AP BIO Unit 5 Review - Heredity
Meiosis
Meiosis occurs twice, leads to 4 haploid cells (daughters with half the amount of chromosomes), and genetically unique cells
In meiosis 1, ^^homologous chromosomes^^ separate
- A pair of chromosomes of the same length, centromere position, and staining pattern that possess genes for the same characters at corresponding loci. One homologous chromosome is inherited from the organism’s father, the other from the mother. Also called homologs, or a homologous pair
In meiosis 2, ^^sister chromatids^^ separate
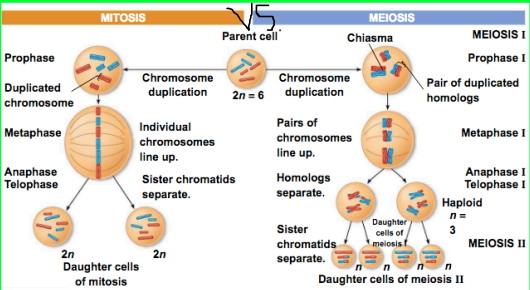
]]Unique cells are made due to]]
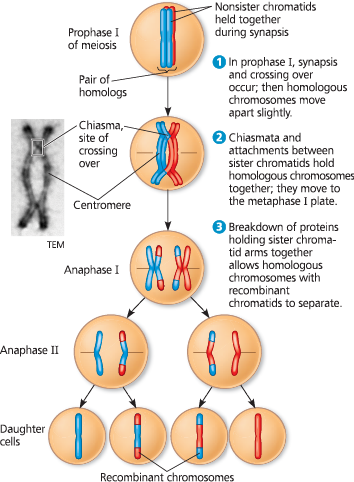
%%crossing over%%
- creates recombinant chromosomes (DNA from two chromosomes into one chromosome)
- synapsis - The pairing and physical connection of duplicated homologous chromosomes during prophase I of meiosis
- Chiasma - The X-shaped, microscopically visible region where crossing over has occurred earlier in prophase I between homologous nonsister chromatids. Chiasmata become visible after synapsis ends, with the two homologs remaining associated due to sister chromatid cohesion
- create new combinations of maternal and paternal alleles
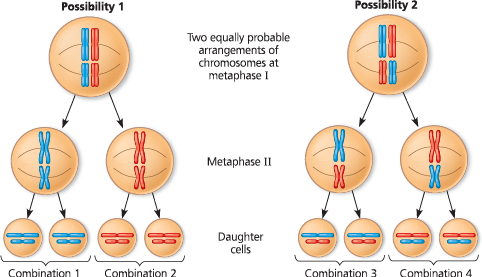
%%independent assortment%%
- along the metaphase plate, the paternal and maternal chromosomes may orientate in either direction
%%random fertilization%%
- The fusion of a male gamete with a female gamete during fertilization will produce a zygote with any of about 70 trillion (223 × 223) diploid combination
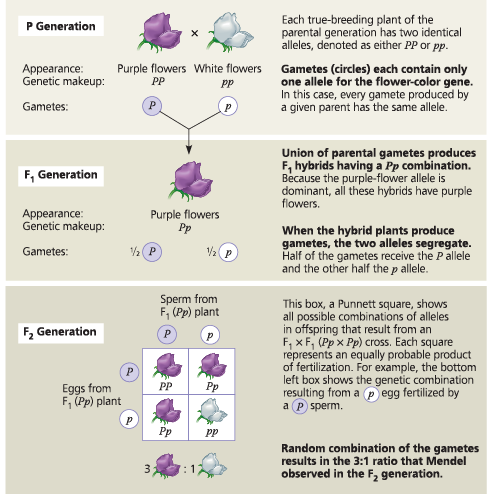
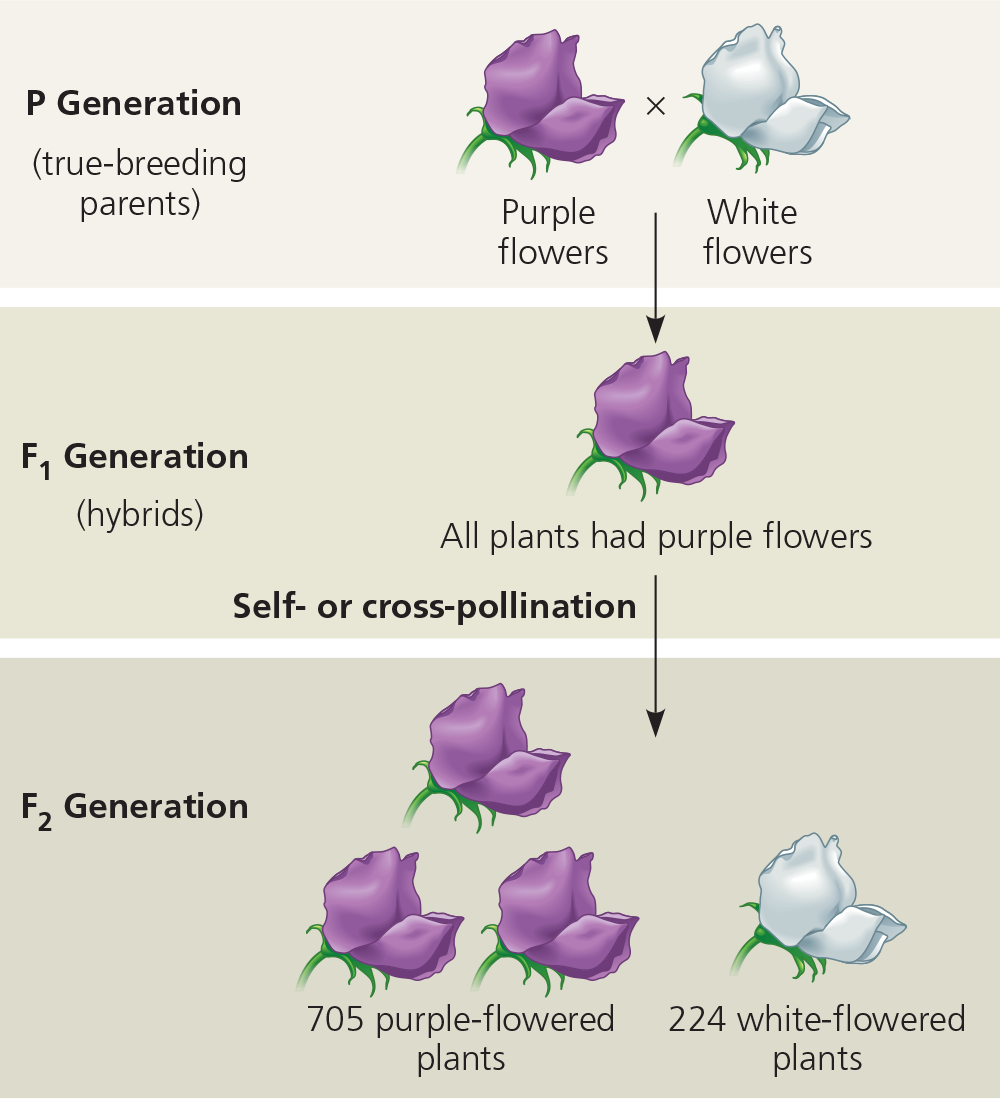
Mendelian Genetics
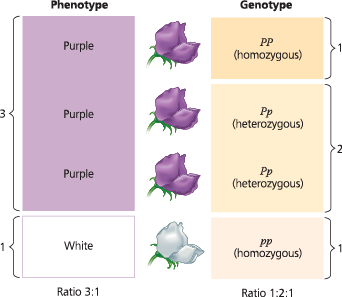
True breeding parents (P)
- 1:1 phenotype and genotype ration
Self fertilizaing hybrid offspring (F1)
- all express dominant trait
Generation after F1 (F2)
3:1 phenotypic ratio
- shows that recessive traits may be based and skip generations
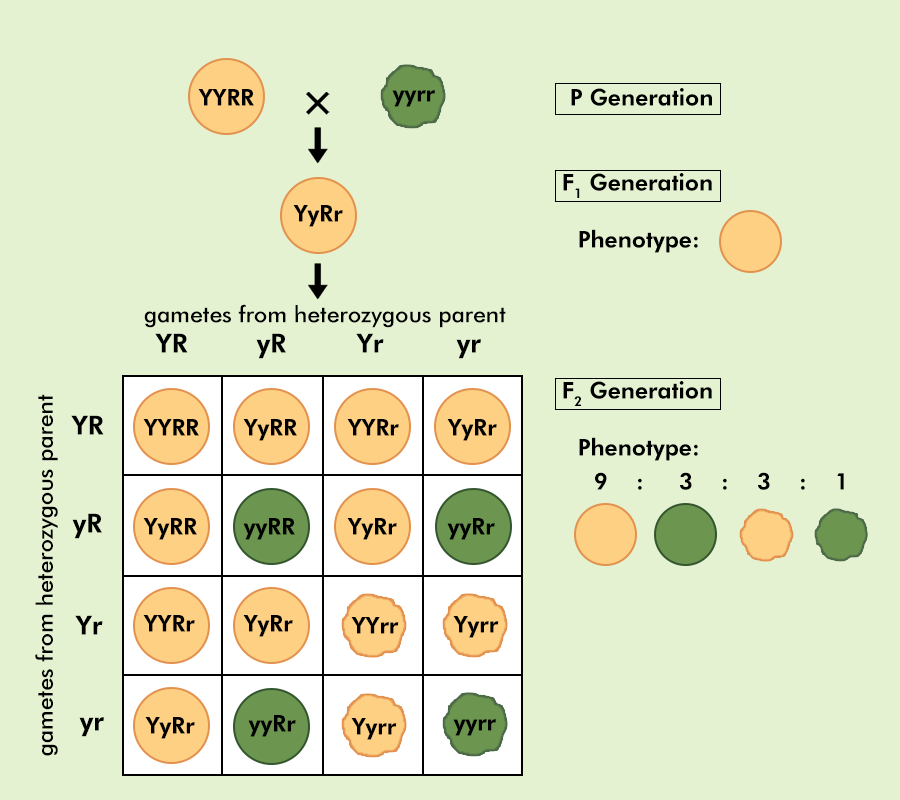
%%Dihybrid Cross%% - 9:3:3:1 ratio
%%Test cross%%: breeding unknown with homozygous recessive genotype
- the phenotype of offspring shows the unknown genotype
Linked Genes
Linked genes
- Genes located close enough together on a chromosome that they tend to be inherited together
Recombinant
- An offspring whose phenotype differs from that of the true-breeding P-generation parents; also refers to the phenotype itself
Examples:
}}In a dihybrid cross with two parents that are heterozygous (AaBb x AaBb) for both traits, these are the offspring}}

@@Does this show linked traits?@@
- The data does not support that the genes are linked. The ratio of 562 : 187 : 189 : 62 is very
similar to 9 : 3 : 3 : 1 which is the phenotypic ratio of two heterozygotes. There isn't enough
deviation between expected and experimental
}}In a dihybrid cross with one wild heterozygous fruit fly and one sepia eyed, apterous fruit fly (AaBb x aabb), these are the offspring}}

@@Does this show linked traits?@@
- Yes, the genes are linked. Because it’s a cross with a recessive parent, the expected results should be 1:1:1:1, but experiment shows a ration closer to 2:1:1:2. The experimental data goes against the theoretical data which indicates they are linked. Also, the genes often show up in the combinations of wild/wild and sp/ap which shows they are linked. Those genes also happen to be the parental phenotype, and since they show up a lot with relatively few recombinants, it also indicates the genes are linked
How many map units apart are the genes?
- number of recombinant offspring / total number of offspring
- recombinant offspring: 125+125=250
- total offspring: 250+125+125+250=750
- 250 / 750 = 33.33 map units apart
Other Effects on Phenotype
Environmental Factors:
Height and weight in humans
- nutrition based
Flower color
- soil pH
Animal fur color
- seasonal camouflage
Melanin expression in humans
- exposure to UV
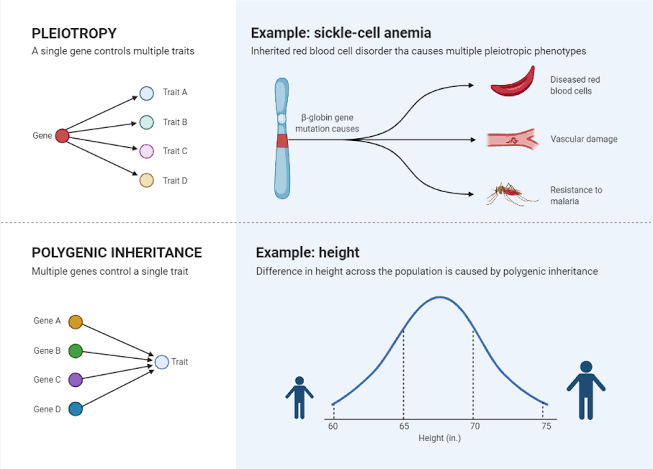
pleiotropy vs polygenic
pleiotropy is when one gene affects multiple characteristics
polygenic inheritance is when one trait is controlled by multiple genes
Looking at Pedigree Trees
[[Pedigrees show if a trait is dominant or recessive[[
- recessive traits may skip a generation
- dominant traits do not skip generations
[[Pedigrees can show if a trait is autosomal or sex linked[[
- X-linked recessive traits, males are much more commonly affected than females
- In autosomal traits, both males and females are equally likely to be affected
{{Example tree 1{{
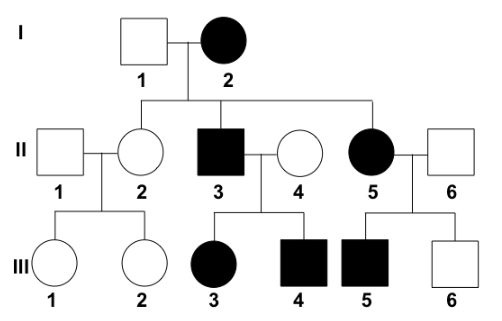
^^What can be determined about the trait?^^
- It is autosomal dominant
^^What is the genotype of I-2?^^
- Ff
- (there are unaffected individuals in II, so I-2 must have one recessive allele)
{{Example tree 2{{
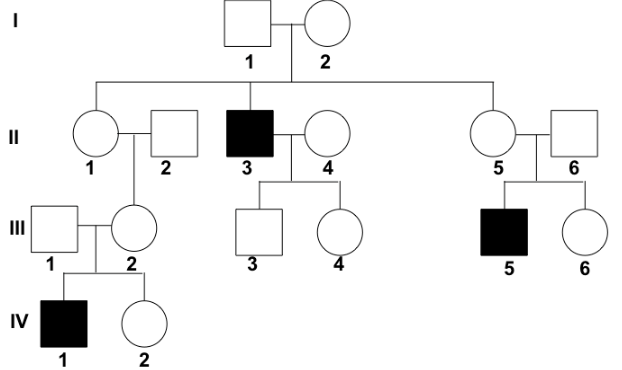
^^What can be determined about the trait?^^
- It is sex-linked recessive
^^What is the genotype of I-2?^^
- XBXb
- She is unaffected, so she must have at least one dominant allele
- her son is affected, and males get their X chromosome from their mothers, so she must have at least one recessive allele