Overview of Transcription
*These notes were taken from LG 2.3
LG:
file:///C:/Users/Valeska/Desktop/Valeska's%20School%20Files/PSHS%20Grade%2010/Biology%202/4th%20Quarter/Learning%20Guides/SLG-BIO2-LG2.3.pdf
Overview of Transcription
Transcription
This is the process by which a segment of DNA directs the synthesis of an RNA.
Here, DNA does not produce proteins directly:
One strand of DNA must be copied first into a complementary RNA sequence.
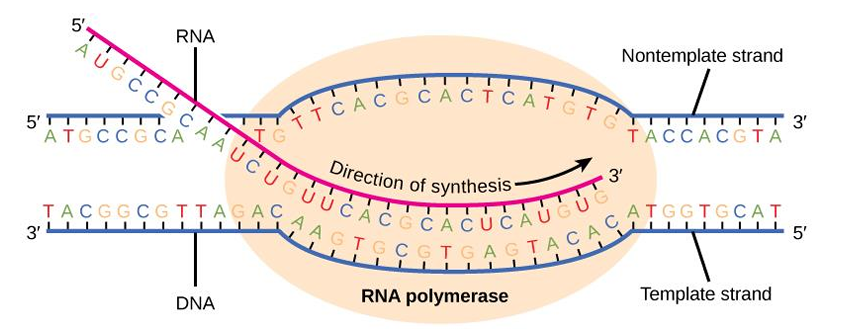
Process of Transcription
In the 5’ to 3’ direction, an enzyme called RNA polymerase unwinds the DNA (responsible for polymerization of RNA).
Like DNA replication, transcription starts with the opening and unwinding of a small portion of the DNA double helix, followed by complementary base pairing between incoming nucleotides and the DNA template.
Unlike DNA replication, the RNA strand does not remain hydrogen bonded to the DNA template strand in transcription.
Also, RNA molecules synthesized during transcription are much shorter than DNA molecules.
A comparison of RNA polymerase and DNA polymerase is presented below.
Table 3.2a. RNA Polymerase vs. DNA Polymerase
RNA Polymerase | DNA Polymerase |
|---|---|
Catalyzes linkage of ribonucleotides | Catalyzes linkage of deoxyribonucleotides |
Can start an RNA chain without a primer | Requires a primer |
Less accurate | High accuracy |
In eukaryotes, the binding of several transcription factors that make the binding of RNA polymerase possible is activated by unique regions in the DNA called promoters (upstream). The major types of RNA are presented below.
Table 3.2b. The Different Types of RNAs
Type of RNA | Function |
|---|---|
mRNAs | Messenger RNAs, code for proteins |
rRNAs | Ribosomal RNAs, form the basic structure of the ribosome and catalyze protein synthesis |
tRNAs | Transfer RNAs, adaptors between mRNA and amino acids |
snRNAs | Small Nuclear RNAs, function in different nuclear process like mRNA splicing |
snoRNAs | Small Nucleolar RNAs, used to process and modify rRNAs |
Stages of Transcription
I. Initiation
Initiation of RNA transcription from one of the two strands of the DNA molecule proceeds as follows:
Prokaryote | Eukaryote |
|---|---|
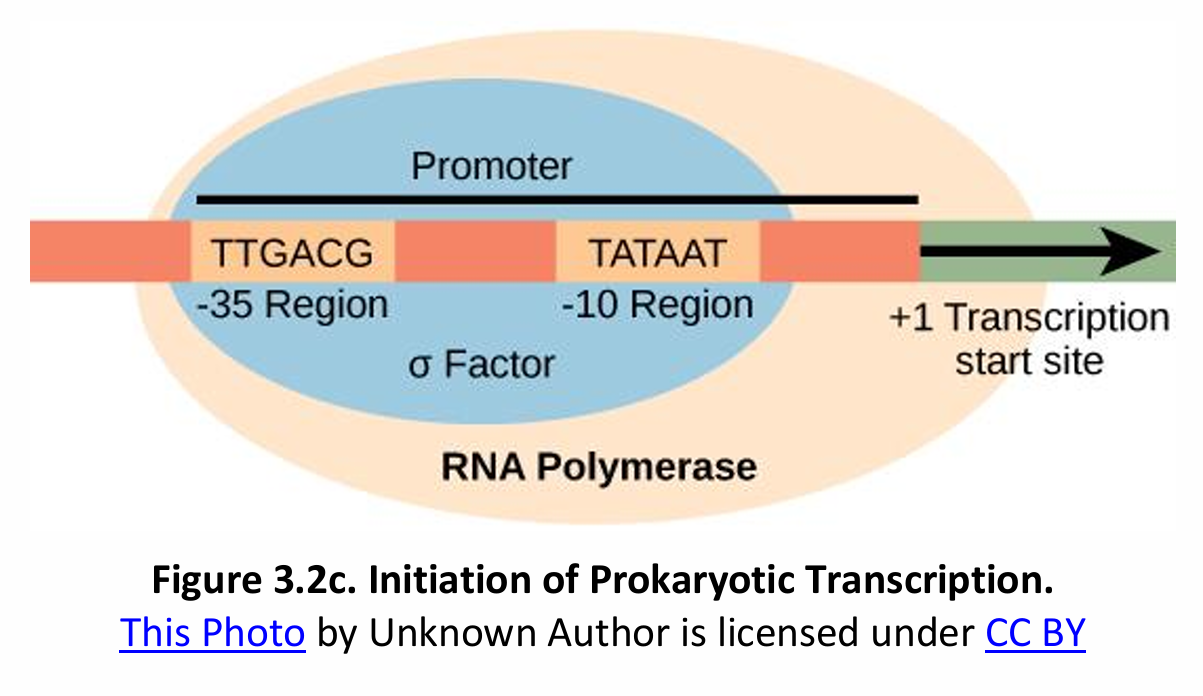 | 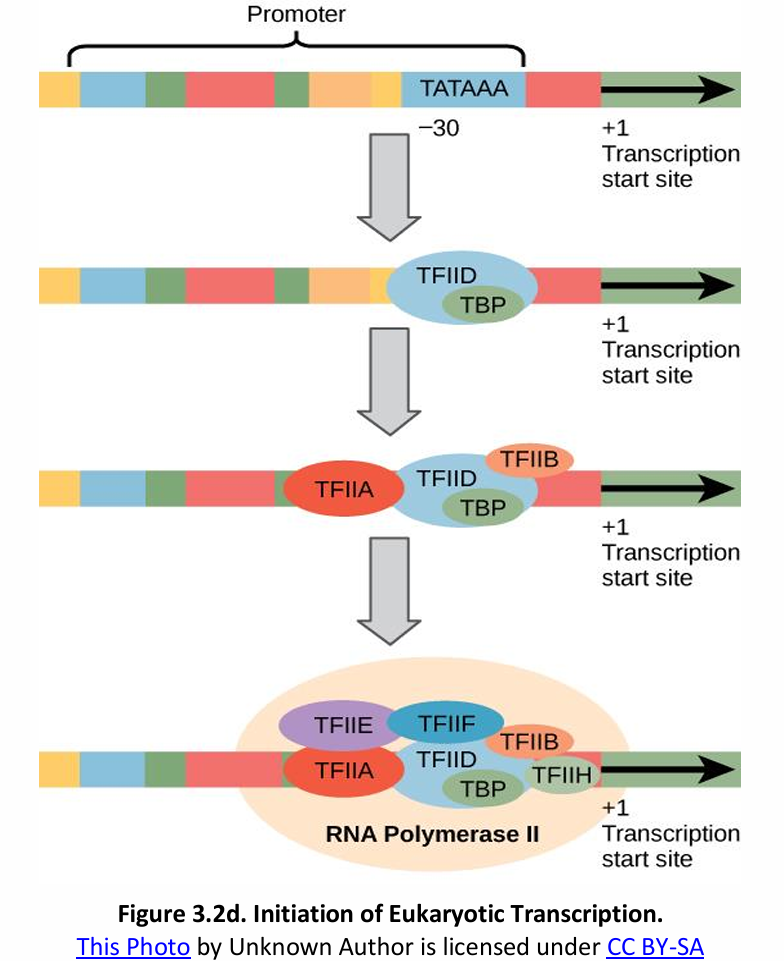 |
RNA polymerase (RNAP) can recognize and bind itself to the promoter (ρ factor) | Transcription factors (TFs) mediate RNA polymerase binding and transcription initiation |
No initiation complex | TFs + RNAP II = Transcription Initiation Complex (TIC) |
Consensus sequence of the promoter (AT-rich regions): Pribnow box – TATAAT | Consensus sequence of the promoter (AT-rich regions): TATA box – TATAAA |
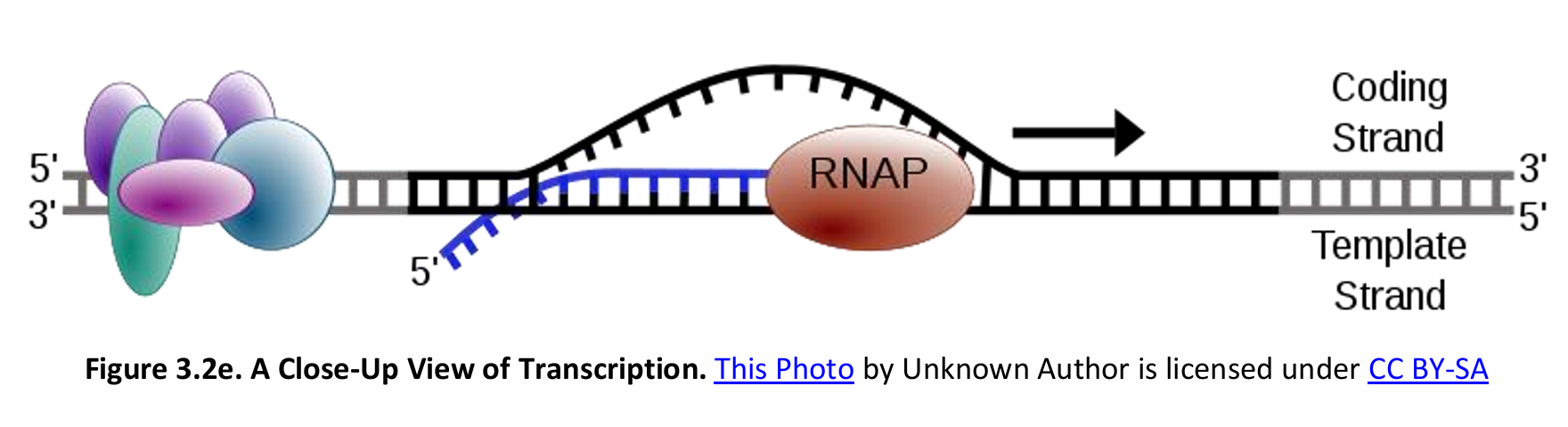
II. Elongation
A single gene can be transcribed simultaneously (i.e. more than one mRNA molecule are being transcribed at the same time).
DNA is double stranded: sense strand (coding strand or non-template strand; and antisense strand (non-coding strand or template strand).
RNA nucleotides complementary to the template strand of the DNA are added to the 3’ end of the growing strand. DNA unwinds 10-20 bases at a time for pairing with RNA nucleotides, then the double helix reforms.
Before the transcription of a gene is completed, a protein called Rho (ρ) factor binds to the termination site and brings about the termination of transcription.
Prokaryote | Eukaryote |
|---|---|
Rho-Dependent Termination 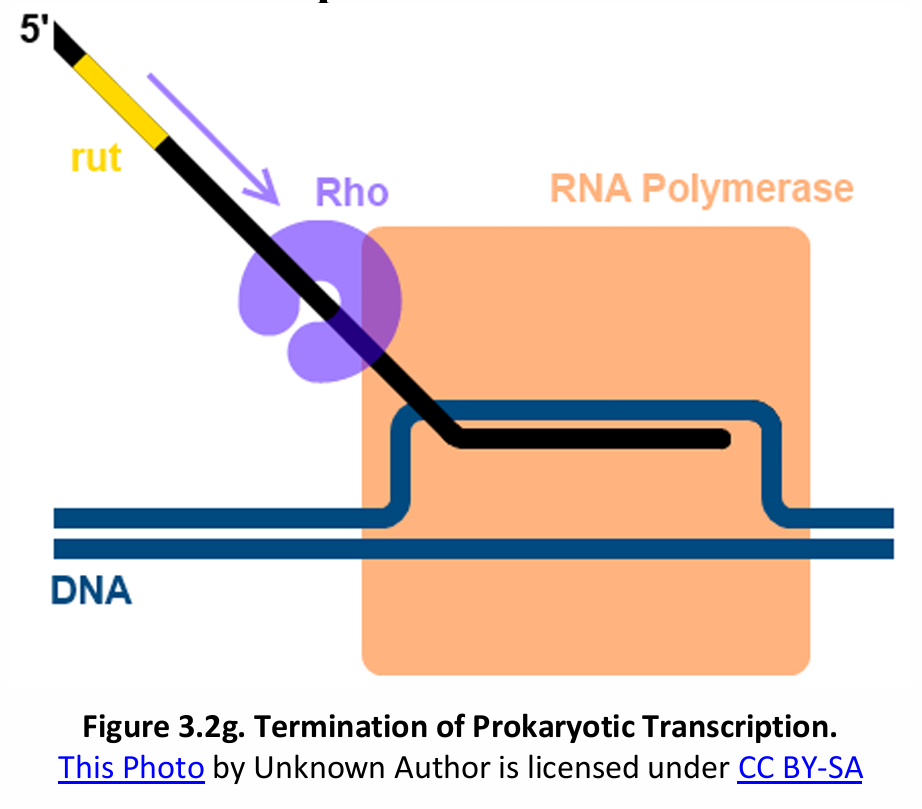 |  |
Rho (ρ) factor recognizes the transcribed RNA rut (C-rich), upstream of the real terminator sequence | RNAP II continues along the DNA strand until it reaches the terminator sequence |
Terminator (RNA sequence) serves as termination signal | Polyadenylation signal sequence transcribes the polyadenylation signal (AAUAAA) |
The mRNA can be translated without further modification | After 10-35 nucleotides downstream from AAUAAA, a pre-mRNA is released |
Rho catches up with RNAP (paused at termination sequence) and allows release | RNAP II continues to transcribe and is eventually dissociated |
Post-Transcriptional Processing in Eukaryotes
The RNAP II does not only transcribes DNA into RNA, but also bears pre-mRNA-processing proteins on its tails, which are then transferred to the nascent RNA at the appropriate time.
The likely order of events in producing a mature mRNA from a pre-mRNA is shown below.
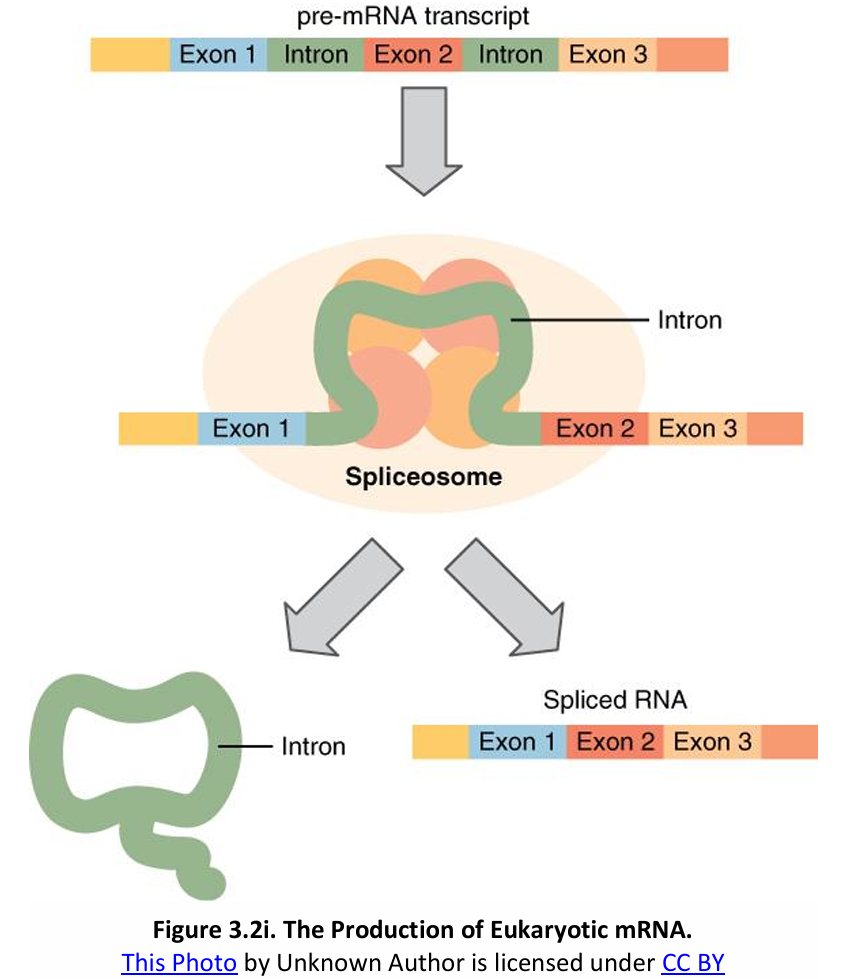
Some regions of the mRNA will not be translated.
Some non-coding sequences called introns are interspersed between coding (expressed) sequences called exons.
The intron loops out as snRNPs (small nuclear ribonucleoprotein particles; complexes of snRNAs and proteins) which bind to signals at the end of each intron.
snRNPs join with other proteins to form a spliceosome. The intron is excised and the exons are then spliced together.
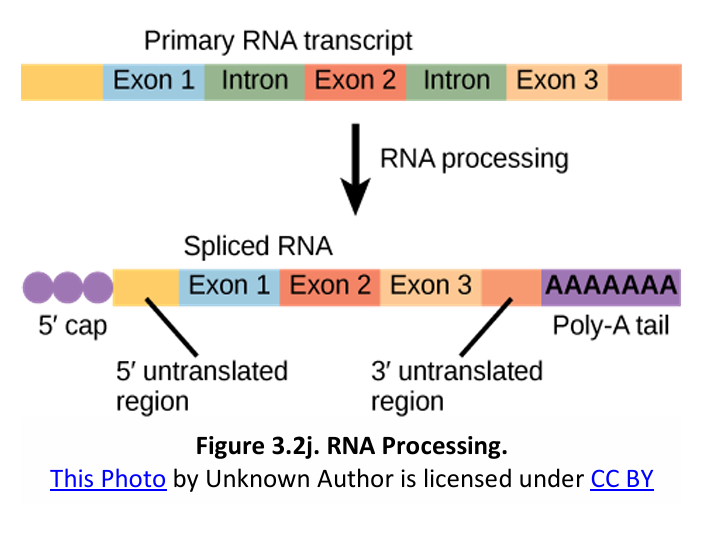
The alteration of mRNA ends includes the following processes:
(1) 5’ capping or the addition of a modified G cap;
(2) 3’ polyadenylation or the addition of a poly-A tail;
(3) UTRs (untranslated regions) for ribosome binding signals.
These modifications share several functions:
(1) to facilitate the export of mRNA from the nucleus to the cytoplasm
(2) to prevent the degradation of mRNA by hydrolytic enzymes; and
(3) serve as signals to the rRNA for attachment to the 5’ end.
The structure of a typical eukaryotic protein coding mRNA including the UTRs is shown on the left in Figure 3.2j.