L8_2025_-_DNA
-Page 3: Lecture Overview
Semester 2 Dates: January 23, 27, 28, 2025
Lecture Topics:
Lecture 8: DNA as a genetic information store
Lecture 9: RNA and gene expression
Lecture 10: Protein synthesis
Page 4: Learning Objectives for Lecture 8 (DNA)
Understand evidence that DNA carries genetic information.
Describe the anti-parallel structure of DNA and the importance of base-pairing.
Explain the Meselson-Stahl experiment demonstrating semi-conservative DNA replication.
Detail how proteins like DNA polymerase facilitate DNA replication.
Discuss the accuracy of DNA replication and the implications of mutations.
Page 5: Historical Context of Protein Phosphorylation
Key Figures:
Philip Cohen (2002) on protein phosphorylation.
Crebs and Fischer (1992) on protein kinase A (PKA).
Carl and Gerty Cori (1947) on glycogen metabolism.
Earl Sutherland (1971) on cAMP signaling.
Gilman and Rodbel (1994) on G proteins.
Scientific inspiration by Lefkowitz/Kobilka (2012) on G protein-coupled receptors.
Page 6: Key Experiments Contributing to DNA Understanding
Mendel (1860s): Genetic traits in peas established the concept of inheritance.
Miescher (1860s): Isolated nuclein from white blood cells – leading to the discovery of DNA.
Morgan (1900s): Confirmed that genes are located on chromosomes via Drosophila studies.
Page 7: Transformation Concept
Definition: Transformation is a change in genotype and phenotype due to assimilation of external DNA by a cell.
At the time of Griffith's work, the inheritance factor's identity remained unknown, mainly attributed to proteins.
Page 8: DNA carries genetic information
Oswald T. Avery: Conducted experiments that systematically destroyed lipids, carbohydrate, proteins and ribonucleic acid of virulent bacteria, but transformation still occurred.
used deoxyribonuclease to destroy DNA and transformation was blocked.
Summary
genetic info is carried in DNA, eve with lipids, carbs, proteins and ribonucleic acid is destroyed, transformation still occurred
Page 9: Confirmatory Evidence for DNA
Chemical evidence confirmed DNA's role as genetic material was supported by viral studies indicating only DNA entered bacterial cells, not proteins (Hershey-Chase experiment).
Page 10: Composition of DNA
Erwin Chargaff (1947): Determined that DNA composition varied between species. (molecular diversity)
within a species the DNA from all cells had the same composition
G=C, A=T
Page 11: Structure of Nucleotides
DNA Composition: A polymer of nucleotides consisting of a sugar (deoxyribose), a phosphate, and nitrogenous bases (A, T, G, C).
A and G are purines (2 fused rings)
C and T are pyrimidines (1 ring)
the sugar is 5-carbon sugar 2 ‘ - deoxyribose
Page 12: Discovery of DNA Structure
Rosalind Franklin (1952): X-ray diffraction revealed DNA's helical nature and base spacing.
this allowed Watson to deduce width of helix and spacing of nitrogenous bases along it
Page 13: Watson and Crick’s Model
Base Pairing: Watson and Crick established specific base pairing rules (A with T, G with C) due to hydrogen bonds, leading to the proposal of double helix structure.
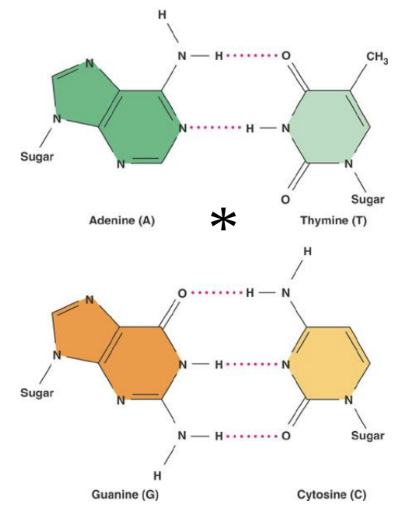
Page 14: Features of the Watson and Crick Model
Key Structural Features:
Sugar-phosphate backbone.
Anti-parallel strands (5’-3’ and 3’-5’).
10 base pairs per helical turn.
Bases are non polar hydrophobic on the inside and the polar hydrophilic phosphate is on the outside
space filling models show a major and minor groove in the double helix
Page 15: DNA Replication Mechanism
Overview: The Watson-Crick model implies strands serve as templates for new strand synthesis via base-pairing.
The two strands need to unwind and separate to expose the bases to allow the synthesis of new DNA to occur.
Page 16: Meselson and Stahl
Concept: DNA replication is semi-conservative; as opposed to ‘conservative’ or ‘dispersive‘ models
verified by Meselson and Stahl through isotopic labelling studies.
Conservative model
the two parental strands re-associate after acting as templates for new strands, restoring the parental double helix
original double helix is intact and unchanged
Semiconservative Model
the two strands of the parental molecule separate, each functions as a template for synthesis of a new complementary strand.
consists of one parental and one newly synthesised strand
Dispersive Model
each strand of both daughter molecules contains a mixture of old and newly synthesised DNA
parental DNA is mot kept intact but dispersed into both strands
Page 17: Experimental Methodology in Replication Studies
Key Features of the Experiment:
Used E. coli as a model organism which could grow on N and C
nitrogen isotopes used for differentiating old and new DNA. N15 N14
Density gradient centrifugation to separate the two types N15 is 1% denser
the results were only consistent with the semi-conservative model
Page 18: Initiation of Replication
DNA replication begins at origins specific to prokaryotes and eukaryotes
proteins recognise the DNA sequence and bind to it, opening up the double helix, in the replication bubble, there are Y-shaped replication forks where the new strands are being elongated
replication starts at multiple sites where the parental strands separate to form replication bubbles
the bubbles expand laterally as DNA replication proceeds in both directions
the replication bubbles fuse and synthesis of the daughter strands are complete
Page 19: Role of DNA Polymerases
Function: DNA polymerases elongate strands by incorporating nucleotides from triphosphate sources (ATP, GTP etc…)
Mechanics of Elongation: A two phosphate unit (pyrophosphate) is split out as the chain is extended by each nucleotide.
nucleotides can ONLY be added to the free 3’ end and never the 5’ end
Page 20: Directionality of DNA Replication
Due to anti-parallel strands and enzymes only being able to extend chains in only one direction (5’→3’), the two trands cannot both be replicated continuously
resulting in a leading strand (continuous replication) and a lagging strand (discontinued replication)
Page 21: Lagging Strand Formation
Okazaki Fragments
typically 100-200 nucleotides in length and are later joined by DNA ligase
DNA polymerase can only extend a pre existing strand, the cell makes a short primer of RNA about 10 nucleotides long, this is then replaced by DNA.
DNA polymerase adds nucleotides to the 3' end of this primer, elongating the new DNA strand. This results in Okazaki fragments.
Page 22: Key Proteins in DNA Replication
Functions of Key Proteins:
Helicase: Unwinds DNA at replication forks.
Single-strand binding proteins: Stabilise unwound DNA.
Topoisomerase: corrects over-winding, swivelling and rejoins
Primase: Synthesises RNA primers at 5’ end of leading strand, synthesises 5’ end of okazaki fragment on lagging strand
DNA polymerases: Elongate leading and lagging strands.
DNA ligase: Joins 3’ end of DNA fragments on leading strand and joins okazaki fragments on lagging strand
Page 23: Lecture 8 Summary
Review discoveries by Mendel, Miescher, Morgan, Griffith, Avery, Hershey-Chase, Chargaff, Meselson-Stahl, Watson-Crick.
Understand DNA polymerase functions on leading and lagging strands.
Memorise names and roles of proteins in replication.
 Knowt
Knowt