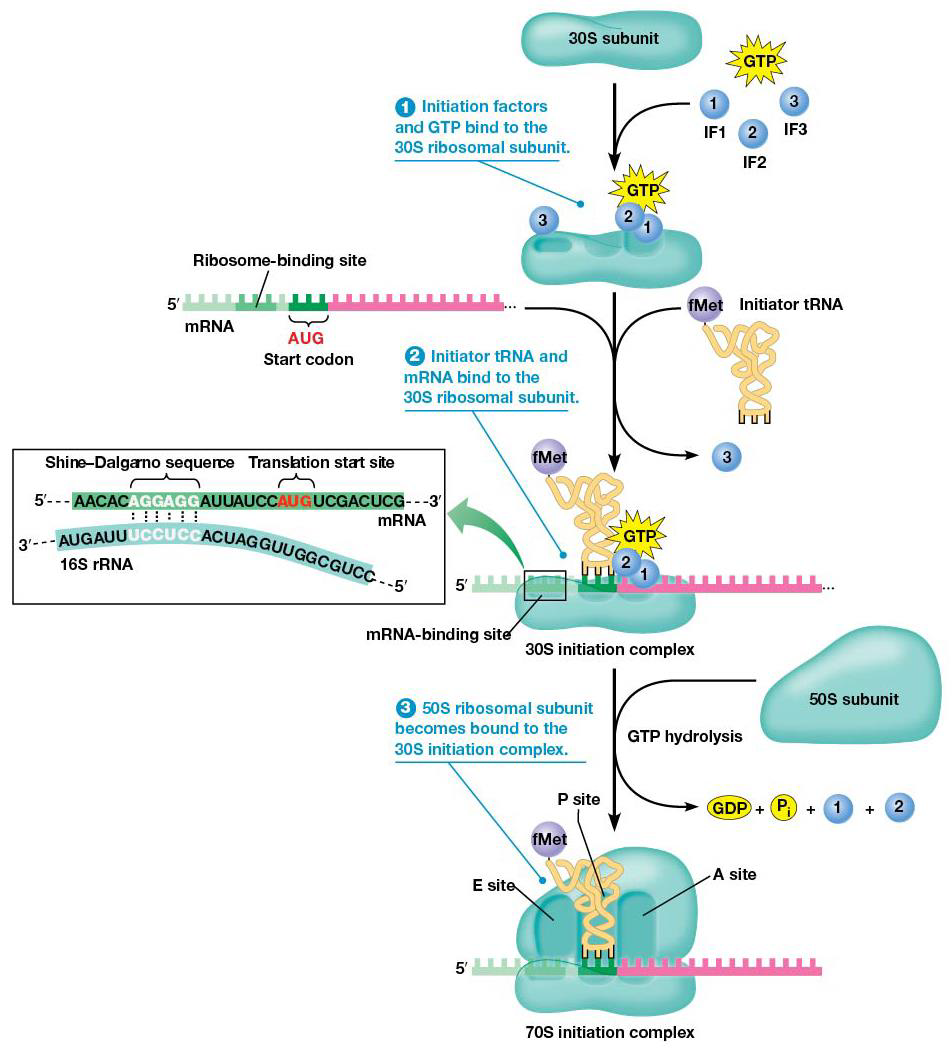
Bacteria Initiation
1. Initiation factors (IF1,IF2,IF3) bind to small (30S) ribosomal subunit, GTP binds to IF2
2. mRNA and tRNA carrying the first amino acid bind to 30S subunit. mRNA binds in the proper orientation due to Shine-Dalgarno sequence, which is made of 3-9 purines located upstream of the start. The purines in mRNA complementarily base pair with a sequence at the 3’ end of 16S rRNA, which forms the ribosome’s mRNA binding site. This places the start codon at the ribosome’s P site, and a special methionine, N-formylmethionine (fMet) is used to start translation (only the carboxyl group can bind to another amino acid). tRNAfMet= initiator tRNA. Initiator tRNA binds the P site of the small subunit by action of IF2 + GTP, making the 30S initiation complex (initiation factors, mRNA, tRNAfMet)
3.IF3 is released, and the 30S initiation complex binds to large (50S) subunit, making the 70S initiation complex. The 50S promotes hydrolysis of IF2-bound GTP→ IF2 and IF1 released, which means all initiation factors are released.
Eukaryotic Initiation Differences
-The start codon in eukaryotes + archaea uses methionine, not N-formylmethionine.
-The initiation factors are called eIFs (eukaryotic initiation factors).
-Different genes encode initiator-tRNAMet and one used during elongation.
-Initiator-tRNAMet favors binding to eIF2 + prevents binding to elongation machinery.
Eukaryotic Initiation Steps
1. eIF2 + GTP binds to initiator-tRNAMet before tRNA binds the small ribosomal subunit, + other initiation factors (eIF1, 3, 5) and eIF1A. (43S preinitiation complex)
2. mRNA is prepped to bind the 43S preinitiation complex, the mRNA 5’ cap is recognized by cap binding initiation factor eIF4RE, which uses eIF4G, which regulates translation efficiency. eIF4A (helicase) and eIF4B join as well.
3. 43S preinitiation complex + the mRNA complex because of interactions between eIF4G and eIF3, and small ribosomal subunit and mRNA.
4. eIF2 hydrolyzes associated GTP, letting several eIFs leave, and eIF5B +GTP binds.
5. After binding to mRNA, the small ribosomal subunit (with tRNAMet) scans along mRNA and begins translation at AUG. The nucleotides on either side of the start codon are used in the start site, [common one is ACCAUGG Kozak sequence.)
6. After initiator-tRNA pairs with start codon, the large ribosomal subunit joins the complex, which is facilitated by eIF5B. When eIF5B hydrolyzes its GTP, the other eIFs dissociate from fully assembled ribosome.
Eukaryotic mRNAs are polyadenylated at the 3’ end. Poly(A)-binding protein (PABP) binds the polyA stretch + bind eIF4G, therefore the 3’ end of mRNA is important for initiation, since it stabilizes the 5’ end.
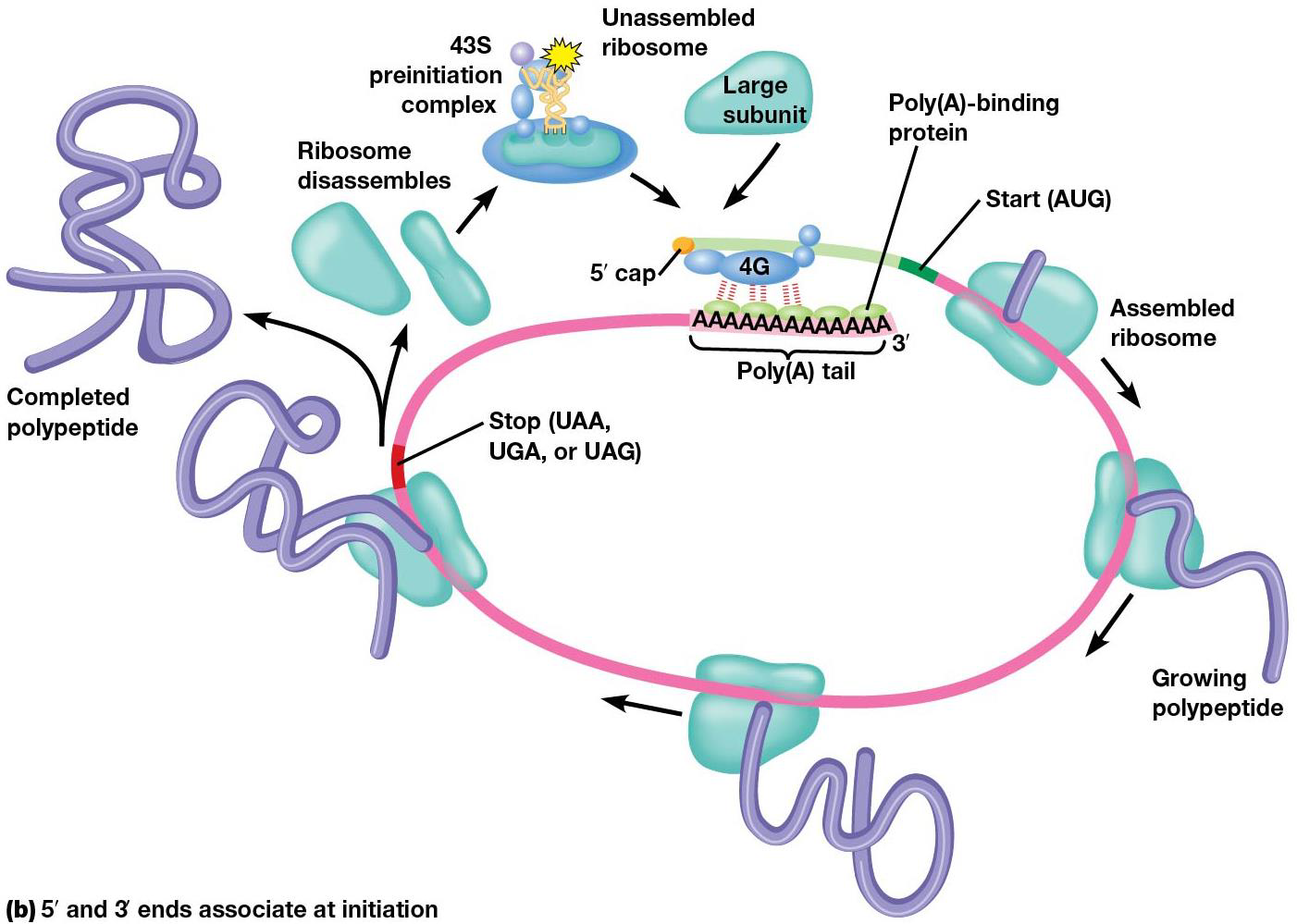
Not all eukaryotic mRNAs must be capped to recruit a ribosome; sometimes, a complex may binds to an internal ribosome entry sequence (IRES) directly the upstream of the start codon. They’re found in some types of mRNA, usually viral ones, and used a lot in molecular bio.
When initiation is done, a polypeptide chain is created. Amino acids are added in sequence to the growing chain (elongation), which involves a repetitive cycle of three steps.
1. Binding of aminoacyl tRNA to ribosome → new amino acid into position to be joined to peptide chain
2.Peptide bond formation links amino acid to growing polypeptide.
3. mRNA is advanced three nucleotides through translocation to bring the next codon into position for translation.
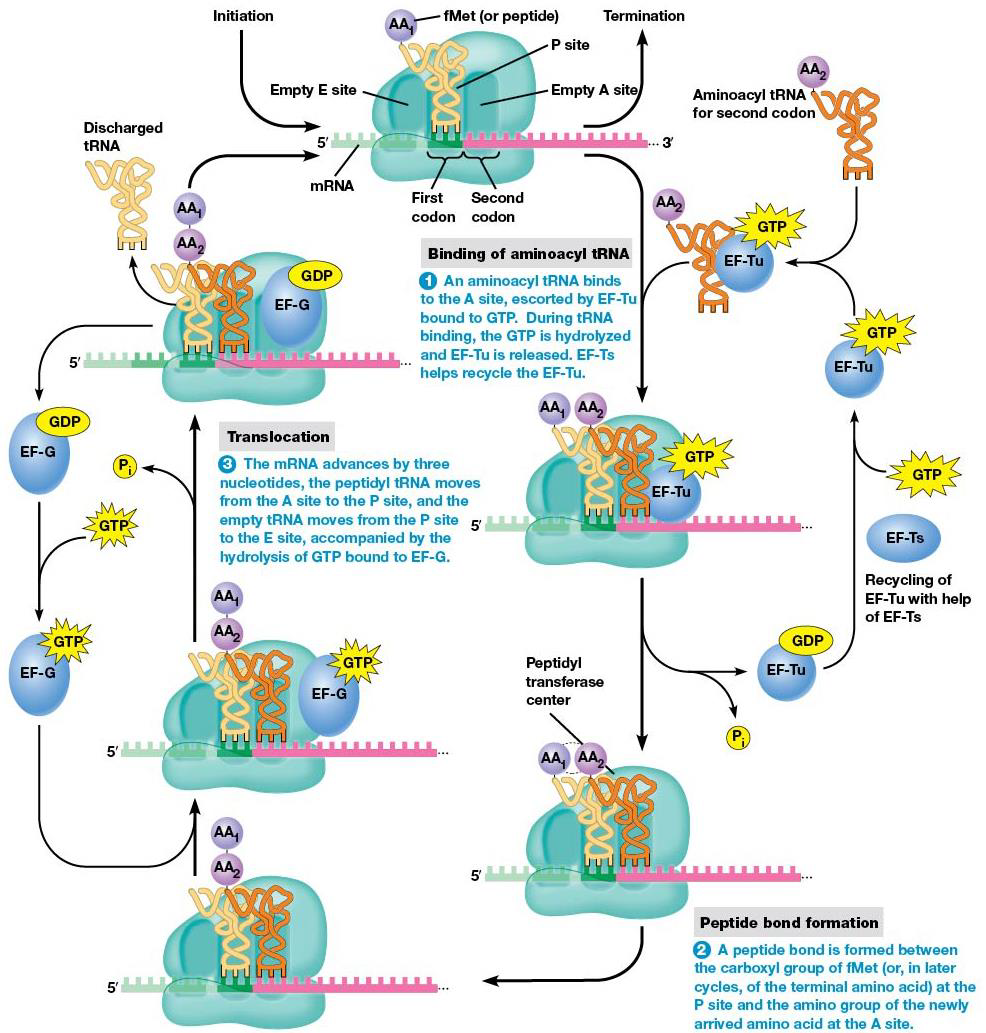
Step 1 of elongation (binding aminoacyl tRNA)
When elongation begins, the start codon is on the P site, the next one on A site. tRNA with anticodon complementary to second codon bonds to A site, which needs two elongation factors, EF-Tu and EF-Ts, and GTP hydrolysis. Every aminoacyl tRNA with bind the A site. EF-Tu-GTP complex moves aminoacyl tRNA to ribosome A site, and while moved, GTP is hydrolyzed, and the complex is released. EF-Ts regens EF-Tu-GDP → EF-Tu-GTP.
Elongation factors don’t recognize particular anticodons, so all types (except initiator-tRNAs) are brought to A site, and only those with anticodon complementary to the codon stay there long enough for GTP hydrolysis to occur. Final error rate in translation is 1/10,000.
Step 2 of elongation (peptide bond formation)
A peptide bond forms between the amino group of the amino acid at the A site, and carboxyl group of amino acid at P site. The growing peptide chain is moved to the tRNA at A site, and no hydrolysis is needed.
For a long time, it was thought a protein ‘peptidyl transferase’ catalyzed the bond formation, but Noller showed that rRNA has the catalytic activity (bacteria = 23S RNA), making it another example of a ribozyme.
Step 3 of elongation (translocation)
After the peptide bond forms, the mRNA forces advances to bring the next codon into proper position. During this translocation, the peptidyl tRNA moves A→ P site, and empty tRNA → E site. Hydrolysis of GTP bound to EF-G triggers a conformational change that completes the movement.
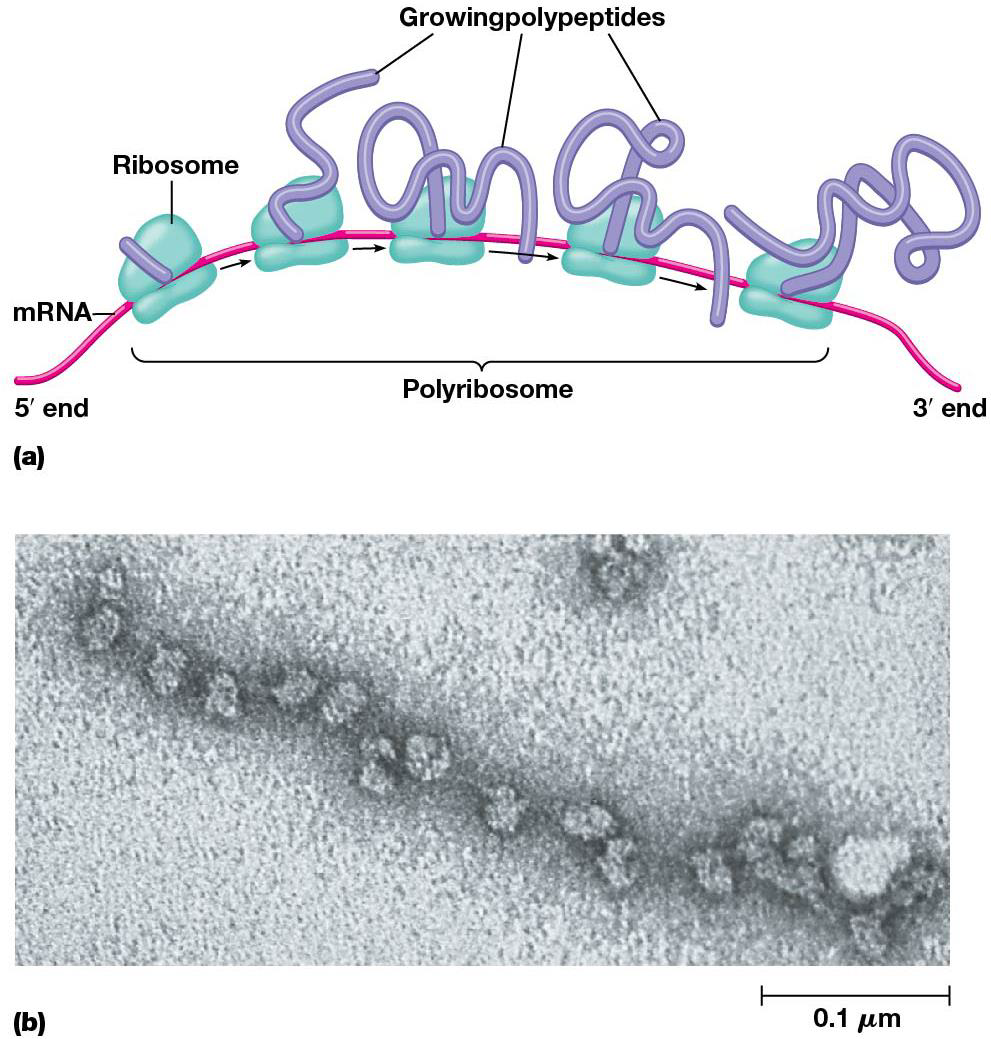
The cell makes good use of each mRNA molecule, many ribosomes translate 1 at the same time. This cluster is called polyribosome/polysome, which maximize the efficiency of mRNA utilization.
Termination of polypeptide synthesis is triggered by release factors that recognize stop codons. Codons are read on mRNA until a stop arrives at A site, and recognized by release factors, rather than tRNAs. They have special regions (polypeptide anticodons) that bind mRNA stop codons at A site. The shape of protein release factors is similar to tRNA (molecular mimicry). After binding to A with GTP, release factors stop translation by triggering release of polypeptide from peptidyl tRNA.
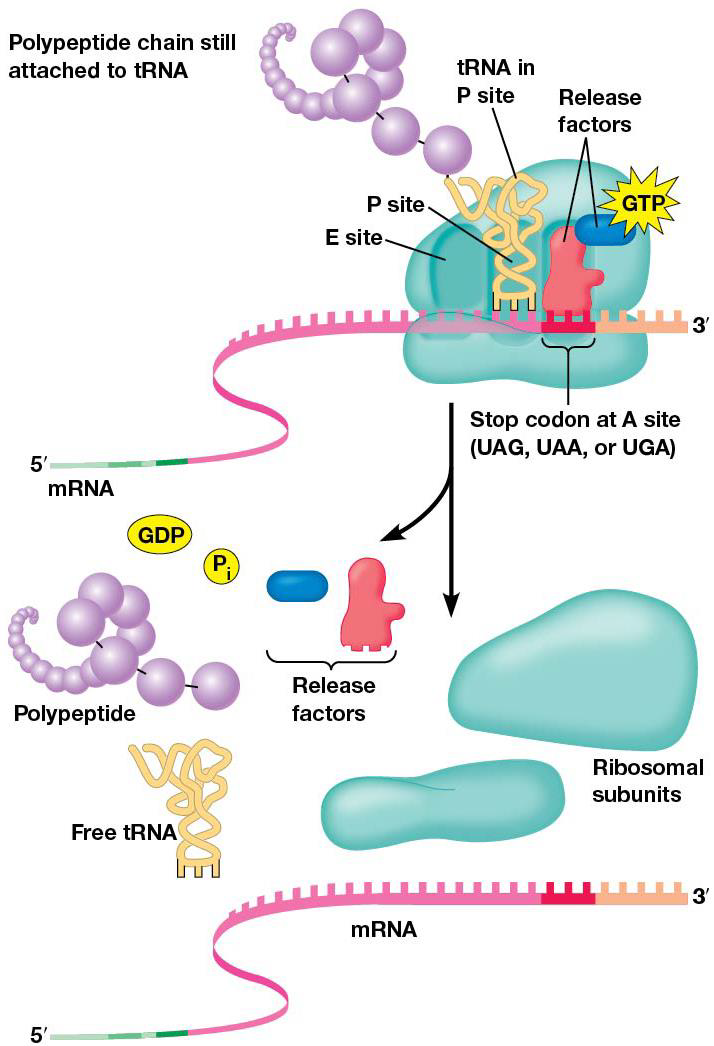
Proteins must fold into their proper 3D shapes before they can function, which is facilitated by chaperone proteins, which begins when the polypeptide chain first emerges from the ribosome exit tunnel.
A key use is to bind polypeptide during the early stages of folding to keep them from binding to other polypeptides. If folding goes wrong, chaperones can fix the proteins and fold them right or destroy them. Some kinds of incorrectly folded proteins bind to each other and make insoluble aggregates in and between cells.
In mature, proper proteins, hydrophobic regions are buried in the interior. In a polypeptide, hydrophobic regions are exposed to water, leading to aggregation. Chaperones detect the hydrophobic regions and repair misfolded proteins by interacting with them in an ATP-dependent manner.
The most common chaperones are Hsp70 and Hsp60. 70 bound to ATP can associate with polypeptides that are still being made or misfolded proteins. ATP hydrolysis is done during changes in protein folding.
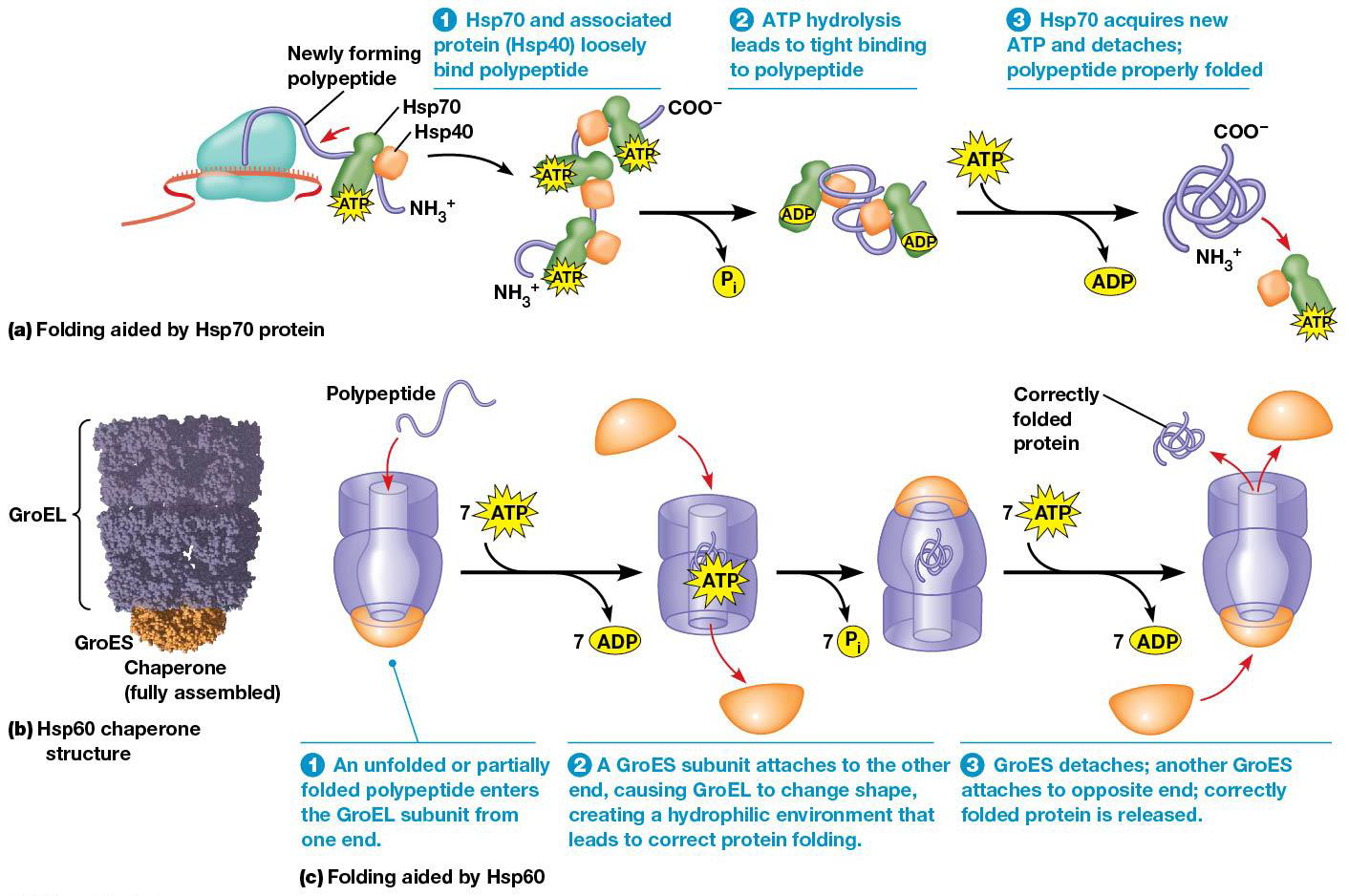
Hsp60 complex (GroEL/GroES complex) in bacteria
1. A partially/misfolded protein enters one end of the complex
2.GroES subunit attaches to the open end, ATP hydrolysis leads to shape change of the GroEL subunit. (creates hydrophilic environment, good for proper folding)
3. The right folded protein is released, and GroES subunit attaches to opposite end.
Protein synthesis uses a good fraction of the cell’s energy. Polypeptide elongation uses hydrolysis of at least 3 ‘high energy’ phosphoanhydride bonds. If each bond has G= 7.3 kcal/mol, total it 29.2 kcal/mol. Additional GTPs are using during formation of initiation complex, the binding of incorrect aminoacyl tRNAs, and termination.
Translation Summary
Translation converts info in mRNAs → chain of amino acids liked by peptide bonds. The ribosome reads the mRNA codon by codon in the 5→3 direction. RNA molecules are important in translation (mRNA, tRNA, rRNA).
19.4 Mutations and Translations
Summary: -Nonsense mutations, which change an amino acid codon to a stop codon, can be overcome by suppressor mutations that allow a tRNA anticodon to read the stop codon as an amino acid.
-Defective mRNAs containing premature stop codons are destroyed by nonsense mediated decay; defective mRNAs containing no stop codon are destroyed by nonstop decay.
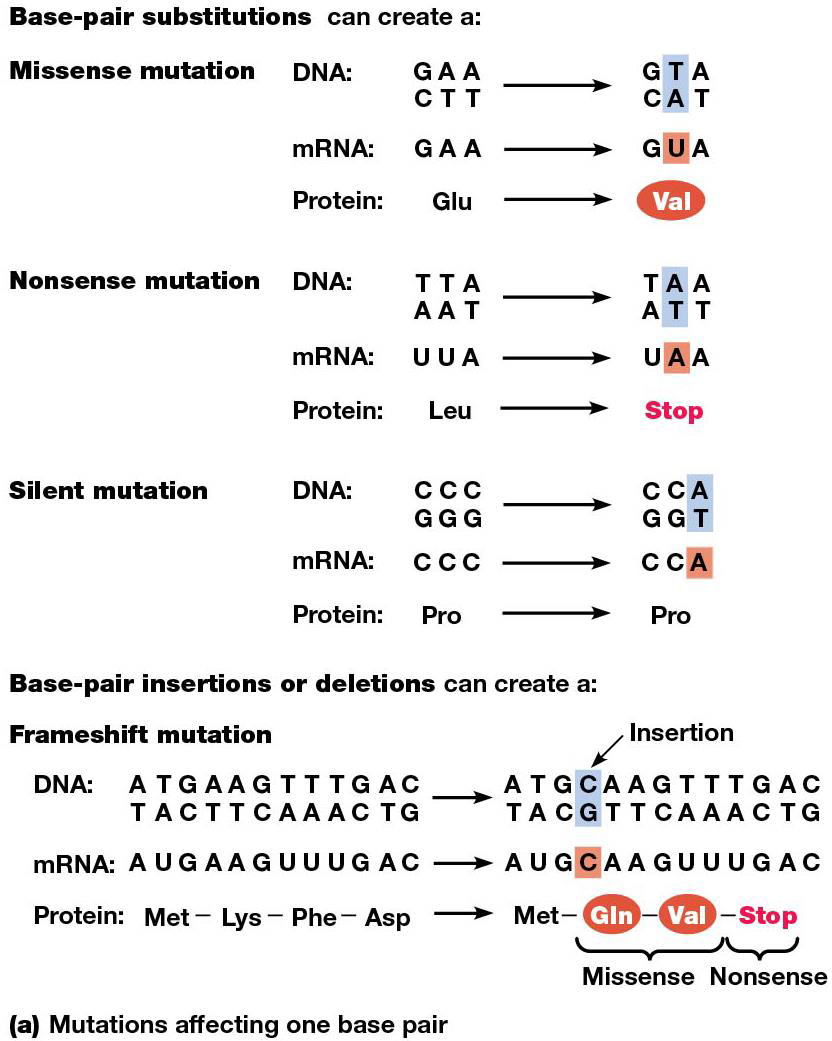
Mutations refers to any change in nucleotide sequence of a genome. mRNAs may contain mutant codons that cause errors in the polypeptide chain synthesized.
Missense mutations
A base-pair substitution can alter a codon. Ex. sickle cell anemia, AT→ TA, so GAA→ GUA in mRNA transcribed from the mutant allele. In the polypeptide, glutamic acid →valine. (missense/nonsynonymous mutation, since it makes the ‘wrong’ amino acid.)
Nonstop/Nonsense Mutations
A base-pair substitution can change a stop codon to an amino acid codon, nonstop mutation.
If it goes from amino acid to stop codon, this is nonsense, leads to a premature stop of translation and a shorter polypeptide. They are typically lethal, but can sometimes be overcome by an independent mutation affecting a tRNA gene (suppressor tRNA.)
Frameshift mutations
mutations can arise from base-pair insertions, deletions, or a combo. This is called indels, and they cause frameshift mutations
Other mutations
Sometimes 1 amino acid change doesn’t cause any function change, if both are similar enough, silent/synonymous.
Some large scale mutations are created by indels with longer DNA segments. Duplications= stretch of DNA is repeated. Inversion = chromosome section is cut out and reinserted, but in reverse. Translocation = movement of a segment from one position to another.
Suppressor tRNAs
They recognize stop codons and insert amino acids, suppressing nonsense mutations. Highly efficient suppressor tRNA may lead to production of abnormal proteins, but at the 3’ ends of mRNAs, release factors bind stop codons better than suppressor tRNAs, which limits the effect of the suppressor to internal locations on the mRNA
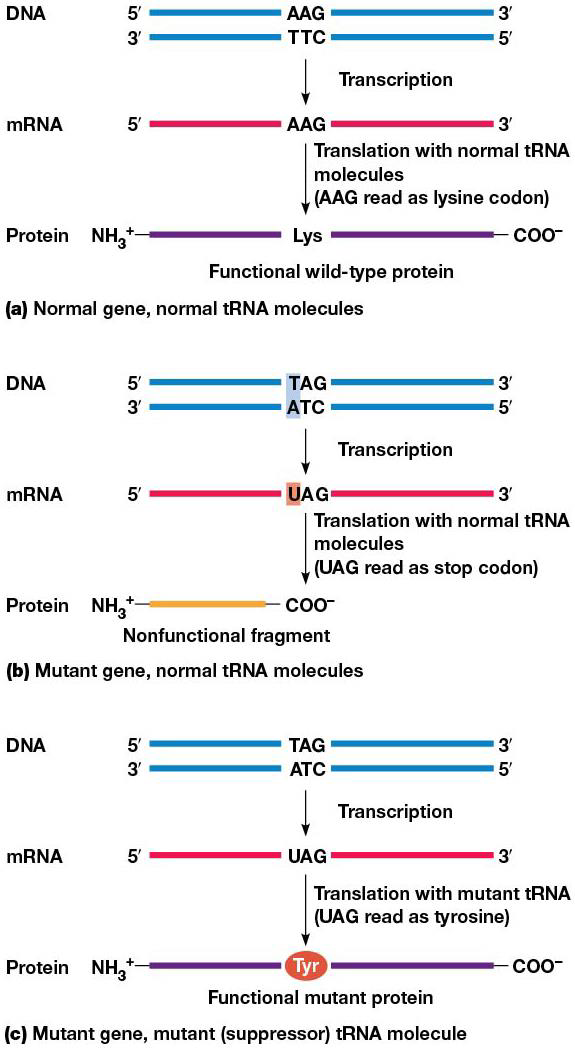
Without suppressor tRNA, a nonsense mutation will cause stopping early and an unfinished polypeptide chain. Eukaryotic cells use nonsense-mediated decay to destroy mRNAs containing premature stop codons. In mammals, the exon junction complex (EJC) is used to detect premature stop codons. An EJC is deposited wherever an intron is removed from pre-mRNA, so each spliced mRNA has 1 complex bound to it. If an mRNA has a stop codon before final EJC, translation is stopped. EJCs still associated with the mRNA target it for degradation.
In eukaryotes, translation is stalled when a ribosome reaches the end of a transcript that doesn’t have a stop codon. An RNA-degrading enzyme binds the empty A site of the ribosome and degrades the bad mRNA via nonstop decay. In bacteria, the transfer messenger RNA (tmRNA) binds the A site and directs addition of amino acids that target the protein for destruction.
19.5 Posttranslational processing
Summary: -Newly made polypeptide chains often require chemical modification before they can function properly. Such modifications include cleavage of peptide bonds, phosphorylation, acetylation, ubiquitination, methylation, glycosylation, and protein splicing.
After polypeptide chains are made, they need to go through posttranslational modification before they can function. In bacteria, the N-formyl group is ALWAYS removed, the methionine at the N-terminus is usually removed. In eukaryotes, the methionine at the N-terminus is often released as well.
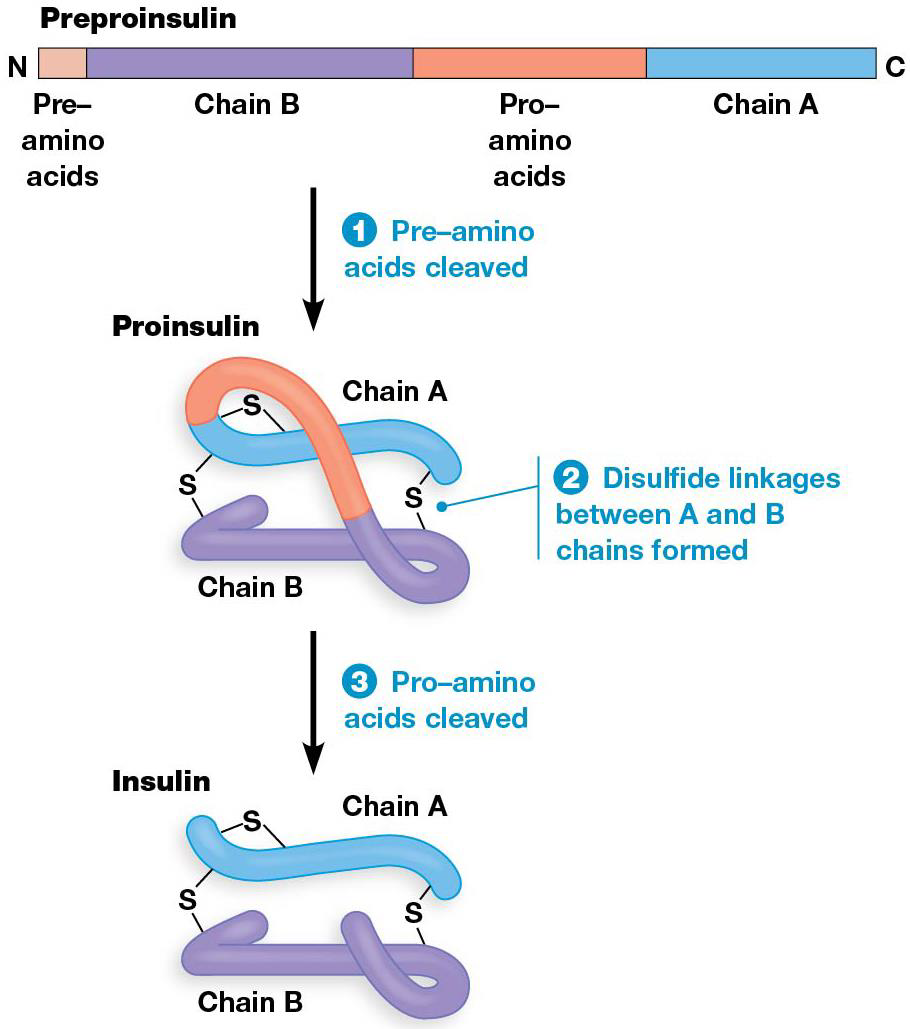
Sometimes whole blocks of amino acids are removed from the polypeptide, ex. enzymes made by inactive precursors. They are activated by removing a sequence from one end of the protein, and the transport may need removal of a terminal signal sequence. Some proteins have internal amino acids that must be removed to be active, like insulin.
Other types of processing events include chemical modification of amino acids—methylation, phosphorylation, acetylation, or ubiquitination. Peptides can be glycosylated or have prosthetic groups. If it is a multisubunit protein, the subunits must be bound to one another.
Some proteins go under a rare process called protein splicing. Similair to RNA splicing, protein sequences called inteins are removed, and the remaining sequences exteins are spliced together.
Posttranslational processing of Insulin
2 subunits of mature insulin come together via disulfide bonds from a single, longer protein (preproinsulin), which has the N-terminal “pre” amino acids removed from proinsulin. Then disulfide bonds form between what will be the A + B subunits of the mature protein. Finally, the intervening “pro” amino acids are removed to form mature insulin