Enzymes, Metabolism, & ATP Test
Adi’s version
Protein Levels of Structure:
What forces/bonds hold a protein together at each level of structure? And these occur between what parts of the protein?
Primary: Peptide bond
Secondary: Hydrogen bonds
Tertiary: ionic, disulfide, hydrophilic, hydrogen
Quaternary: ionic, disulfide, hydrophilic, hydrogen
Primary: Amino acids (amino group + carboxyl group bonding together)
Secondary: 2 primary structures bonded together – the O and H of amino acid – can take form as either an alpha-helix or a pleated sheet
Tertiary: R group of amino acids – different from bonds that stabilize secondary structure – 1 polypeptide chain
Quaternary: 3 polypeptide chains
How does each level of structure contribute to the overall shape of a protein?
When is the polypeptide backbone formed?
When does the polypeptide backbone first fold or bend?
What level of structure is not found in all proteins?
In what level are alpha helices and beta pleated sheets formed?
A. polypeptide backbone forms during primary structure (peptide bonds hold it together)
B. polypeptide first folds/bends during secondary structure
C. Quaternary structure not found in all proteins
D. secondary structure
Given a polypeptide with certain properties/amino R groups, in a given environment (water, oil) can you predict how this protein might behave/fold?
In water, Polar R groups are found on the outside of the folded protein OR hydrogen bonding to another Polar R group
In water, Nonpolar R groups are found in the inside of the folded protein often in a hydrophobic interaction with another nonpolar R group
In water, Charged R groups are found on the outside of the folded protein OR ionic bonding to another oppositely Charged R group
How would all of the above R groups behave if the protein was in oil?
Polar: would be found on the inside of oil
Nonpolar: would be found on the outside of the oil (oil is non polar)
Charged R groups: would be located on the inside of the R groups (polar)
What kinds of changes may cause a protein to denature/unfold? List all the possible environmental changes that may cause a protein to denature. Be specific - don’t just say a change in “x” - say HOW “x” changes (increased,decreases, etc)
Changes: If enzyme is boiled, if strong acid or base is added
How it affects:
pH
What is the pH scale and what is it a measure of?
- how acidic/basic a substance is
What does it mean for a solution to be at pH 7? How would you draw a model of this?
- pH 7=neutral
- Drawing: lots of water molecules, equal number of hydrogen (H+) ions and hydroxide ions (OH-)
What is an acid? Base? Buffer?
- acid: chemical compounds that increase the H+ concentration of a solution
- base: chemical compounds that increase the OH- concentration of a solution
- buffer: keeps pH level of solution constant– chemicals that add/take away H+ from a solution
How would you draw a model of a slightly basic or acidic solution?
- slightly basic: more hydroxide (OH-) ions than hydrogen ions (H+)
- slightly acidic: more hydrogen ions (H+) than hydroxide ions (OH-)
What are some examples of common acids and bases?
Common acids: hydrochloric acid, lemon juice, vinegar, cola, beer
Common bases: sodium hydroxide, oven cleaner, household bleach
How does a change in ph away from optimum potentially affect protein structure?
- loss of ionic bonds/hydrogen bonds
- can denature the enzyme
- can disrupt the active site
Chemical reactions and the ATP cycle
Exergonic V Endergonic reactions – what will the Potential Energy graph look like for each?
Exergonic Reaction: reactions which release energy
Examples: burning of cable, (metabolism)-- food breaking
down
Endergonic reaction: reaction which absorbs energy
(energy consuming)
Ex: resting, moving, building up muscles
What is activation energy? What does it look like in a potential energy graph?
Activation energy: how much energy is needed to make the reaction occur
What is the transition state? Is this more or less stable than reactants? Does this have a high or lower potential energy than the reactants?
Transition state: step in the middle where substance isn’t a product or a reaction
- has a higher potential energy than the reactants
What is the difference in an energy graph with/without an enzyme
With enzyme: much faster rate
Without enzyme: slower (normal)
Endergonic process: requires energy use by cell; energy is typically found in the form of ATP.
ATP phosphorylates another molecule, resulting ADP & Pi
Exergonic process: produces energy use by cell respiration: ADP & Pi bonded again, forming ATP
Why are exergonic and endergonic reactions frequently coupled (found paired together) in metabolism?
- it lets cells use energy efficiently
Enzyme structure & activity:
How do enzymes affect chemical reactions?
- they decrease the amount of activation energy needed for chemical reaction to occur by stabilizing the transition state
- “speed up” reactions
How does an enzyme bind to its substrate? What attractions hold these together?
- substrates attach to enzymes active site to form enzyme substrate complex
- enzymes can be reused
Vocab to think about: Active site, induced fit vs lock and key, enzyme specificity
Active site: where substrate binds to enzyme – needs to be right shape and chemistry (-) binds to (+)
Induced fit: enzyme hugs to better bind substrate and help reaction happen – active site slightly changes as substrate binds
Lock and Key: shape of active site is designed for one substrate to fit
Why/how does an enzyme only bind to one type of substrate (two factors)
- active site has specific type of substrate it needs (lock and key, induced fit, specificity)
- ensures accuracy in cell processes
Factors affecting enzyme-catalyzed reactions:
What would prevent an enzyme from binding to its substrate? (3-4 things)
Enzyme inhibitors bind to active site instead of substrate
Non competitive: inhibitor binds to site on the enzyme other than the active site – called allosteric site
Competitive inhibitor: competes with substrate to bind to the active site – blocks reaction
Changes in pH (denatured enzyme) – can’t bind with substrate
Changes in temperature (boiling) – can’t bind with substrate
Substrate concentration: if there is not enough substrate it won’t bind with enzyme
How do the following factors affect reaction rates of enzyme-catalyzed reactions? WHY?
Substrate concentration
Temperature
pH
presence/absence of
competitive inhibitor
Non-competitive inhibitor
Activator
Substrate concentration:
High: rate increases:
Low: slows down
Temperature:
High: denatured enzyme
Low: slows down enzyme
pH:
high/low – denatures enzyme
Competitive inhibitor: if it wins, stops reaction
Noncompeitive: attaches to part of enzymes other than active site → stops reaction
Activator: makes the enzyme work better (allosteric activator bind away from the active site)
Metabolic pathway control mechanisms:
Pathway: a series of chemical reactions that result in a final product
Intermediate molecule: a molecule that is both the product of the reaction and the substrate for the next reaction in its pathway
Enzymes are often acting where you see arrows in these pathway
Inhibitors & activators bind to enzymes, turning them “off” or “super on” (temporarily)
Inhibiting key enzymes can turn pathways off or super on as needed by body or cell in moment
What is the purpose and advantage of each of these? Why do they regulate metabolic pathways?
Negative feedback
Positive feedback
Feedback: when a molecule that is produced later in a pathway affects (negatively or positively) a molecule earlier in pathway
Negative feedback: pathway which is inhibited by the final product of the pathway
Positive feedback: product of the pathway which accels/enhances the enzyme in the beginning of the pathway
Negative control results from inhibition
Positive results from activation
Which type of feedback will help maintain homeostasis?
Negative feedback
Question 1
What forces/bonds hold a protein together at each level of structure? And these occur between what parts of the protein?
Primary: In the primary structure, bonds that hold a protein together are the peptide bonds. These occur between the carboxyl group of one amino acid and the amino group of the next amino acid in the chain.
Secondary: In the secondary structure, hydrogen bonds are holding the structure. This occurs between an H atom of one amino acid and O atom from another amino acid in the chain.
aTertiary: In the tertiary structure, there are four types of bonds that hold this structure together, ionic bonds, hydrogen bonds, disulfide bonds, and hydrophobic interactions. Ionic bonds occur between oppositely charged atoms. A hydrogen bond, as explained earlier, is a bond between a H atom of one amino acid and an O atom of another amino acid (an attraction between polar and polar). A disulfide bond occurs between two sulfur bonded covalently. Hydrophobic interactions are dispersion forces attractions between non-polar groups
Quaternary: The same as Tertiary
Question 2
How does each level of structure contribute to the overall shape of a protein?
When is the polypeptide backbone formed?
The polypeptide backbone is formed in the primary structure when a sequence of amino acids are linked in by the peptide bonds.
When does the polypeptide backbone first fold or bend?
The polypeptide backbone first bends in the secondary structure.
What level of structure is not found in all proteins?(Only found in proteins with more than one polypeptide chain)
The quaternary structure
In what level are alpha helices and beta pleated sheets formed?
The secondary structure of the protein
Question 3
Given a polypeptide with certain properties / amino acids R-groups , in a given environment (water, oil) can you predict how this protein might behave / fold.
in water, Polar R groups are found on the surface of the folded protein OR hydrogen bonding to another polar R group. Since its hydrophilic
in water, nonpolar R groups are found on the interior of the folded protein, often in a hydrophilic interaction with another non-polar R group
in water, Charged R groups found on surface of the folded protein OR ionic bonding to an oppositely charged R group. Since its hydrophilic
How would all of the above R groups behave if the protein was in oil?
Polar R groups
In oil, they would be hydrophobic and prefer to be on the interior of the protein to minimize contact with the nonpolar oil molecules
Nonpolar R groups
In oil, they would be more exposed on the surface of the protein, as they would be more compatible with the nonpolar oil environment.
Charged R groups
In oil, they would be less stable and might disrupt the protein's structure. This is because charged groups typically require a polar environment to stabilize their charges. In a nonpolar environment like oil, they would lack the necessary interactions to maintain their stability.
Question 4
What kinds of changes may cause a protein to denature/unfold? List all the possible environmental changes that may cause a protein to denature. Be specific - don’t just say a change in “x” - say HOW “x” changes (increase, decrease, etc).
pH Levels: An increase or decrease in pH can disrupt the ionic bonds and interactions between charged R groups, leading to protein unfolding.
Temperature: An increase in temperature typically increases molecular movement, potentially disrupting hydrogen bonds and other interactions that maintain protein structure.
Detergents: The presence of detergents can disrupt hydrophobic interactions and lead to denaturation by binding to nonpolar regions of the protein.
Organic Solvents: An increase in organic solvents can affect the hydrophobic interactions within the protein, pushing nonpolar R groups towards the surface and increasing unfolding.
pH
What is the pH scale and what is it a measure of?
pH is a measure of H+ concentration in a solution
It quantifies how acidic for basic a solution is, when pH levels are low, its acidic, and if pH levels are high, it’s basic
What does it mean for a solution to be at pH 7? How would you draw a model of this?
When a solution is at a pH of 7, this means that the solution is neutral
In a model, pH 7 would be drawn in the middle of the scale
What is an acid? Base? Buffer?
Acid
Substances that release hydrogen ions (H+) when they’re dissolved in water
Bases
Substances that release hydroxide ions (OH-) when they’re dissolved in water
Buffer
Buffers are mixtures that can react with acids or bases to keep the pH within a particular range
How would you draw a model of slightly basic or an acidic solution?
For a acidic solution, anything in the ranges of 0 to 6.9 pH, are acidic
For a basic solution, anything that ranges from 7.1 to 14 pH, are basic
This means that to draw a acidic solution, you would draw it towards the right of the scale, and if you wanted to draw a basic solution, you draw it to the left
What are some examples of common acids & bases
Acids
Vinegar, toilet bowl cleaners, lemon juice, and soda
Bases
Egg whites, spinach, and chlorine bleach
How does a change in pH from optimum potentially affect protein structure?
A change in pH from optimum can disrupt the ionic bonds and interactions between charged R groups in a protein. This disruption can lead to protein denaturation or unfolding, which affects its overall shape and functionality. Specifically, if the pH increases or decreases significantly from its optimal range, it can destabilize the protein's structure, impacting its ability to perform biological functions.
Protein Folding & pH Vocabulary
Primary: The sequence of amino acids in a polypeptide chain.
Secondary: The local folding of the polypeptide chain into structures like alpha-helices and beta-sheets, stabilized by hydrogen bonds.
Tertiary: The overall three-dimensional shape of a polypeptide chain, determined by interactions between side chains (R groups).
Quaternary: The arrangement of multiple polypeptide chains to form a functional protein.
Alpha-helix: swirly thing
Beta-sheet: pleated sheet
Covalent: A covalent bond is a chemical bond that involves the sharing of electron pairs between atoms
Ionic bond: An attraction between positive ca\hared and negative charged
Hydrogen-bonds: an attraction between polar and polar
Hydrophobic interactions: Dispersion Forces attractions between nonpolar groups
Disulfide bridge: A covalent bond between 2 sulfur atoms
Denature/denaturation: The process of altering the structure of a protein, leading to loss of function.
H+ ions/OH- ions: Ions that determine the acidity or basicity of a solution.
Acid: A substance that donates H+ ions in solution.
Base: A substance that accepts H+ ions in solution.
Buffer: A solution that resists changes in pH.
Solution: A homogeneous mixture of two or more substances.
Solvent: The substance that dissolves another substance (usually a liquid).
Solute: The substance that is dissolved in a solvent.
Chemical Reactions & the ATP Cycle
Exergonic v Endergonic reactions what will Potential Energy graph look like for each?
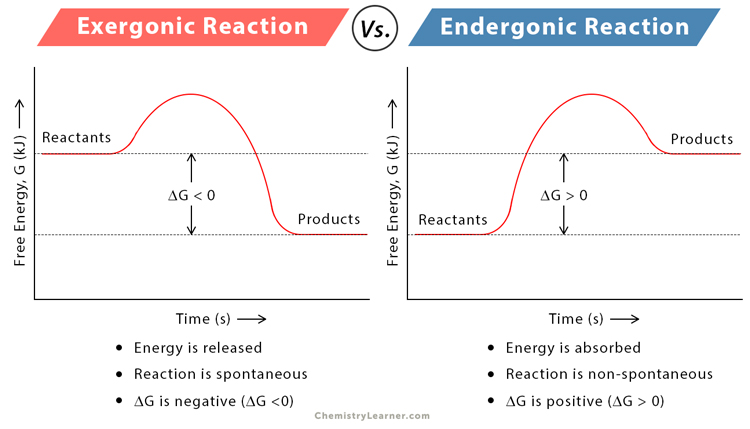
What is activation energy? What does it look like in a Potential energy graph?
The minimum amount of energy needed for reactions to form products in a chemical reaction

What is transition state? Is this more or less stable than reactants? Does this have a higher or lower potential energy than the reactants?
A transition state is a high-energy, unstable state that molecules must pass through during a chemical reaction.It's the point of maximum energy in a reaction pathway.
Higher potential energy than reactants
What is the difference in an energy graph with / without an enzyme
With enzyme
Lower activation energy
Without enzyme
Higher activation energy
Endergonic process: requires energy use by cell; energy is typically in the form of ATP
ATP phosphorylates another molecule, resulting ADP & Pi
Exergonic process: produces energy for use by cell; energy typically in form of ATP.
ADP is “renewed” using energy from cell respiration: ADP & Pi bonded again, forming ATP
Why are exergonic and endergonic reactions frequently coupled (found paired together) in metabolism?
It allows cells to harness the energy released from one reaction to drive another that requires energy input.
Enzyme Structure & Activity
How do enzymes affect chemical reactions?
Enzymes speed up reactions without being consumed, they are reusable
How does an enzyme bind its substrate? What attractions hold these together?
Substrates attach to an enzyme’s active site to form the Enzyme substrate complex
Hydrogen Bonds
An active site needs to be the right shape & chemistry to attach to the substrate models
look back to the first enzyme notes at the diagram
Vocab to think about: Active site, induced fit vs lock and key, enzyme specificity
look at notes
Why/how does an enzyme only bind one type of substrate? (two factors)
It’s because of the shape and chemical properties of the enzyme.
Factors Affecting Enzyme-Catalyzed
What could prevent an enzyme form binding its substrate? (name 3-4 things)
Inhibitors
pH
Temperature
Substrate concentration
How do the following factors affect reaction rates of enzyme-catalyzed reactions? WHY?
Substrate concentration
Increased Rate: As substrate concentration increases, the rate of reaction increases up to a certain point. This is because more substrate molecules are available to collide with the enzyme's active site, forming more enzyme-substrate complexes.
Saturation Point: However, when all enzyme active sites are saturated with substrate, increasing the substrate concentration further will not significantly increase the reaction rate.
Temperature
Increased Rate: As temperature increases, the kinetic energy of molecules increases, leading to more frequent collisions between enzyme and substrate.
Denaturation: At extremely high temperatures, the enzyme's protein structure can denature, losing its shape and function, and thus reducing the reaction rate.
pH
Optimal pH: Enzymes have an optimal pH range at which they function best.
Denaturation: Extreme pH values can denature the enzyme, altering its shape and function
Presence/absence of:
Competitive inhibitor
Reduced Rate: Competitive inhibitors compete with the substrate for the active site, reducing the enzyme's ability to bind the substrate and slowing the reaction rate.
Non-competitive inhibitor
Reduced Rate: Non-competitive inhibitors bind to a site other than the active site, changing the enzyme's shape and reducing its ability to bind the substrate or catalyze the reaction.
Activator
Increased Rate: Activators bind to the enzyme, increasing its activity by changing its shape or enhancing its affinity for the substrate.
Metabolic Pathway Control Mechanisms
What is the purpose and advantage of each of these? Why and how do they regulate metabolic pathways?
Negative feedback
Purpose: To maintain homeostasis or a steady state. Advantage: It ensures that systems remain stable and balanced. How it regulates:
A stimulus triggers a response that counteracts the initial stimulus.
This counteraction reduces the effect of the original stimulus.
The system returns to its original state.
Example: Blood sugar regulation.
When blood sugar rises, the pancreas releases insulin, which stimulates cells to take up glucose from the blood, lowering blood sugar levels.
Positive feedback
Purpose: To amplify a process and bring it to completion. Advantage: It can be used to rapidly initiate or complete a process. How it regulates:
A stimulus triggers a response that amplifies the original stimulus.
This amplification leads to a further increase in the response.
The process continues in a cycle until a specific endpoint is reached.
Example: Fever
A fever can be initiated by a bacterial or viral infection.
The body's temperature-regulating system responds by increasing body temperature.
This higher temperature can help fight off the infection, but if it becomes too high, it can be dangerous.
Which type of feedback will help maintain homeostasis
Negative Feedback
Chemical Reactions/Enzyme Vocabulary
Chemical Potential Energy: The energy stored in the chemical bonds of molecules.
Reactant: A substance that undergoes a chemical change.
Product: A substance formed as a result of a chemical reaction.
Activation Energy: The minimum amount of energy required for a reaction to occur.
Exergonic Reaction: A reaction that releases energy.
Endergonic Reaction: A reaction that absorbs energy.
ATP: Adenosine triphosphate, an energy-carrying molecule.
ADP: Adenosine diphosphate, a lower-energy molecule formed from ATP.
Inorganic Phosphate Group: A phosphate group that is not part of an organic molecule.
Phosphorylation: The addition of a phosphate group to a molecule.
Catalyst: A substance that speeds up a chemical reaction without being consumed in the process.
Enzyme: A biological catalyst, typically a protein.
Substrate: The specific molecule on which an enzyme acts.
Active Site: The region of an enzyme where the substrate binds.
Induced Fit: A model of enzyme-substrate interaction where the enzyme's shape changes slightly to accommodate the substrate.
Lock and Key: A simpler model of enzyme-substrate interaction where the substrate fits perfectly into the active site.
Enzyme Specificity: The ability of an enzyme to catalyze a specific reaction with a specific substrate.
-ase: A common suffix used to denote enzymes.
Saturation of Enzyme: The point at which all enzyme active sites are occupied by substrate molecules, limiting the reaction rate.
Denatured/Denaturation: The loss of an enzyme's shape and function due to factors like temperature or pH.
Optimal Temperature: The temperature at which an enzyme functions best.
Optimal pH: The pH at which an enzyme functions best.
Inhibition: A process that slows down or stops an enzyme's activity.
Competitive Inhibition: A type of inhibition where a molecule competes with the substrate for the active site.
Non-competitive Inhibition: A type of inhibition where a molecule binds to a site other than the active site, changing the enzyme's shape and reducing its activity.
Activation: The process of turning on or increasing the activity of an enzyme.
Metabolic Pathways: A series of interconnected chemical reactions that occur within a cell.
An intermediate is a molecule that is both a substrate and a product in a metabolic pathway
Negative Feedback: A regulatory mechanism where the product of a pathway inhibits an earlier step in the pathway, maintaining homeostasis.
Positive Feedback: A regulatory mechanism where the product of a pathway stimulates further production of the product, leading to an amplified response.