🤣AP BIO Chapter 14 - Gene Expression: From Gene to Protein
Overview
14.1 - Genes specify proteins via transcription and translation
14.2 - Transcription is the DNA-directed synthesis of RNA: a closer look
14.3 - Eukaryotic cells modify RNA after transcription
14.4 - Translation is the RNA-directed synthesis of a polypeptide: a closer look
14.5 - Mutations of one or a few nucleotides can affect protein structure and function
BIG IDEAS: The highly conserved processes of gene transcription and translation (Big Idea 3) shared by all domains (Big Idea 1) result from molecular interactions that include specific enzymes (Big Idea 4) and transcription factors that act as control mechanisms (Big Idea 2)
Given that polypeptides are the link between genotype and phenotype, how could this be used to support the hypothesis that RNA, not DNA, evolved first, sometimes referred to as the RNA World hypothesis? (Big Ideas 1 & 4)
What is responsible for the phenotype of the albino donkey, strikingly different from its pigmented relative?
The albino donkey has a faulty version of a key protein, an enzyme required for pigment synthesis, and this protein is faulty because the gene that codes for it contains incorrect information
Albino Donkey

The DNA inherited by an organism leads to ___ by dictating the synthesis of proteins and of RNA molecules involved in protein synthesis
- specific traits
What is the link between genotype and phenotype?
Proteins
{{Gene Expression:{{
The process by which information encoded in DNA directs the synthesis of proteins or, in some cases, RNAs that are not translated into proteins and instead function as RNAs
What are the two stages of gene expression?
Transcription and translation
@@14.1 - Genes specify proteins via transcription and translation@@
Beadle and Tatum’s studies of mutant strains of Neurospora led to the one gene–one polypeptide hypothesis. During gene expression, the information encoded in genes is used to make specific polypeptide chains (enzymes and other proteins) or RNA molecules
Transcription is the synthesis of RNA complementary to a template strand of DNA. Translation is the synthesis of a polypeptide whose amino acid sequence is specified by the nucleotide sequence in mRNA
Genetic information is encoded as a sequence of nonoverlapping nucleotide triplets, or codons. A codon in messenger RNA (mRNA) either is translated into an amino acid (61 of the 64 codons) or serves as a stop signal (3 codons). Codons must be read in the correct reading frame
What did Archibald Garrod suggest?
Genes dictate phenotypes through enzymes; symptoms of an inherited disease reflect an inability to make a particular enzyme
{{Enzyme:{{
Proteins that catalyze specific chemical reactions in the cell
What did George Beadle and Boris Ephrussi suggest?
Each mutation affecting eye color blocks pigment synthesis at a specific step by preventing production of the enzyme that catalyzes that step
Why did they work with the bread mold Neurospora crassa?
It is a haploid species and only needs one allele of a protein-coding gene disabled; it has modest food requirements
{{Minimal Medium:{{
A culture containing minimal nutrients for the growth of wild-type cells
{{Complete Medium:{{
Contains all nutrients needed for growth, including any that a mutant cell can’t synthesize
The experimental approach of Beadle and Tatum - To obtain nutritional mutants, Beadle and Tatum exposed Neurospora cells to X-rays to induce mutations. They then screened mutants with new nutritional requirements, such as arginine, as shown here
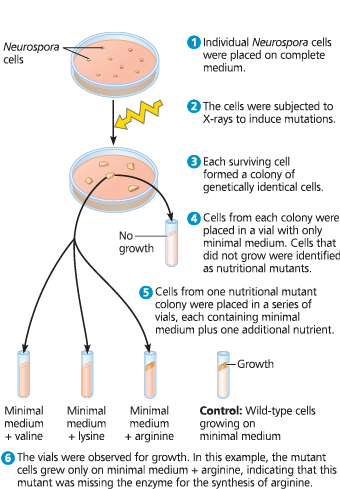
The one gene–one protein hypothesis - Based on results from work in their lab on nutritional mutants, Beadle and Tatum proposed that the function of a specific gene is to dictate production of a specific enzyme that catalyzes a particular reaction. The model shown here for the arginine-synthesizing pathway illustrates their hypothesis
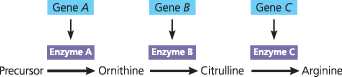
{{One gene–one enzyme hypothesis:{{
The function of a gene is to dictate the production of a specific enzyme
Genes act by regulating definite ___ events
- chemical
Explain to a degree how accurate the term one gene-one protein is
It is accurate because not all proteins are enzymes but are still gene products (Keratin - animal hair, insulin). However, many proteins are constructed from two or more different polypeptide chains, and each polypeptide is specified by its own gene.
Is one gene-one polypeptide an accurate term?
Not entirely. A eukaryotic gene can code for a set of closely related polypeptides via a process called alternative splicing, which you will learn about later in this chapter. Plus, quite a few genes code for RNA molecules that have important functions in cells even though they are never translated into protein
The bridge between DNA and protein synthesis is ___
- nucleic acid RNA
Explain the differences between RNA and DNA
RNA contains ribose instead of deoxyribose as its sugar and has the nitrogenous base uracil rather than thymine
What is the protein’s primary structure?
Each polypeptide of a protein also has monomers arranged in a particular linear order
{{Transcription:{{
The synthesis of RNA using a DNA template
A DNA strand provides a template for ___
- complementary DNA strands and complementary sequence of RNA nucleotides
{{Messenger RNA (mRNA):{{
A type of RNA, synthesized using a DNA template, that attaches to ribosomes in the cytoplasm and specifies the primary structure of a protein. (In eukaryotes, the primary RNA transcript must undergo RNA processing to become mRNA)
{{Translation:{{
The synthesis of a polypeptide using the genetic information encoded in an mRNA molecule. There is a change of “language“ from nucleotides to amino acids
{{Ribosome:{{
A complex of rRNA and protein molecules that functions as a site of protein synthesis in the cytoplasm; consists of a large subunit and a small subunit. In eukaryotic cells, each subunit is assembled in the nucleolus
{{Nucleolus:{{
A specialized structure in the nucleus consisting of chromosomal regions containing ribosomal RNA (rRNA) genes along with ribosomal proteins imported from the cytoplasm; site of rRNA synthesis and ribosomal subunit assembly
The sites of translation are ___
- ribosomes
Bacterial DNA and mRNA from ribosomes and the other protein-synthesizing equipment are not ___
- separated by a nuclear membrane
{{Primary Transcript:{{
An initial RNA transcript form any gene; also called pre-mRNA when transcribed from a protein-coding gene
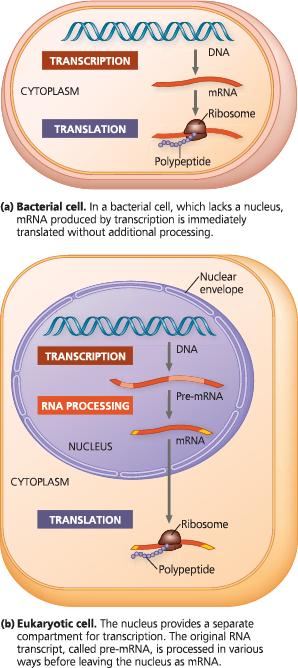
Overview: the roles of transcription and translation in the flow of genetic information - In a cell, inherited information flows from DNA to RNA to protein. The two main stages of information flow are transcription and translation. A miniature version of part (a) or (b) accompanies several figures later in the chapter as an orientation diagram to help you see where a particular figure fits into the overall scheme of gene expression
Genes program protein synthesis via genetic messages in the form of ___
- messenger RNA
Molecular chain of command with the directional flow of genetic information

Some enzymes exist that use ___ as templates for DNA synthesis, but, in general, genetic information flows from DNA to RNA to protein
- RNA molecules
{{Triplet Code:{{
A genetic information system in which a series of three-nucleotide-long words specifies a sequence of amino acids for a polypeptide chain
The triplet code - For each gene, one DNA strand functions as a template for the transcription of RNAs, such as mRNA. The base-pairing rules for DNA synthesis also guide transcription, except that uracil (U) takes the place of thymine (T) in RNA. During translation, the mRNA is read as a sequence of nucleotide triplets, called codons. Each codon specifies an amino acid to be added to the growing polypeptide chain. The mRNA is read in the 5′ → 3′ direction
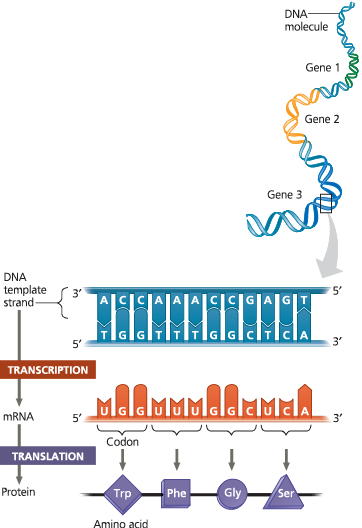
Compare the sequence of the mRNA to that of the nontemplate DNA strand, in both cases reading from 5′ → 3′
The mRNA sequence (5′-UGGUUUGGCUCA-3′) is the same as the nontemplate DNA strand sequence (5′-TGGTTTGGCTCA-3′), except there is a U in the mRNA wherever there is a T in the DNA
{{Template Strand:{{
The DNA strand that provides the pattern, or template, for ordering, by complementary base pairing, the sequence of nucleotides in an RNA transcript
{{Codon:{{
A three-nucleotide sequence of DNA or mRNA that specifies a particular amino acid or termination signal; the basic unit of the genetic code
Why is the nontemplate DNA strand is often called the coding strand?
The term codon is also used for the DNA nucleotide triplets along the nontemplate strand. These codons are complementary to the template strand and thus identical in sequence to the mRNA, except that they have T wherever there is a U in the mRNA
The codon table for mRNA - The three nucleotide bases of an mRNA codon are designated here as the first, second, and third bases, reading in the 5′ → 3′ direction along the mRNA. The codon AUG not only stands for the amino acid methionine (Met) but also functions as a “start” signal for ribosomes to begin translating the mRNA at that point. Three of the 64 codons function as “stop” signals, marking where ribosomes end translation
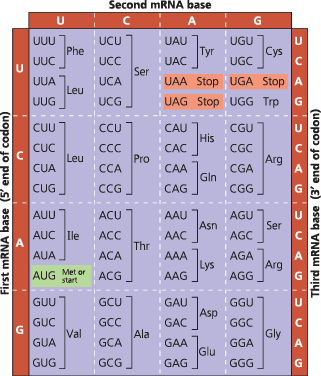
{{Reading Frame:{{
On an mRNA, the triplet grouping of ribonucleotides used by the translation machinery during polypeptide synthesis
Expression of genes from different species - Because diverse forms of life share a common genetic code, one species can be programmed to produce proteins characteristic of a second species by introducing DNA from the second species into the first

@@In a research article about alkaptonuria published in 1902, Garrod suggested that humans inherit two “characters” (alleles) for a particular enzyme and that both parents must contribute a faulty version for the offspring to have the disorder. Today, would this disorder be called dominant or recessive?@@
Recessive
@@What polypeptide product would you expect from a poly-G mRNA that is 30 nucleotides long?@@
A polypeptide made up of 10 Gly (glycine) amino acids
@@The template strand of a gene contains the sequence 3′-TTCAGTCGT-5′. Suppose that the nontemplate sequence could be transcribed instead of the template sequence. Draw the nontemplate sequence in 3′ to 5′ order. Then draw the mRNA sequence and translate it@@

@@Predict how well the protein synthesized from the contemplate strand would function if at all@@
If the nontemplate sequence could have been used as a template for transcribing the mRNA, the protein translated from the mRNA would have a completely different amino acid sequence and would most likely be nonfunctional. (It would also be shorter because of the UGA stop signal shown in the mRNA sequence above—and possibly others earlier in the mRNA sequence.)
@@Describe the process of gene expression, by which a gene affects the phenotype of an organism@@
A gene contains genetic information in the form of a nucleotide sequence. The gene is first transcribed into an RNA molecule, and a messenger RNA molecule is ultimately translated into a polypeptide. The polypeptide makes up part or all of a protein, which performs a function in the cell and contributes to the phenotype of the organism
@@14.2 - Transcription is the DNA-directed synthesis of RNA@@
RNA synthesis is catalyzed by RNA polymerase, which links together RNA nucleotides complementary to a DNA template strand. This process follows the same base-pairing rules as DNA replication, except that in RNA, uracil substitutes for thymine
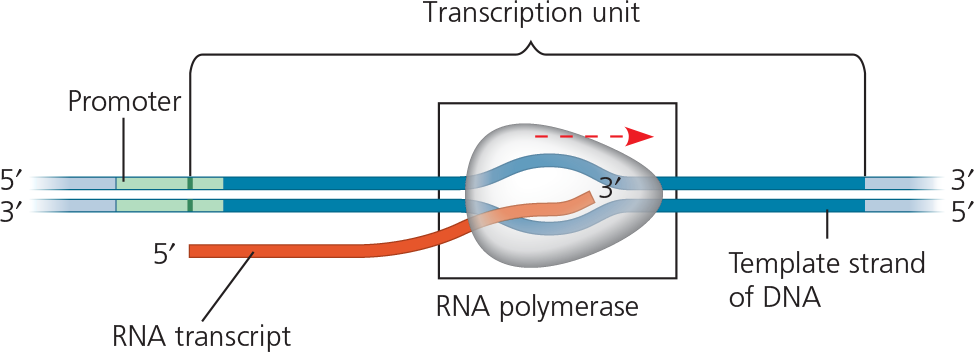
The three stages of transcription are initiation, elongation, and termination. A promoter, often including a TATA box in eukaryotes, establishes where RNA synthesis is initiated. Transcription factors help eukaryotic RNA polymerase recognize promoter sequences, forming a transcription initiation complex. Termination differs in bacteria and eukaryotes
{{RNA Polymerase:{{
An enzyme that links ribonucleotides into a growing RNA chain during transcription, based on complementary binding to nucleotides on a DNA template strand
Compare DNA and RNA polymerases
They both function in replication and can assemble a polynucleotide only in its 5’ → 3’ direction. However, RNA polymerases are able to start a chain from scratch; they don’t need a primer
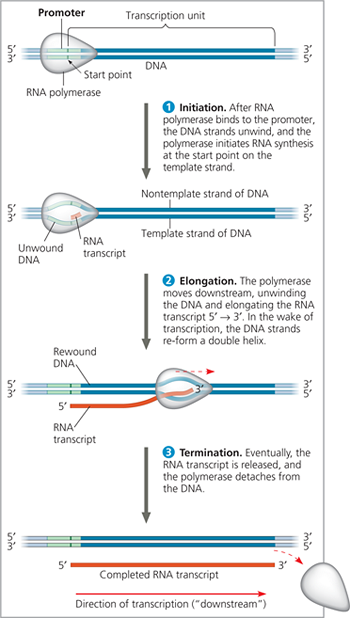
The stages of transcription: initiation, elongation, and termination - This general depiction of transcription applies to both bacteria and eukaryotes, but the details of termination differ, as described in the text. Also, in a bacterium, the RNA transcript is immediately usable as mRNA; in a eukaryote, the RNA transcript must first undergo processing
Compare the use of a template strand during transcription and replication
The processes are similar in that polymerases form polynucleotides complementary to an antiparallel DNA template strand. In replication, however, both strands act as templates, whereas in transcription, only one DNA strand acts as a template
{{Promoter:{{
A specific nucleotide sequence in the DNA of a gene that binds RNA polymerase, positioning it to start transcribing RNA at the appropriate place
{{Terminator:{{
In bacteria, a sequence of nucleotides in DNA that makes the end of a gene and signals RNA polymerase to release the newly made RNA molecule and detach from the DNA
The direction of transcription is ___ and the other direction is _
- downstream; upstream
{{Transcription Unit:{{
A region of DNA that is transcribed into an RNA molecule
Where are promoters, terminators, and transcription units in relation to each other?
The promoter sequence in DNA is upstream from the terminator, and the transcription unit is downstream from the promoter
{{Start Point:{{
In transcription, the nucleotide position on the promoter where RNA polymerase begins synthesis of RNA
The ___ typically extends several dozen or more nucleotide pairs upstream from the start point
- start point
RNA polymerase binds in a precise location and orientation on the promoter, thereby determining ___
- where transcription starts and which of the two strands of the DNA helix is used as the template
{{Transcription Factor:{{
A regulatory protein that binds to DNA and affects the transcription of specific genes
Compare RNA polymerase binding in prokaryotes and eukaryotes
In bacteria, part of the RNA polymerase itself specifically recognizes and binds to the promoter. In eukaryotes, a collection of proteins called transcription factors mediate the binding of RNA polymerase and the initiation of transcription. Only after transcription factors are attached to the promoter does RNA polymerase II bind to it
{{Transcription Initiation Complex:{{
The completed assembly of transcription factors and RNA polymerase bound to a promoter
The initiation of transcription at a eukaryotic promoter - In eukaryotic cells, proteins called transcription factors mediate the initiation of transcription by RNA polymerase II
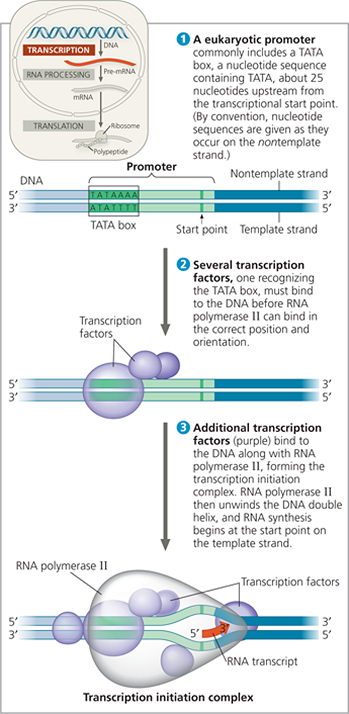
Explain how the interaction of RNA polymerase with the promoter would differ if the figure showed transcription initiation for bacteria
The RNA polymerase would bind directly to the promoter, rather than being dependent on the previous binding of other factors
Transcription elongation - RNA polymerase moves along the DNA template strand, joining complementary RNA nucleotides to the 3′ end of the growing RNA transcript. Behind the polymerase, the new RNA peels away from the template strand, which re-forms a double helix with the nontemplate strand
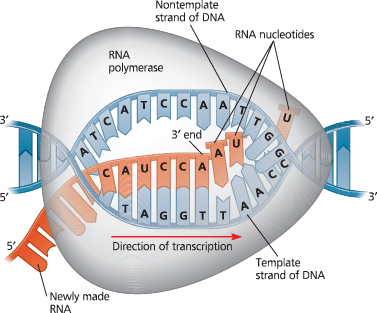
Describe transcription termination in bacteria
Transcription proceeds through a terminator sequence in the DNA. The transcribed terminator (an RNA sequence) functions as the termination signal, causing the polymerase to detach from the DNA and release the transcript, which requires no further modification before translation
Describe transcription termination in eukaryotes
RNA polymerase II transcribes a sequence on the DNA called the polyadenylation signal sequence, which specifies a polyadenylation signal (AAUAAA) in the pre-mRNA. Then, at a point about 10–35 nucleotides downstream from the AAUAAA signal, these proteins cut the RNA transcript free from the polymerase, releasing the pre-mRNA. The pre-mRNA then undergoes processing, the topic of the next section
What makes a “signal“?
When this stretch of six RNA nucleotides appears it is immediately bound by certain proteins in the nucleus
@@What is a promoter? Is it located at the upstream or downstream end of a transcription unit?@@
A promoter is the region of DNA to which RNA polymerase binds to begin transcription. It is at the upstream end of the gene (transcription unit)
@@What enables RNA polymerase to start transcribing a gene at the right place on the DNA in a bacterial cell? In a eukaryotic cell?@@
In a bacterial cell, part of the RNA polymerase recognizes the gene’s promoter and binds to it. In a eukaryotic cell, transcription factors mediate the binding of RNA polymerase to the promoter. In both cases, sequences in the promoter bind precisely to the RNA polymerase, so the enzyme is in the right location and orientation
@@Suppose X-rays caused a sequence change in the TATA box of a particular gene’s promoter. How would that affect the transcription of the gene?@@
The transcription factor that recognizes the TATA sequence would be unable to bind, so RNA polymerase could not bind and transcription of that gene probably would not occur
@@What are the similarities and differences in the initiation of gene transcription in bacteria and eukaryotes?@@
Both bacterial and eukaryotic genes have promoters, regions where RNA polymerase ultimately binds and begins transcription. In bacteria, RNA polymerase binds directly to the promoter; in eukaryotes, transcription factors bind first to the promoter, and then RNA polymerase binds to the transcription factors and promoter together
@@14.3 - Eukaryotic cells modify RNA after transcription@@
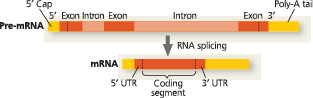
Eukaryotic pre-mRNAs undergo RNA processing, which includes RNA splicing, the addition of a modified nucleotide 5′ cap to the 5′ end, and the addition of a poly-A tail to the 3′ end. The processed mRNA includes an untranslated region (5′ UTR or 3′ UTR) at each end of the coding segment
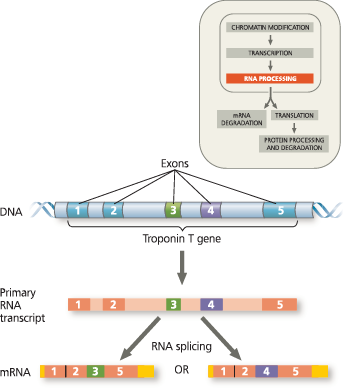
Most eukaryotic genes are split into segments: They have introns interspersed among the exons (regions included in the mRNA). In RNA splicing, introns are removed and exons joined. RNA splicing is typically carried out by spliceosomes, but in some cases, RNA alone catalyzes its own splicing. The catalytic ability of some RNA molecules, called ribozymes, derives from the properties of RNA. The presence of introns allows for alternative RNA splicing
{{RNA Processing:{{
Modification of RNA primary transcripts, including splicing out of introns, joining together of exons, and alteration of the 5’ and 3’ ends
The ___ end is synthesized first
- 5’
{{5’ Cap:{{
A modified form of guanine nucleotide added onto the end of a pre-mRNA molecule
{{Poly-A Tail:{{
A sequence of 50-250 adenine nucleotides added onto the 3’ end of a pre-mRNA molecule
What are the functions of the 5’ cap and poly-A tail?
Facilitate the export of the mature mRNA from the nucleus, help protect the mRNA from degradation by hydrolytic enzymes, and help ribosomes attach to the 5’ end of mRNA
RNA processing: Addition of the 5′ cap and poly-A tail - Enzymes modify the two ends of a eukaryotic pre-mRNA molecule. The modified ends may promote the export of mRNA from the nucleus, and they help protect the mRNA from degradation. When the mRNA reaches the cytoplasm, the modified ends, in conjunction with certain cytoplasmic proteins, facilitate ribosome attachment. The 5′ cap and poly-A tail are not translated into protein, nor are the regions called the 5′ untranslated region (5′ UTR) and 3′ untranslated region (3′ UTR). The pink segments will be described shortly
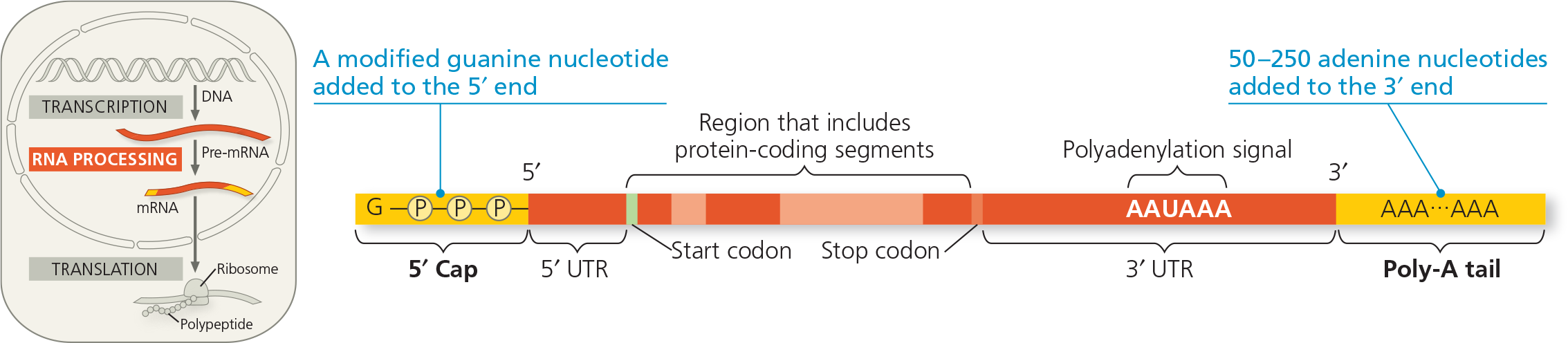
{{RNA Splicing:{{
After synthesis of a eukaryotic primary RNA transcript, the removal of portions and the joining of the remaining portions
{{Intron:{{
A noncoding, intervening sequence within a primary transcript that s removed from the transcript during RNA processing; also refers to the region of DNA from which this sequence was transcribed
{{Exon:{{
A sequence within a primary transcript that remains in the RNA after RNA processing; also refers to the region of DNA from which this sequence was transcribed
Why are the UTRs an exception to the expression rule?
They make up part of the mRNA as an exon and exit the nuclease, but are not translated into a protein
RNA polymerase II transcribes ___ from the DNA, but the mRNA molecule that enters the cytoplasm is _
- both introns and exons; shortened
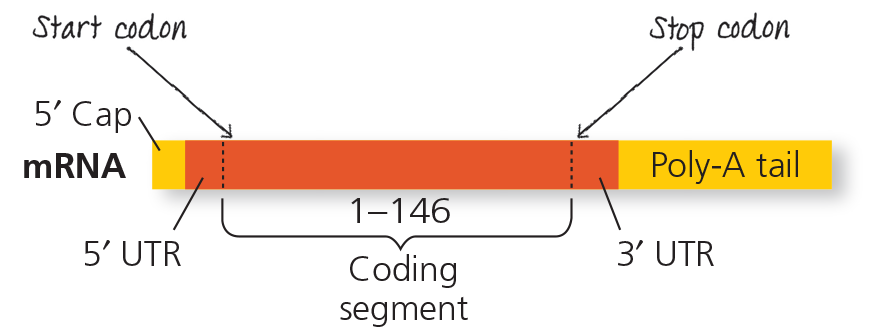
RNA processing: RNA splicing - The RNA molecule shown here codes for β-globin, one of the polypeptides of hemoglobin. The numbers under the RNA refer to codons; β-globin is 146 amino acids long. The β-globin gene and its pre-mRNA transcript have three exons, corresponding to sequences that will leave the nucleus as mRNA. (The 5′ UTR and 3′ UTR are parts of exons because they are included in the mRNA; however, they do not code for protein.) During RNA processing, the introns are cut out and the exons spliced together. In many genes, the introns are much larger than the exons
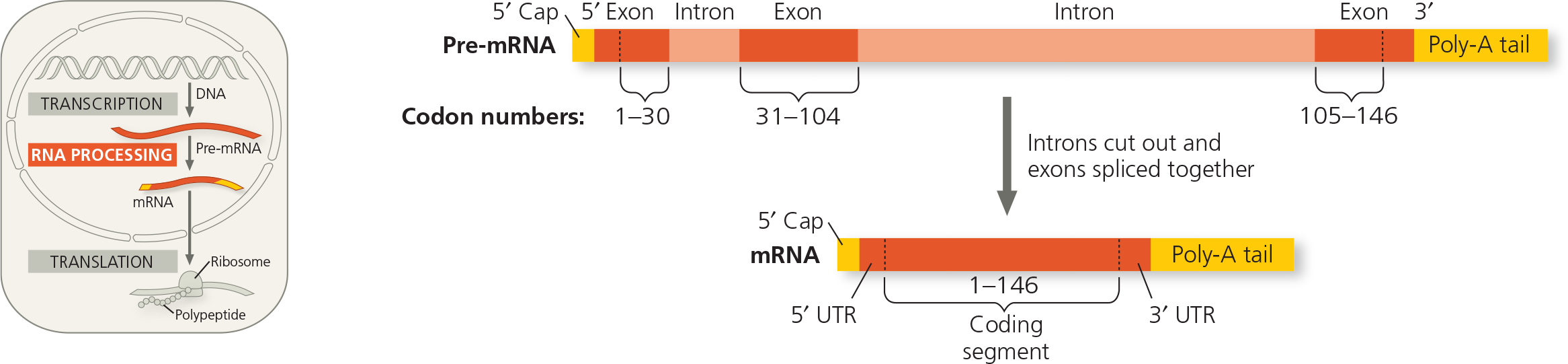
On the mRNA, indicate the sites of the start and stop codons
{{Alternative RNA Splicing:{{
A type of eukaryotic gene regulation at the RNA-processing level in which different mRNA molecules are produced from the same primary transcript, depending on which RNA segments are treated as exons and which as introns
On the mRNA, indicate the sites of the start and stop codons - On the mRNA, indicate the sites of the start and stop codons
{{Spliceosome:{{
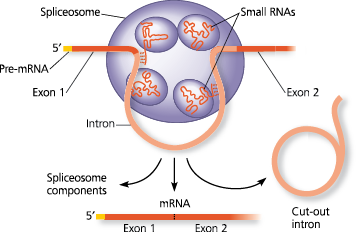
A large complex made up of proteins and RNA molecules that splices RNA by interacting with the ends of an RNA intron, releasing the intron and joining the two adjacent exons
What happens when the spliceosome binds to several short nucleotide sequences along the intron?
The intron is released (and rapidly degraded), and the spliceosome joins together the two exons that flanked the intron
RNAs in the spliceosome not only participate in ___ but also catalyze _
- the assembly of the spliceosome and recognition of the splice site; the splicing process
A spliceosome splicing a pre-mRNA - The diagram shows a portion of a pre-mRNA transcript, with an intron (pink) flanked by two exons (red). Small RNAs within the spliceosome base-pair with nucleotides at specific sites along the intron. Next, small spliceosome RNAs also catalyze cutting of the pre-mRNA and the splicing together of the exons, releasing the intron for rapid degradation
{{Ribozymes:{{
An RNA molecule that functions as an enzyme, such as an intron that catalyzes its own removal during RNA splicing
The intron RNA functions as a ___ and _ its own excision
- ribozyme; catalyzes
Describe how some RNA molecules function as enzymes
RNA is single-stranded, a region of an RNA molecule may base-pair, in an antiparallel arrangement, with a complementary region elsewhere in the same molecule which provides a 3d structure, RNA bases can have functional groups (that partake in catalysis) like amino acids in enzymes, and RNA can hydrogen-bond to other nucleic acids which adds specificity to its catalytic activity
@@Given that there are about 20,000 human genes, how can human cells make 75,000–100,000 different proteins?@@
Due to alternative splicing of exons, each gene can result in multiple different mRNAs and can thus direct the synthesis of multiple different proteins
@@How is RNA splicing similar to how you would watch a TV show recorded earlier using a DVR? In what ways is it different? What would introns correspond to in this analogy?@@
In watching a show recorded with a DVR, you watch segments of the show itself (exons) and fast-forward through the commercials, which are thus like introns. However, unlike introns, commercials remain in the recording, while the introns are cut out of the RNA transcript during RNA processing
@@What would be the effect of treating cells with an agent that removed the cap from mRNAs?@@
Once the mRNA has exited the nucleus, the cap prevents it from being degraded by hydrolytic enzymes and facilitates its attachment to ribosomes. If the cap were removed from all mRNAs, the cell would no longer be able to synthesize any proteins and would probably die
@@What function do the 5′ cap and the poly-A tail serve on a eukaryotic mRNA?@@
Both the 5′ cap and the 3′ poly-A tail help the mRNA exit from the nucleus and then, in the cytoplasm, help ensure mRNA stability and allow it to bind to ribosomes
@@14.4 - Translation is the RNA-directed synthesis of a polypeptide@@
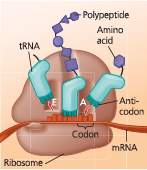
A cell translates an mRNA message into protein using transfer RNAs (tRNAs). After being bound to a specific amino acid by an aminoacyl-tRNA synthetase, a tRNA lines up via its anticodon at the complementary codon on mRNA. A ribosome, made up of ribosomal RNAs (rRNAs) and proteins, facilitates this coupling with binding sites for mRNA and tRNA
Ribosomes coordinate the three stages of translation: initiation, elongation, and termination. The formation of peptide bonds between amino acids is catalyzed by ribosomal RNAs as tRNAs move through the A and P sites and exit through the E site
After translation, modifications to proteins can affect their shape. Free ribosomes in the cytosol initiate synthesis of all proteins, but proteins with a signal peptide are synthesized on the ER
A gene can be transcribed by multiple RNA polymerases simultaneously. Also, a single mRNA molecule can be translated simultaneously by a number of ribosomes, forming a polyribosome. In bacteria, these processes are coupled, but in eukaryotes they are separated in time and space by the nuclear membrane
In the process of translation, the message is ___, and the translator is _
- a series of codons along an mRNA molecule; tRNA
{{Transfer RNA (tRNA):{{
An RNA molecule that functions as a translator between nucleic acid and protein languages by picking up a specific amino acid and carrying it to the ribosome, where the tRNA recognizes the appropriate codon in the mRNA
Translation: the basic concept - As a molecule of mRNA is moved through a ribosome, codons are translated into amino acids, one by one. The translators, or interpreters, are tRNA molecules, each type with a specific nucleotide triplet called an anticodon at one end and a corresponding amino acid at the other end. A tRNA adds its amino acid cargo to a growing polypeptide chain after the anticodon hydrogen-bonds to the complementary codon on the mRNA. The figures that follow show some of the details of translation in a bacterial cell
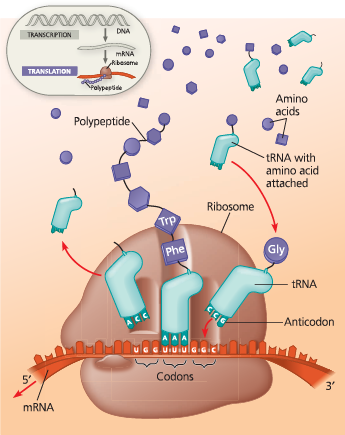
How does tRNA enable translation of an amino acid from a mRNA codon?
tRNA bears a specific amino acid at one end of its three-dimensional structure, while at the other end is a nucleotide triplet that can base-pair with the complementary codon on mRNA
{{Anticodon:{{
A nucleotide triplet at one end of a tRNA molecule that base-pairs with a particular complementary codon on an mRNA molecule
The structure of transfer RNA
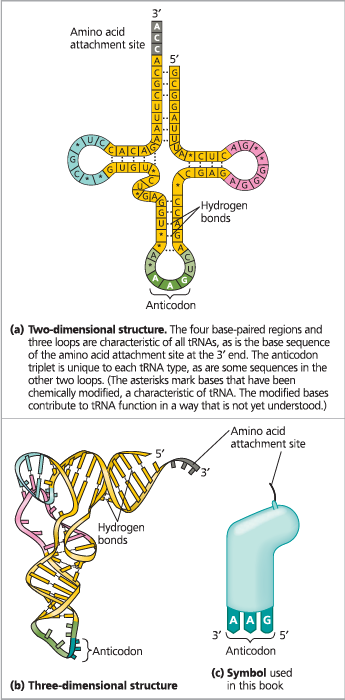
Anticodons are conventionally written ___ to align properly with codons written _
- 3′ → 5′; 5′ → 3′
In a eukaryotic cell, tRNA, like mRNA, is made in the ___ and then travels from _, where translation occurs
- nucleus; the nucleus to the cytoplasm
In both bacterial and eukaryotic cells, each tRNA molecule is used ___
- repeatedly
What are the two instances of molecular recognition for an accurate translation?
First, a tRNA that binds to an mRNA codon specifying a particular amino acid must carry that amino acid, and no other, to the ribosome. The second instance of molecular recognition is the pairing of the tRNA anticodon with the appropriate mRNA codon
{{Aminoacyl-tRNA Synthase:{{
An enzyme that joins each amino acid to the appropriate tRNA
Aminoacyl-tRNA synthetases provide specificity in joining amino acids to their tRNAs - Linkage of a tRNA to its amino acid is an endergonic process that occurs at the expense of ATP (which loses two phosphate groups, becoming AMP)
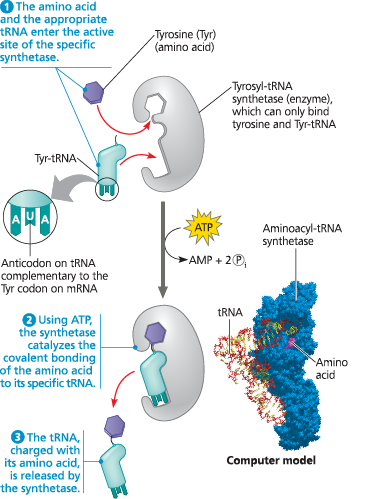
{{Wobble:{{
Flexibility in the base-pairing rules in which the nucleotide at the 5’ end of a tRNA anticodon can form hydrogen bonds with more than one kind of base in the third position (3’ end) of a codon
{{Ribosomal RNA (rRNA):{{
RNA molecules that, together with proteins, makeup ribosomes; the most abundant type of RNA
In eukaryotes, the subunits are made in the ___
- nucleolus
In both bacteria and eukaryotes, a large and a small subunit join to form a functional ribosome only when attached to ___
- an mRNA molecule
About one-third of the mass of a ribosome is made up of proteins; the rest consists of rRNAs, either ___ molecules in bacteria or _ in eukaryotes
- three; four
The anatomy of a functioning ribosome
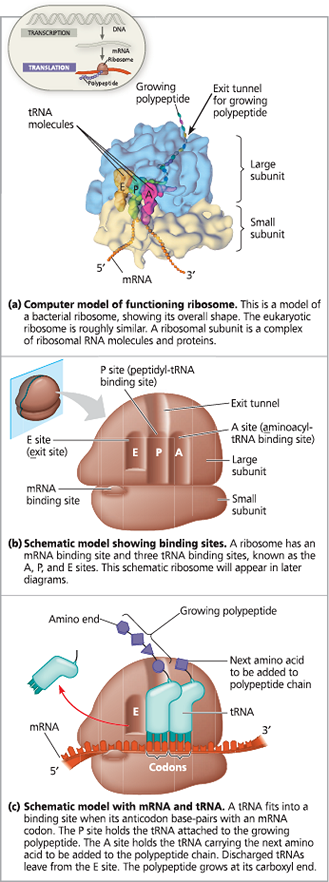
Certain antibiotic drugs can inactivate ___ ribosomes without affecting _ ribosomes to make proteins
- bacterial; eukaryotic
{{P Site:{{
Holds the tRNA carrying the growing polypeptide chain (P stands for peptidyl tRNA)
{{A Site:{{
Holds the tRNA carrying the next amino acid to be added to the polypeptide chain (A stands for aminoacyl tRNA)
{{E Site:{{
The place where discharged tRNAs leave the ribosome (E stands for exit)
The ribosome positions the new amino acid to be added to the ___ end of the polypeptide and catalyzes a _ bond
- carboxyl; polypeptide
{{Guanosine Triphosphate (GTP):{{
Hydrolysis provides energy for initiation, elongation, and formation of peptide bonds in protein synthesis
The initiation of translation
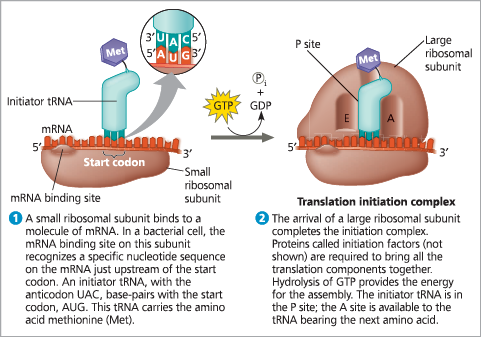
{{Translation Initiation Complex:{{
The union of mRNA, initiator tRNA, and a small ribosomal subunit is followed by the attachment of a large ribosomal subunit
{{Initiation Factors:{{
Proteins required to bring the translation initiation complex together
{{N-Terminus:{{
initial methionine at the amino end
{{C-Terminus:{{
Final amino acid at the carboxyl end
What direction is a polypeptide synthesized in?
From N to C terminus
Amino acids are added one by one to the previous amino acid at the ___ of the growing chain
- C-terminus
{{Elongation Factors:{{
Set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide
What results from the hydrolysis of GTP?
It increases the accuracy and efficiency of codon recognition and provides energy for the translocation step
The elongation cycle of translation - The hydrolysis of GTP plays an important role in the elongation process. Not shown are the proteins called elongation factors
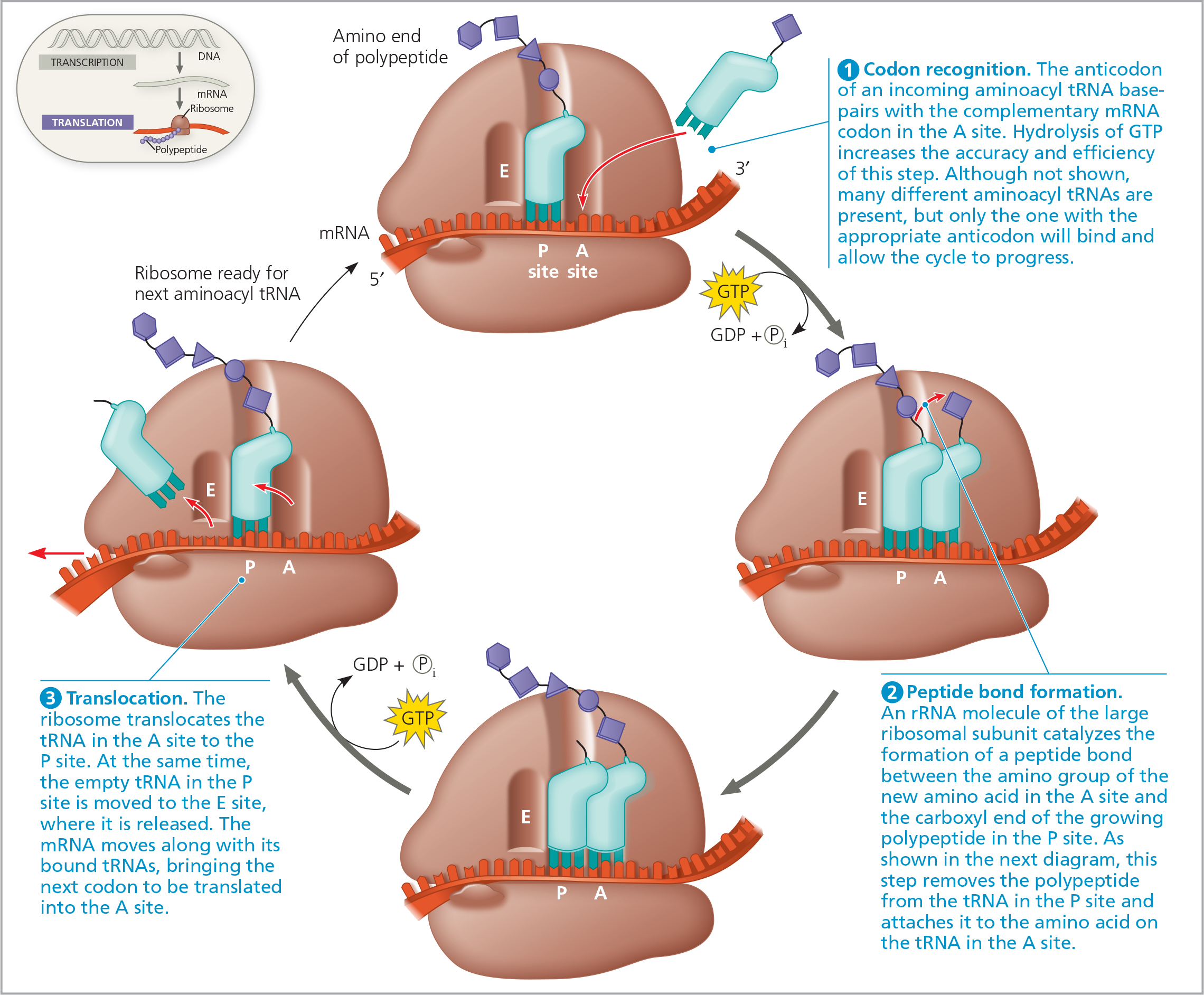
The mRNA is moved through the ribosome in one direction only, ___ end first; this is equivalent to the ribosome moving _ on the mRNA. The important point is that the ribosome and the mRNA move relative to each other, unidirectionally, codon by codon
- 5’; 5’ → 3’
Elongation continues until a stop codon (UAG, UAA, UGA) in the mRNA ___
- reaches the A site
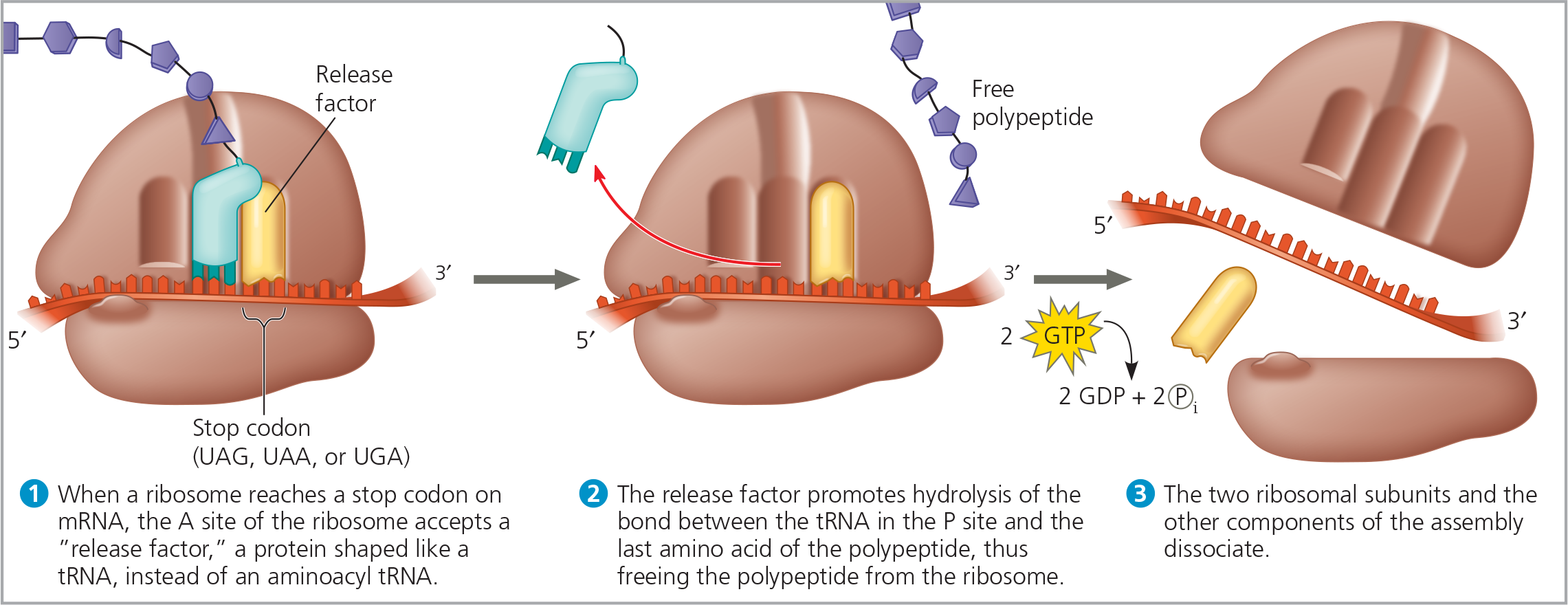
{{Release Factor:{{
A protein shaped like an aminoacyl tRNA that binds directly to the stop codon in the A site
How do release factors help the polypeptide exit the ribosome?
It causes the addition of a water molecule instead of an amino acid to the polypeptide chain. This reaction breaks (hydrolyzes) the bond between the completed polypeptide and the tRNA in the P site, releasing the polypeptide through the exit tunnel of the ribosome’s large subunit
The termination of translation - Like elongation, termination requires GTP hydrolysis as well as additional protein factors, which are not shown here
Describe a polypeptide’s primary structure and 3D shape
Genes determine the amino acid sequence/primary sequence which fold and coil with each other
{{Post-Translational Modifications:{{
Additional steps that alter a protein to do its job
Polypeptide synthesis always begins in the cytosol as a free ribosome starts to translate an mRNA molecule. There the process continues to completion—unless ___
- the growing polypeptide itself cues the ribosome to attach to the ER
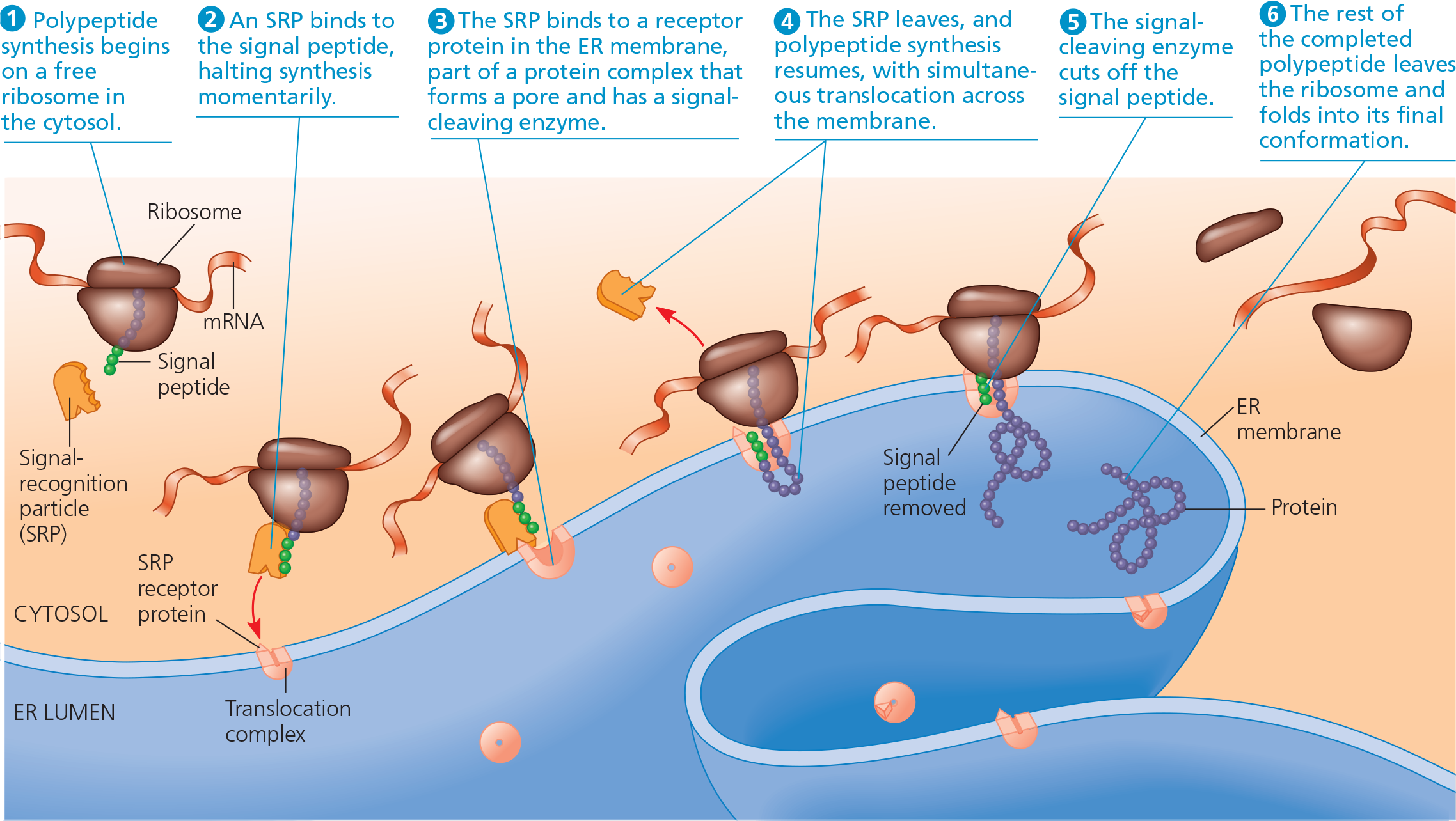
{{Signal Peptide:{{
A sequence of about 20 amino acids at or near the leading (amino) end of a polypeptide that targets it to the endoplasmic reticulum or other organelles in a eukaryotic cell
{{Signal-Recognition Particle (SRP):{{
A protein-RNA complex that recognizes a signal peptide as it emerges from a ribosome and helps direct the ribosome to the endoplasmic reticulum by binding to a receptor protein on the ER
The signal mechanism for targeting proteins to the ER
In both bacteria and eukaryotes, ___ ribosomes translate an mRNA at the same time
- multiple
{{Polyribosomes (Polysomes):{{
String of ribosomes making many copies of a polypeptide simultaneously
Polyribosomes
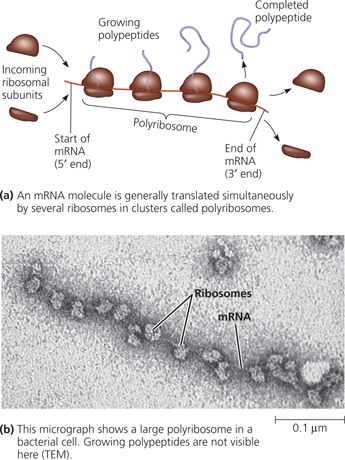
Both bacteria and eukaryotes augment the number of copies of a polypeptide by transcribing ___ mRNAs from the same gene
- multiple
How does a lack of compartmentalization impact bacteria in transcription and translation?
Like a one-room workshop, a bacterial cell ensures a streamlined operation by coupling the two processes. In the absence of a nucleus, it can simultaneously transcribe and translate the same gene and the newly made protein can quickly diffuse to its site of function. The eukaryotic cell’s nuclear envelope segregates transcription from translation and provides a compartment for extensive RNA processing. This processing stage includes additional steps whose regulation can help coordinate the eukaryotic cell’s elaborate activities
Coupled transcription and translation in bacteria - In bacterial cells, the translation of mRNA can begin as soon as the leading (5′) end of the mRNA molecule peels away from the DNA template. The micrograph (TEM) shows a stretch of E. coli DNA being transcribed by RNA polymerase molecules. Attached to each RNA polymerase molecule is a growing strand of mRNA, which is already being translated by ribosomes. The newly synthesized polypeptides are not visible in the micrograph but are shown in the diagram
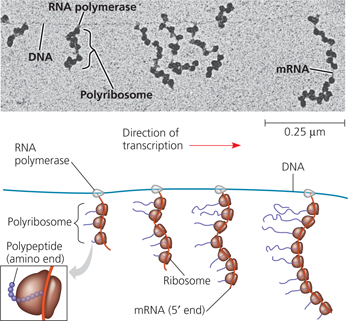
Which one of the mRNA molecules started being transcribed first? On that mRNA, which ribosome started translating first?
The mRNA farthest to the right (the longest one) started being transcribed first. The ribosome at the top, closest to the DNA, started translating first and thus has the longest polypeptide
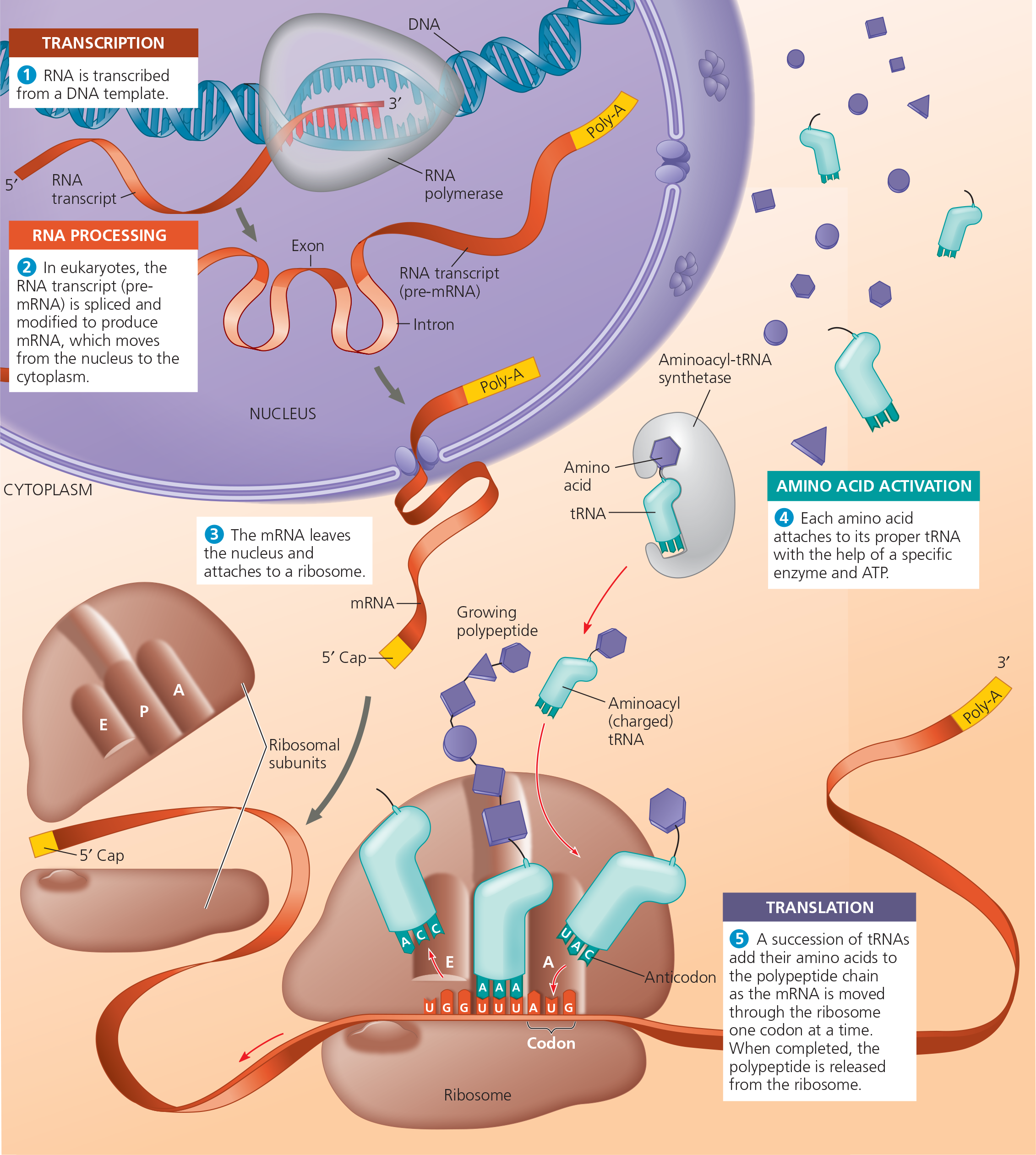
A summary of transcription and translation in a eukaryotic cell - This diagram shows the path from one gene to one polypeptide. Keep in mind that each gene in the DNA can be transcribed repeatedly into many identical RNA molecules and that each mRNA can be translated repeatedly to yield many identical polypeptide molecules. (Also, remember that the final products of some genes are not polypeptides but RNA molecules that don’t get translated, including tRNA and rRNA.) In general, the steps of transcription and translation are similar in bacterial, archaeal, and eukaryotic cells. The major difference is the occurrence of RNA processing in the eukaryotic nucleus. Other significant differences are found in the initiation stages of both transcription and translation and in the termination of transcription
@@What two processes ensure that the correct amino acid is added to a growing polypeptide chain?@@
First, each aminoacyl-tRNA synthetase specifically recognizes a single amino acid and attaches it only to an appropriate tRNA. Second, a tRNA charged with its specific amino acid binds only to an mRNA codon for that amino acid
@@Discuss the ways in which rRNA structure likely contributes to the ribosomal function@@
The structure and function of the ribosome seem to depend more on the rRNAs than on the ribosomal proteins. Because it is single-stranded, an RNA molecule can hydrogen-bond with itself and with other RNA molecules. RNA molecules make up the interface between the two ribosomal subunits, so presumably RNA-RNA binding helps hold the ribosome together. The binding site for mRNA in the ribosome includes rRNA that can bind the mRNA. Also, complementary hydrogen bonding within an RNA molecule allows it to assume a particular three-dimensional shape and, along with the RNA’s functional groups, presumably enables rRNA to catalyze peptide bond formation during translation
@@Describe how a polypeptide to be secreted is transported to the endomembrane system@@
A signal peptide on the leading end of the polypeptide being synthesized is recognized by a signal-recognition particle that brings the ribosome to the ER membrane. There the ribosome attaches and continues to synthesize the polypeptide, depositing it in the ER lumen
@@Draw a tRNA with the anticodon 3′-CGU-5′. What two different codons could it bind to? Draw each codon on an mRNA, labeling all 5′ and 3′ ends, the tRNA, and the amino acid it carries@@
Because of wobble, the tRNA could bind to either 5′-GCA-3′ or 5′-GCG-3′, both of which code for alanine (Ala). Alanine would be attached to the tRNA (see diagram, upper right) 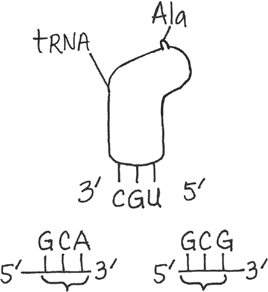
@@What function do tRNAs serve in the process of translation?@@
In the context of the ribosome, tRNAs function as translators between the nucleotide-based language of mRNA and the amino-acid-based language of polypeptides. A tRNA carries a specific amino acid, and the anticodon on the tRNA is complementary to the codon on the mRNA that codes for that amino acid. In the ribosome, the tRNA binds to the A site. Then the polypeptide being synthesized ( currently on the tRNA in the P site) is joined to the new amino acid, which becomes the new (C-terminal) end of the polypeptide. Next, the tRNA in the A site moves to the P site. After the polypeptide is transferred to the new tRNA, thus adding the new amino acid, the now empty tRNA moves from the P site to the E site, where it exits the ribosome
@@14.5 - Mutations of one or a few nucleotides can affect protein structure and function@@
Small-scale mutations include point mutations, changes in one DNA nucleotide pair, which may lead to production of nonfunctional proteins. Nucleotide-pair substitutions can cause missense or nonsense mutations. Nucleotide-pair insertions or deletions may produce frameshift mutations
Spontaneous mutations can occur during DNA replication, recombination, or repair. Chemical and physical mutagens cause DNA damage that can alter genes
{{Mutation:{{
A change in the nucleotide sequence of an organism’s DNA or in the DNA or RNA of a virus
{{Point Mutations:{{
A change in a single nucleotide pair of a gene
{{Large-Scale Mutations:{{
Chromosomal rearrangements that affect long segments of DNA
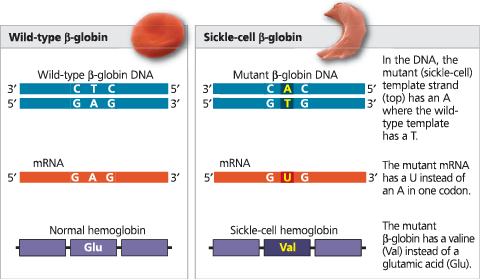
The molecular basis of sickle-cell disease: a point mutation - The allele that causes sickle-cell disease differs from the wild-type (normal) allele by a single DNA nucleotide pair. The micrographs are SEMs of a normal red blood cell (on the left) and a sickled red blood cell (right) from individuals homozygous for wild-type and mutant alleles, respectively
Small-scale mutations within a gene can be divided into two general categories:
(1) single nucleotide-pair substitutions and (2) nucleotide-pair insertions or deletions. Insertions and deletions can involve one or more nucleotide pairs
{{Nucleotide-Pair Substitution:{{
A type of point mutation in which one nucleotide in a DNA strand and its partner in the complementary strand are replaced by another pair of nucleotides
A change in a nucleotide pair may transform one codon into another that is translated into ___
- the same amino acid
{{Silent Mutation:{{
A nucleotide-pair substitution that has no observable effect on the phenotype
{{Missense Mutation:{{
A nucleotide-pair substitution that results in a codon that codes for a different amino acid
Types of small-scale mutations that affect mRNA sequence
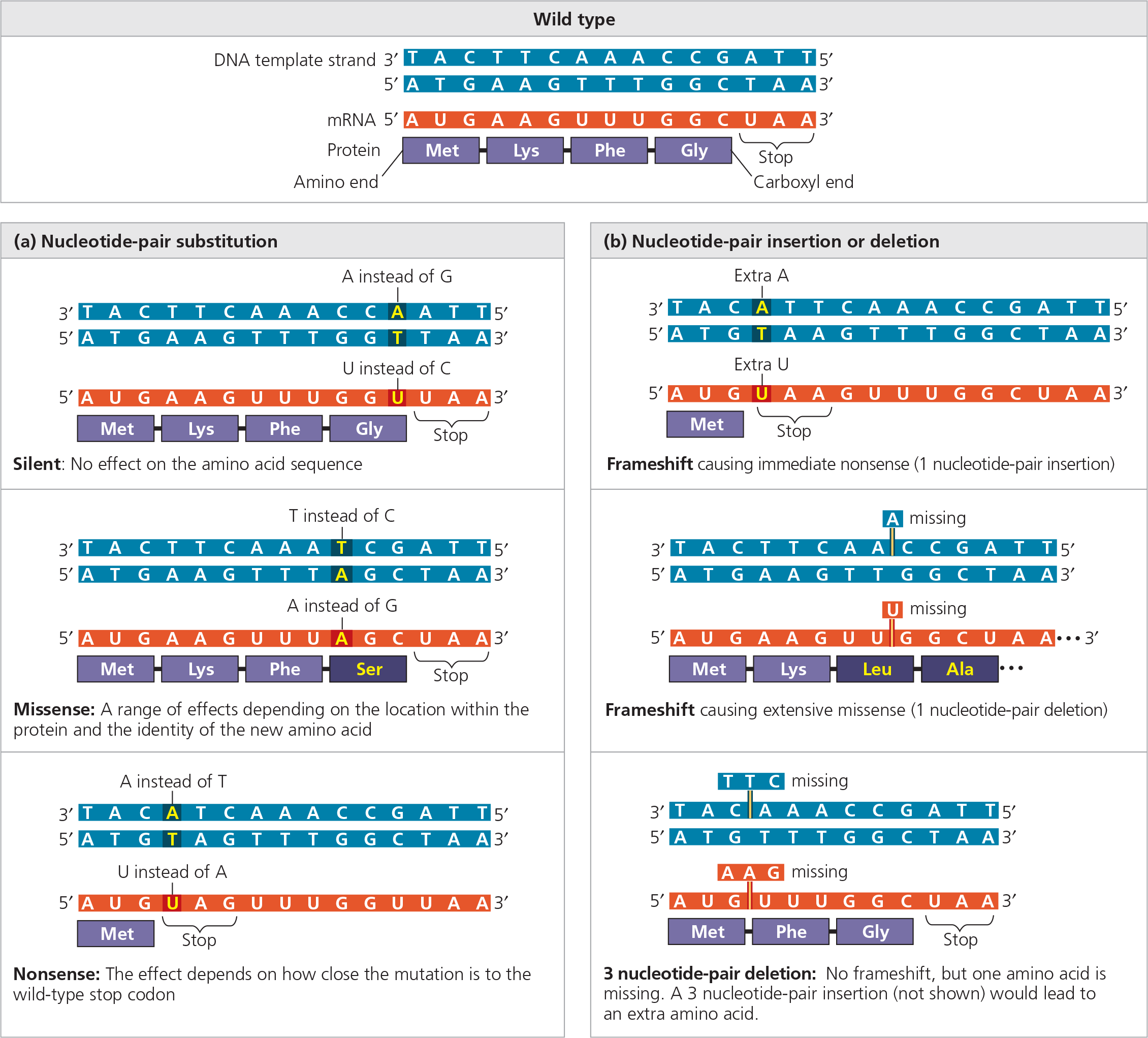
{{Nonsense Mutation:{{
A mutation that changes an amino acid codon to one of the three stop codons, resulting in a shorter and usually nonfunctional protein
{{Insertion:{{
A mutation involving the addition of one or moe nucleotide pairs to a gene
{{Deletion:{{
A deficiency in a chromosome resulting from the loss of a fragment through breakage; a mutational loss of one or more nucleotide pairs from a gene
___ of nucleotides have a greater effect on the resulting protein and may alter the reading frame of the genetic message, the triplet grouping of nucleotides on the mRNA that is read during translation
- Insertion or deletion
{{Frameshift Mutation:{{
A mutation occurring when nucleotides are inserted in or deleted from a gene and the number inserted or deleted is not a multiple of three, resulting in the improper grouping of the subsequent nucleotides into codons
{{Spontaneous Mutations:{{
Mutation from an incorrect base used as a template in the next round of replication
{{Mutagens:{{
A chemical or physical agent that interacts with DNA and can cause a mutation
{{Nucleotide Analogs:{{
Chemicals that are similar to normal DNA nucleotides but that pair incorrectly during DNA replication
Describe a gene
A gene is a discrete unit of inheritance that affects a phenotypic character. They are assigned to specific loci on chromosomes. They are viewed as a region of specific nucleotide sequences along the length of the DNA molecules of a chromosome. A gene functions as a DNA sequence that codes for a specific polypeptide chain
{{Gene:{{
A region of DNA that can be expressed to produce a final functional product that is either a polypeptide or an RNA molecule
@@What happens when one nucleotide pair is lost from the middle of the coding sequence of a gene?@@
In the mRNA, the reading frame downstream from the deletion is shifted, leading to a long string of incorrect amino acids in the polypeptide, and in most cases, a stop codon will arise, leading to premature termination. The polypeptide will most likely be nonfunctional
@@Individuals heterozygous for the sickle-cell allele show effects of the allele under some circumstances. Explain in terms of gene expression@@
Heterozygous individuals, said to have sickle-cell trait, have a copy each of the wild-type allele and the sickle-cell allele. Both alleles will be expressed, so these individuals will have both normal and sickle-cell hemoglobin molecules. Apparently, having a mix of the two forms of β-globin has no effect under most conditions, but during prolonged periods of low blood oxygen (such as at higher altitudes), these individuals can show some signs of sickle-cell disease
@@The template strand of a gene includes this sequence: 3′-TACTTGTCCGATATC-5′. It is mutated to 3′-TACTTGTCCAATATC-5′. For both versions, draw the DNA, the mRNA, and the encoded amino acid sequence. What is the effect on the amino acid sequence?@@
No effect: The amino acid sequence is Met-Asn-Arg-Leu both before and after the mutation because the mRNA codons 5′-CUA-3′ and 5′-UUA-3′ both code for Leu. (The fifth codon is a stop codon.) 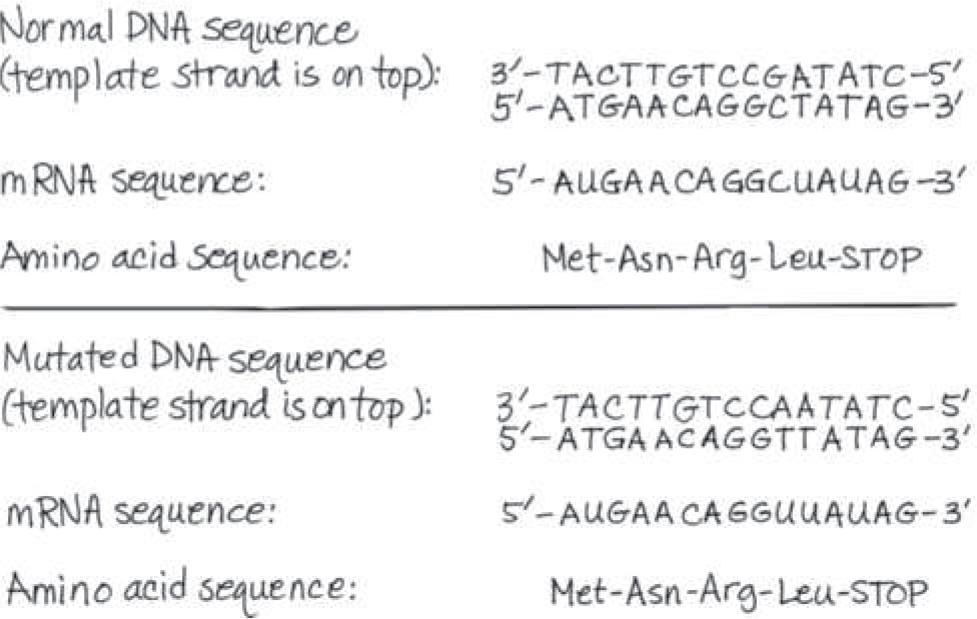
@@What will be the results of chemically modifying one nucleotide base of a gene? What role is played by DNA repair systems in the cell?@@
When a nucleotide base is altered chemically, its base-pairing characteristics may be changed. When that happens, an incorrect nucleotide is likely to be incorporated into the complementary strand during the next replication of the DNA, and successive rounds of replication will perpetuate the mutation. Once the gene is transcribed, the mutated codon may code for a different amino acid that inhibits or changes the function of a protein. If the chemical change in the base is detected and repaired by the DNA repair system before the next replication, no mutation will result