trafficking_2_NOTES(1)
Four Principal Modifications in the ER and Golgi:
Glycosylation (covalent addition and processing of carbohydrates).
Disulfide bond formation in the ER.
Proper protein folding and multi-subunit assembly in the ER.
Specific proteolytic cleavages in various compartments (ER, Golgi, vesicles).
Quality Control in the ER
Glycoprotein modification retaining a single glucose moiety.
Glucose binding by ER chaperones (calnexin).
Glucosidase II removes remaining glucose.
Conformational-sensing enzyme UGGT evaluates folding integrity:
Yes: Adds glucose.moeity; continues the cycle.
No: Leads to degradation via proteasome.
Unfolded Protein Response (UPR)
Protein Sensors:
PERK and ATF6 activate response pathways.
Roles include chaperones, transporters, degradation processes.
Controlled by BiP endoribonuclease.
Activation leads to transcription factors regulation and translation inhibition.
Protein Targeting Signals
Signals: Encoded in amino acid sequences of protein (signal sequence) or attached signals like (carbohydrates)
Recognition by specific receptors:
Soluble proteins: Receptive integral membrane proteins.
Transmembrane proteins: receptors can be Coat components on the cytosolic surface of the budding transport vesicle
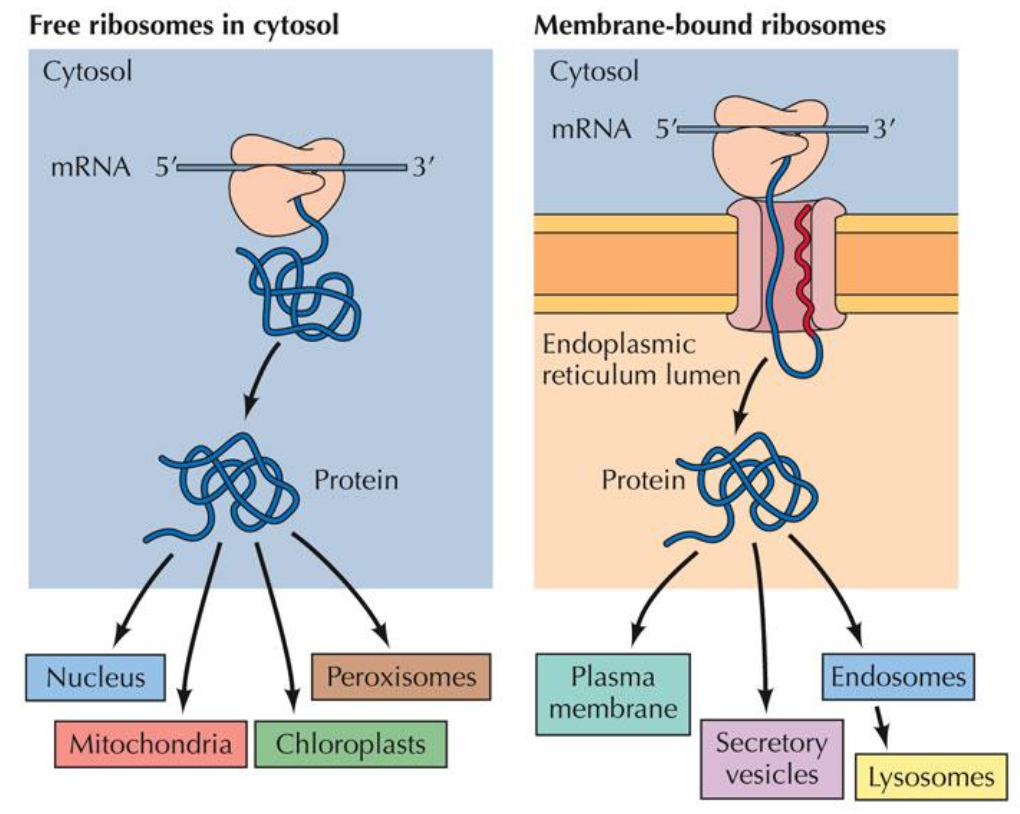
Pathways and Destinations of Protein Sorting
Sources:
Free ribosomes (cytosol) and membrane-bound ribosomes.
Destinations:
Cytosol, nucleus, mitochondrial, chloroplast, peroxisomes, lysosomes, plasma membrane.
Uniqueness of Import Process for Peroxisomal& mitochondrial protein import
Proteins imported through:
Outer boundary membrane.
import occurs Post-translationally .
Proteins in native folded state for peroxisomes.
Mitochondria! Structural Features and Functions
Site of aerobic respiration and ATP production
High abundance in eukaryotic cells (up to 25% of cytoplasmic volume).
Inner membrane features:
Cristae increase surface area for ATP production.
many cristae infoldings into the central aqueous matrix compartment
Outer membrane: large pores facilitate molecule transfer to move from the cytosol to the intermembrane space.
Major site for aerobic respiration and ATP generation using glucose.
Mitochondrial Protein Synthesis Overview
Majority synthesized by cytosolic ribosomes (over 1000 different proteins).
mtDNA encodes only 13 proteins;
Most mitochondrial proteins imported from cytoplasm.
Proteins can reside in:
Mitochondrial matrix (e.g., citrate synthase).
Inner membrane (e.g., ATP synthase subunits).
Outer membrane (e.g., mitochondrial porins).
Intermembrane space (e.g., cytochrome C).
Proteins reach Mitochondrial Matrix via Signal Sequences
many mitochondrial proteins are synthesized by ribosomes in the cytoplasm and contains a matrix targeting signal sequence.
Matrix targeting signal sequence:
Found at N-terminus (20-50 amino acids long).
Contains hydrophobic amino acids and positively charged residues (Arg and Lys).
Forms an amphipathic alpha-helix for function.
positively charged hydrophilic amino acids on one side and hydrophobic amino acids on the other
amphipathicity of matric targeting sequence is important for its function
Key Signals:
ER: NH3+ end with 6-12 hydrophobic aa.
Mitochondria: NH3+ end with 3-5 non-consecutive Arg, Lys.
Peroxisome: COO- end with Ser-Lys-Leu.
Nucleus: Internal 5 basic aa/2 small clusters.
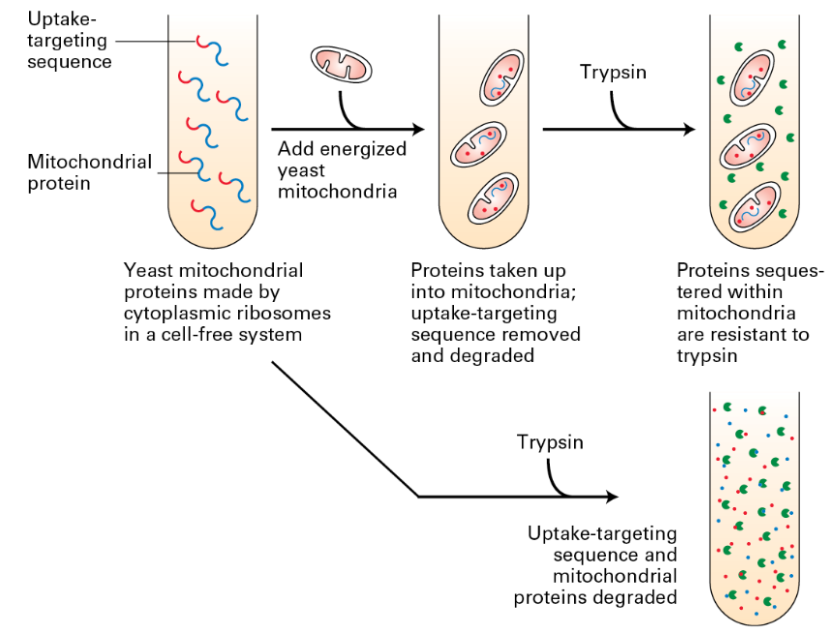
Post-Translational vs Co-Translational
Experiments highlight:
Uptake-targeting sequence removal results in degradation.
Resistance to trypsin when imported successfully;
Significance of targeting sequence.
Mitochondrial Matrix Protein Targeting
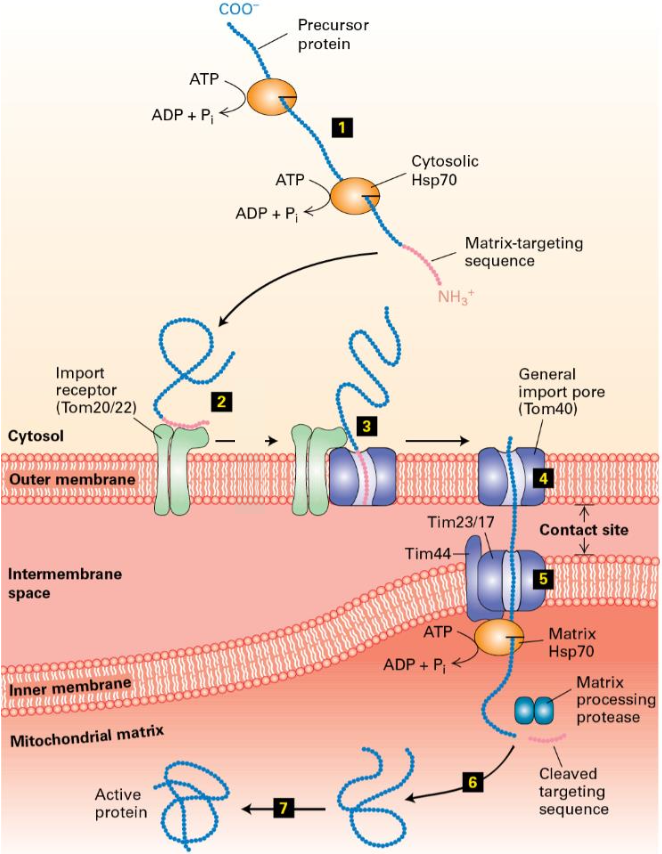
Import Mechanism Steps
Proteins synthesized by cytosolic ribosomes.
mitochondrial proteins can be imported only in an unfolded state. maintained in this state by cytosolic chaperones (Hsp70 & Hsp90) with help of ATP hydrolysis
Proteins bind to outer membrane import receptor (Tom20/22) via matrix targeting sequence.
MTS Insertion into outer membrane translocon (Tom40).
Translocation protein moves through Tom40 and inserts into inner membrane translocon complex(Tim23/17).
Protein translocates through Tim23/17. Binding by matrix Hsp70 helps drive import into the matrix. MTS removed by a matrix protease
Hsp70 ATP hydrolysis releases newly imported protein
Protein folds into its mature, active conformation within the matrix (many require chaperonins to fold properly)
Page 19: Energy Inputs for Import
Requirements for Protein Import into Mitochondria
Cytosolic ATP hydrolysis by Hsp70 maintains unfolded proteins.
Sequential binding by matrix Hsp70 facilitates import. subsequent ATP hydrolysis driven release matrix Hsp70 acts a as a rachet to pull the polypeptide into the mitochondrial matrix
– Hsp70/Tim44 in mitochondria is similar to BiP/Sec63 in the ER (post-
translational translocation)
H+ electrochemical gradient (proton-motive force) across the inner
membrane
Protons are pumped into the inter-membrane space from the matrix
Generates a proton motive force across the inner membrane in actively respiring mitochondria
The positive charges on the amphipathic matrix-targeting sequence is pulled
into the matrix by the inside negative-membrane potential
Inner Membrane Protein Import
Requires N-terminal matrix targeting sequence plus an internal hydrophobic stop-transfer sequence.
Similar transfer mechanisms as matrix proteins via Tom40 and Tim23/17.
Hydrophobic stop-transfer anchor sequence blocks transfer throughTim 23/17
The peptide is laterally transferred into the inner membrane bilayer
• E.g., Cytochrome oxidase subunit CoxVa
Import of Multipass Transmembrane Proteins
Lacks N-terminal targeting sequence; uses internal sequences.
Incorporated through multiple translocons and assisted by intermembrane chaperones.
Page 22: Targeting to Intermembrane Space
Use of matrix-targeting sequence and hydrophobic blocking sequence.
Intermediate is released in intermembrane space after cleavage.
Polypeptides have two different targeting sequences (both are ultimately cleaved)
matrix-targeting sequence
a hydrophobic segment that blocks complete translocation of the protein across the inner membrane
The matrix targeting sequence takes the protein to the matrix and the matrix targeting sequence is cleaved.
2nd targeting sequence inserts into Tim23/17 translocon
Prevents translocation into the matrix
The resulting membrane-embedded intermediate diffuses laterally away from Tim23/17
Hydrophobic segment is removed by an inner membrane protease
Soluble protein is released into the inter-membrane
Peroxisomes
Overview and Functions
Single membrane-bound organelles involved in oxidative metabolism (e.g., fatty acid oxidation).
Catalase: enzyme that breaks down hydrogen peroxide.
Associated disorders: Zellweger syndrome and Adrenoleukodystrophy.
Peroxisome Signaling Features
Key Signals Identified:
ER and mitochondrial signals as before.
Peroxisome: COO- end with Ser-Lys-Leu at C-terminus.
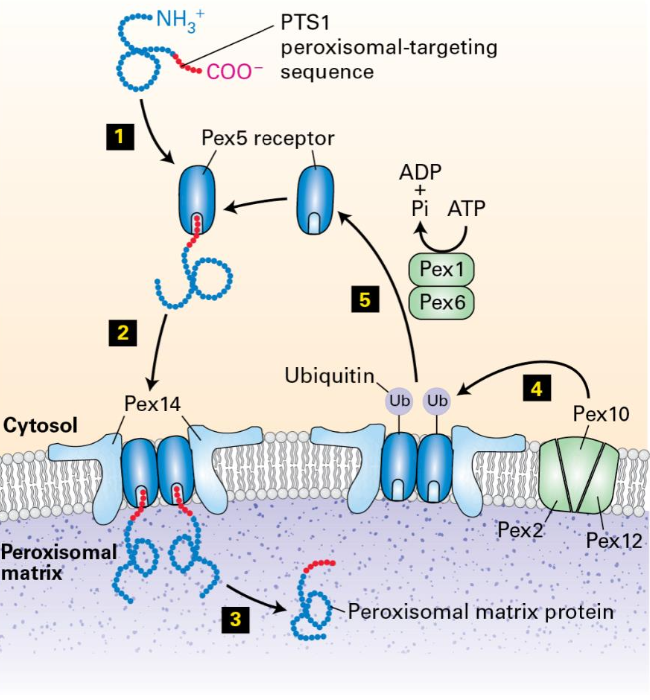
Signal sequence: peroxisomal targeting sequence (PTS1): S-K-L at the C terminus receptor
The Transient Pore Model
PTS-1 in the protein that is targeted to the peroxisome binds to Pex5
Pex5 with the bound matrix protein forms a multimeric complex Pex14
Forming transient pore
After assembly of the matrix protein-Pex5-Pex14 complex, the matrix protein dissociates from Pex5 and is released into the peroxisomal matrix
Pex5 is ubiquitinylated by the membrane proteins Pex 2, Pex 10, and Pex 12
Ubiquitinated Pex 5 is recognized by Pex1 and Pex6
uses ATP hydrolysis to remove Pex 5 from the peroxisome membrane
Pex5 is deubiquitinated and is ready for the next round of protein transfer
General mechanism in targeting proteins through Signal Sequences
Target destination information: Signal sequence within the polypeptide chain
Receptor to bind signal sequence: Present in the target organelle. Ensures specificity of targeting
Translocation channel: After signal sequence binds its receptor, polypeptide is transferred to a translocation channel. This allows the polypeptide to pass through the membrane bilayer of that organelle
Mechanism to ensure unidirectional transfer of proteins:Achieved by coupling translocation to an energetically favorable