BIO 151 Notes.docx
BIO 151
CHAPTER 1
Skin
- first line of defense
- epithelium = layers of keratinized cells
- lines organs and inner cavities
- Respiratory tract
- GI tract
- Urogenital tract
Mucosal surface
- Secretes mucus
- Beating cilia removes mucus and unwanted material
Sebaceous glands
- In hair follicles
- Secretes sebum to inhibit bacterial growth
Antimicrobial peptides
- Produced by all epithelia
- Perturbs pathogen membranes
Tears and saliva
- Contains lysozyme that degrades bacterial cell walls
Acidic environments
- Stomach, vagina, skin
Innate Immune System
- If skin is breached, infection remains localized and are extinguished within a few days without illness
- Genetically programmed
- In place even before infection
- Respond in the same way to repeated exposures
- Specific to structures shared by groups of microbes = cannot distinguish microbes
- Recognition
- soluble proteins and cell-surface receptors bind to pathogens or to altered cells and serum
- peptides, proteins, glycoproteins, proteoglycans, peptidoglycans, nucleic acids
- Recruitment
- effector mechanisms provided by effector cells to kill and eliminate pathogen
- complement serum proteins to mark pathogens

- Serum proteins of complement system recognizes pathogen
- Activation of serum proteins = fragment binds to pathogen and acts as marker
- Fragment recruits leukocytes with surface receptors to site
- Leukocyte binds to fragment and engulfs it
Cytokines
- Soluble secreted proteins
- Trigger and regulate innate immune response
- Induces vasodilation of the endothelium
- Leakage of blood plasma
- Expansion of fluid volume = edema or swelling = pressure on nerve endings causes pain
- Alters adhesive properties of endothelium
- WBCs attach to it to enter the inflamed tissue = inflammatory cells
- Increases swelling and contribute to pain
- Enables cells and molecules of the immune system to be brought rapidly and in large numbers into the site of infection

Adaptive Immune System
- Adds/ enhances innate immune response
- Innate immune response slows spread of infection and recruits lymphocytes
- Slow response (days to weeks)
- Adapts to the nuances of the infecting pathogen
- Specificity and diversity
- cell-surface receptors recognize specific antigens and different portions (epitopes) of polysaccharide and macromolecules
- huge lymphocyte repertoire due to variability in structures of antigen-binding sites
- different clones differ in their receptors and therefore specificity for antigens
- Clonal selection
- Selection of lymphocytes by antigens for activation to produce specific antibody
- specific antigen receptors exist on lymphocytes before they are presented with an antigen due to random mutations during initial maturation and proliferation
- Selection of lymphocytes by antigens for activation to produce specific antibody
- Clonal expansion
- Lymphocyte specific for an antigen undergoes proliferation after exposure
- Increase in number of cells that express identical receptors for the antigen
- Keeps pace with rapidly dividing pathogens
- Immunological memory
- To elicit a stronger and faster adaptive immune response
- Exposure to antigen generates long-lived memory cells specific for the antigen
- Also called acquired or protective immunity
- Primary immune response: first exposure to pathogen
- Secondary immune response: subsequent exposures with more rapid, larger, and qualitatively different from primary response
Adaptive Immunity is better understood than innate immunity
- Infections are overcome before they cause any symptoms
- Inherited deficiencies and impairment of function are rare
- Clinicians work together with adaptive response for a cure = more favored than innate
Hematopoiesis
- Leukocyte are derived from pluripotent hematopoietic stem cell
- Gives rise to precursors of lymphoid, myeloid, and erythroid lineages
- Sources: yolk sac > fetal liver > spleen > bone marrow
- Occurs throughput life due to short-lived nature of blood cells
Erythroid Lineage
- Gives rise to erythrocytes and megakaryocytes
- Megakaryocyte
- Fusion of multiple precursor cells
- Permanent residents of the bone marrow
- has small, non-nucleated fragments = platelets
- maintain integrity of vessels
- blood clotting
Myeloid Lineage
- granulocytes
- kill pathogens and enhance inflammation
- aka polymorphonuclear leukocytes
- neutrophil
- capture, engulfment and killing via phagocytosis
- effector cells of innate
- shortly-lived = pus
- eosinophil
- against parasites and worms
- basophil
- regulates response to parasites
- Monocytes
- Circulates in blood
- Recruited by macrophages into infected tissues = matures into macrophages
- In the presence of infection and extensive tissue damage, macrophages may be overwhelmed and die due to the number of dead pathogens and dead neutrophils
- Macrophage
- Derived from embryonic stem cell
- Becomes resident tissue macrophages
- First detects infections
- Becomes activated and recruit neutrophils and other leukocytes to the site of infection for the innate response by secreting cytokines
- General scavenger cells
- Dendritic cells
- Determines whether and when the innate response needs support from adaptive response
- Carry intact and degraded pathogen to lymphoid organs = initiates adaptive response
- Mast Cells
- Resident in all connective tissue
- Plays a role in inflammation
- Violent spasms of smooth muscle to eject parasites from respiratory and GI tracts
Lymphoid Lineage
- Natural killer cells
- Defense against viral infections
- Recruited by macrophages
- Secretes cytokines that impede viral replication
- Small lymphocytes circulate in inactive forms
- B cells
- Cell surface receptors: immunoglobulins
- Effector cells: plasma cells = soluble antibodies
- Can recognize antigen in epitopes
- T cells
- T-cell receptors
- Cannot recognize antigen alone
- Antigen must be bound to major histocompatibility complex molecule
- “presents” antigen to the T cell receptor
Effector Cells of Adaptive
- Plasma cells
- Production of antibodies
- Circulate in blood
- Enters tissue
- Binds to antigen
- Cytotoxic T cells
- Expresses CD8 receptors on its surface
- Kill cells infected with intracellular pathogen
- Helper cells
- Expresses CD4 receptors
- Secrete cytokines to help activate other cells
- Regulatory T cells
- Inhibit immune response by closing CD4 and CD8 responses
- Prevents unnecessary tissue damage
Humoral Immunity
- Immunity due to antibodies
- For extracellular microbes
- Facilitate engulfment and destruction of microbes and toxins by phagocytes
- Neutralization: binds to pathogen and inhibit growth, replication, or interaction with human cell receptors
- Phagocytes can bind to antibody molecules
- Opsonization: Antibody can coat bacteria to enhance phagocytosis
Lymphoid Tissues
- Major lymphoid organs: bone marrow, thymus, spleen, adenoids, tonsils, appendix, lymph nodes, and Peyer’s patches
- Primary lymphoid tissues: where lymphocytes develop and mature
- Secondary lymphoid tissues: where mature lymphocytes become stimulated
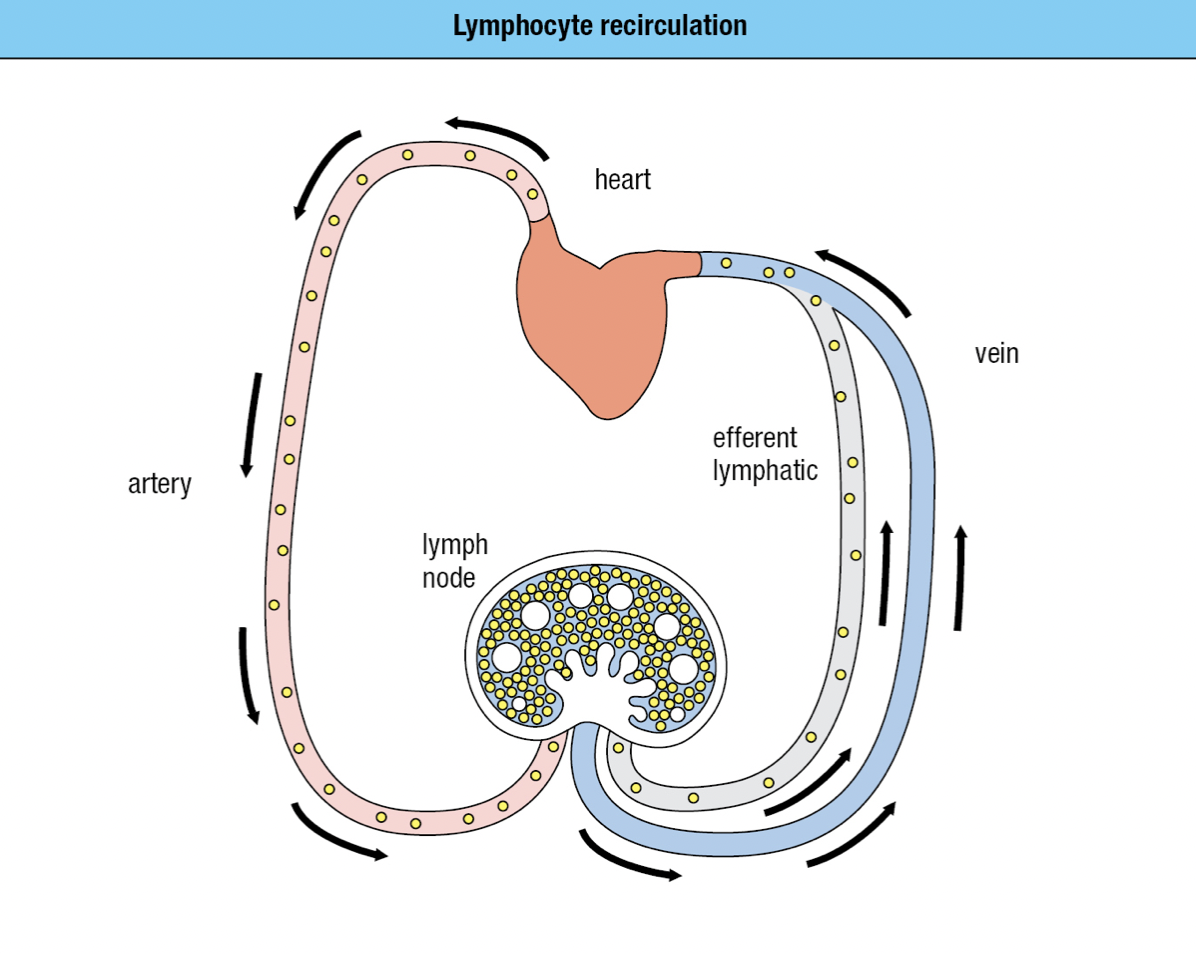
- Forms a network of lymphatics and collect the leaked plasma from the blood and returns it to the circulation as lymph via the thoracic duct and empties into the left subclavian vein
- No pump = sluggish flow = away from peripheral tissues = driven by continual movements
- Or else edema
- B cells and T cells move through both blood and lymph
- If they become activated by a pathogen, they remain in the lymph node
- Otherwise they leave in the efferent lymph and return to the blood
- Continual state of flux = lymphocyte recirculation
- To monitor the secondary lymphoid tissues for infection
Secondary Lymphoid Tissues
- To establish infection, a microorganism must colonize a tissue and overwhelm the local innate immune response
- Dendritic cells that are either pathogen-infected or loaded with pathogen antigens are carried in the lymph through lymphatics to the nearest lymph node (draining lymph node)
- Prevents potentially dangerous materials from entering circulation
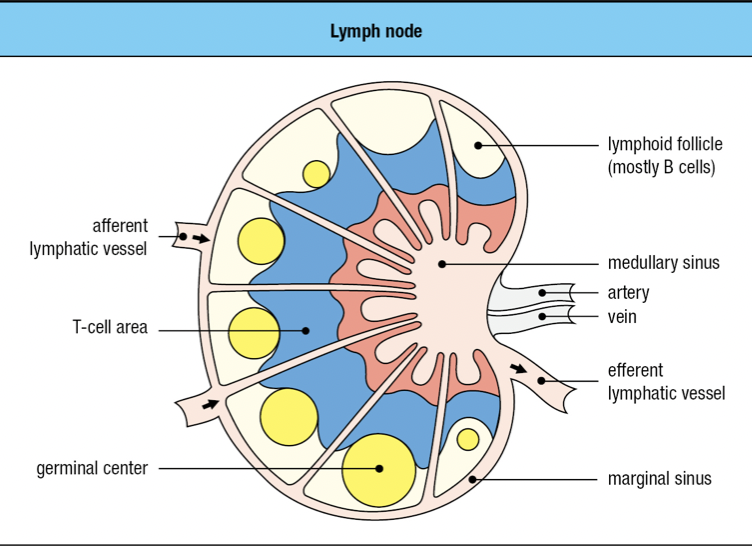
- Lymphoid follicles
- Where lymphocytes are segregated into B cell areas and T cell areas
- Pathogens and antigen-laden DC in lymph enter via afferent lymphatic vessel
- Lymph is drained via the efferent lymphatic vessel
- Dendritic cells stay in the node
- Allows pathogens and other extraneous materials to be extracted by the resident macrophages
- Activated B cells proliferate in the node
- Forms germinal center
- T cells are activated by antigen-bearing DC
- Some become Helper T cells and help B cells activate into plasma cells in the node
- Lymphocyte activation, proliferation, and differentiation lead to swelling of lymph node

ADAPTIVE IMMUNE SYSTEM
- Antibody Responses
- B lymphocytes or B cells
- Plasma cells
- When B cells encounter an antigen, they become plasma cells
- Antibodies/ immunoglobulins
- Cell-mediated immune responses
- T lymphocytes or t cells found in all cells except RBCs
- Major histocompatibility complex and T-Cell receptors
Spleen
- Filters blood to remove damaged and senescent RBCs
- Also a secondary lymphoid tissue that defends against blood-borne pathogens
- Splenic macrophages and DCs
- Stimulation of B cells and T cells from the blood
- Red pulp: blood cells are monitored and elderly or damaged ones are removed
- White pulp: similar to lymph node but without lymphatics
Mucosa-associated Lymphoid Tissue (MALT)
- Secondary LT of mucosal tissues
- Pathogens arrive by direct delivery mediated by M cells of the mucosal epithelium
Gut-associated LT (GALT)
- Tonsils, adenoids, appendix, Peyer’s patches of the GI tract
Bronchial-associated LT (BALT)
- Lines the respiratory epithelium
CHAPTER 2
IMMEDIATE INNATE RESPONSE
- First line of defenses
- Physical barriers
- Molecular mechanisms of innate immunity
- Second line of defense
- Induced mechanisms of innate
- mobilization of cells upon detection of infection
- Third line of defense
- Adaptive immune response
Barriers
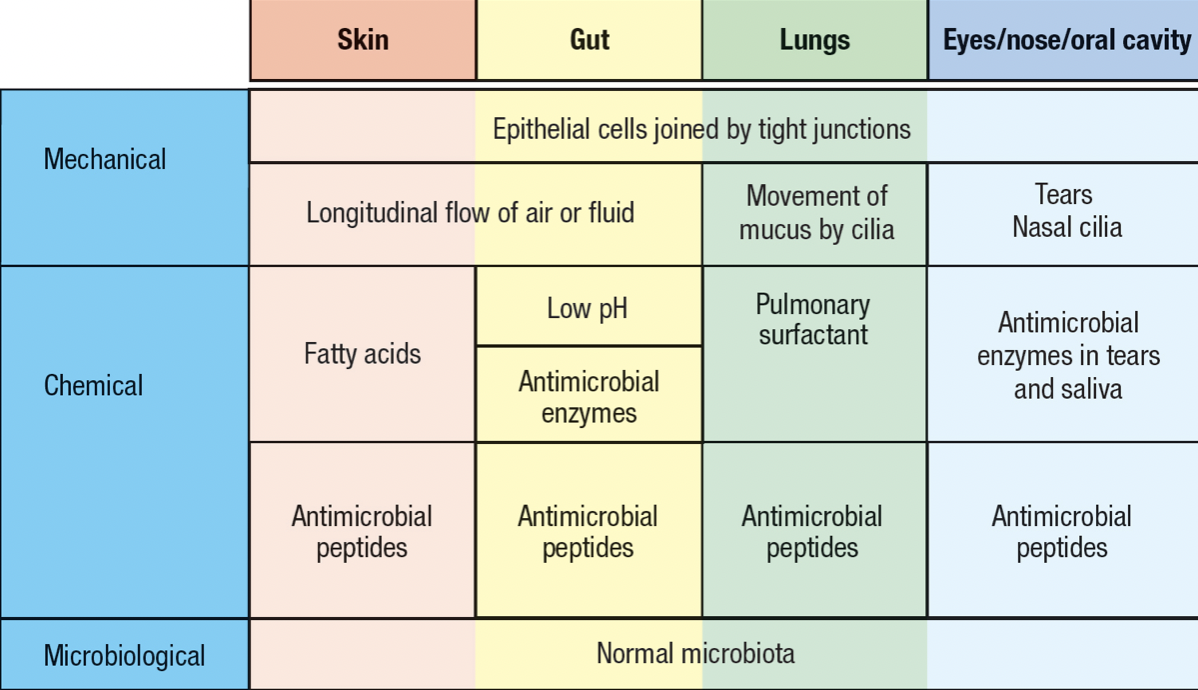
- Skin and mucosal epithelia
- Commensal microorganism = microbiota
- Deters pathogen invasion
- Invading pathogen must compete with the resident commensals for nutrients and space
- Extracellular pathogens
- Accessible to soluble secreted molecules of immune system
- Intracellular pathogens
- to combat, human cells are killed to interfere with the pathogen’s life cycle
- expose any pathogen released from dead cells to soluble molecules
Complement System
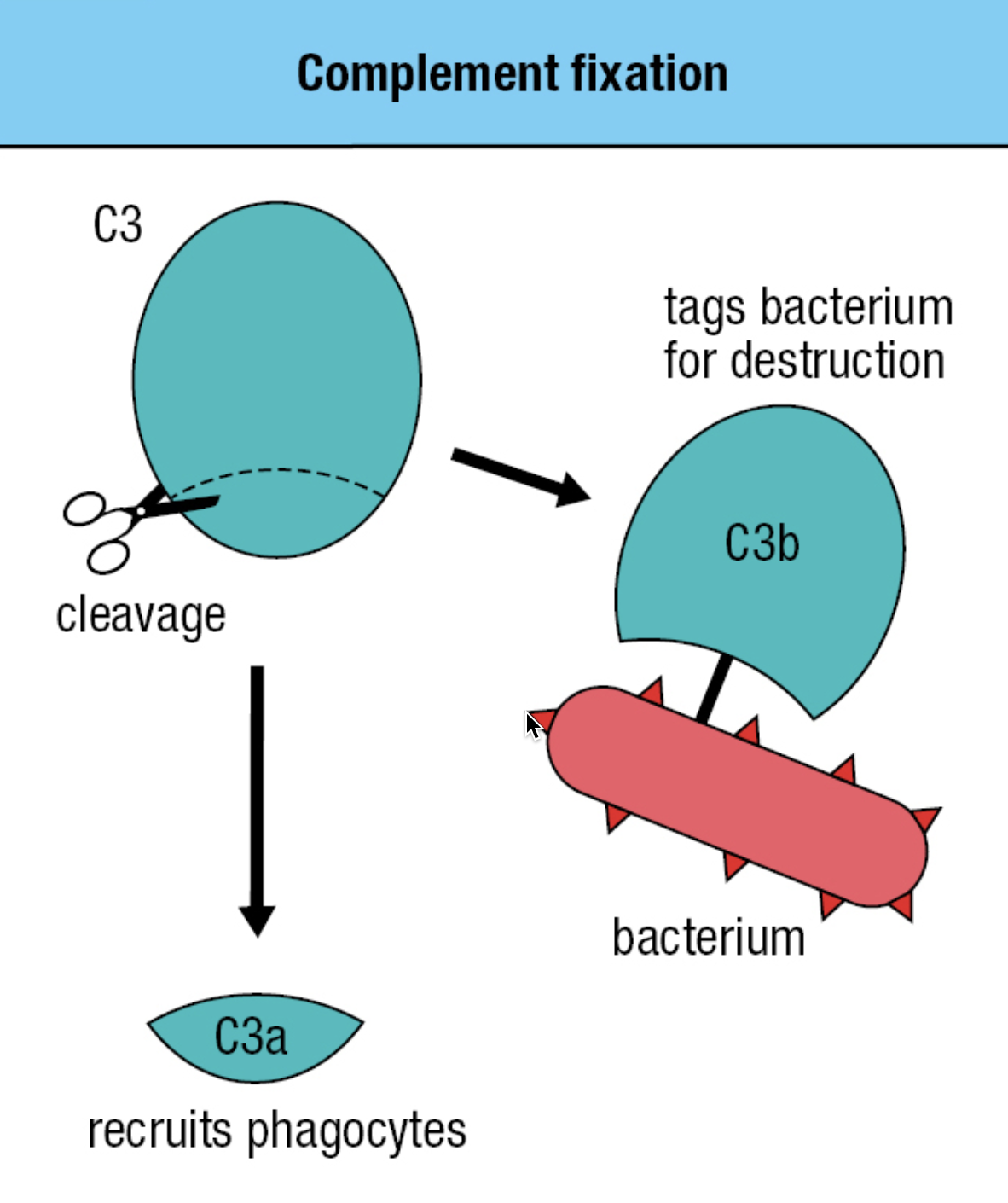
- plasma proteins made in the liver, present in blood
- induced when tissue becomes infected
- opsonizes bacteria and extracellular virus particles
- has proteases that circulate in the blood, lymph, and tissues
- inactive form = zymogens
- Protease cleaves and activates the next protease
- C3 most important proteins of the complement
- Those lacking C3 are prone to severe infections
- Has thioester bond with glycoprotein
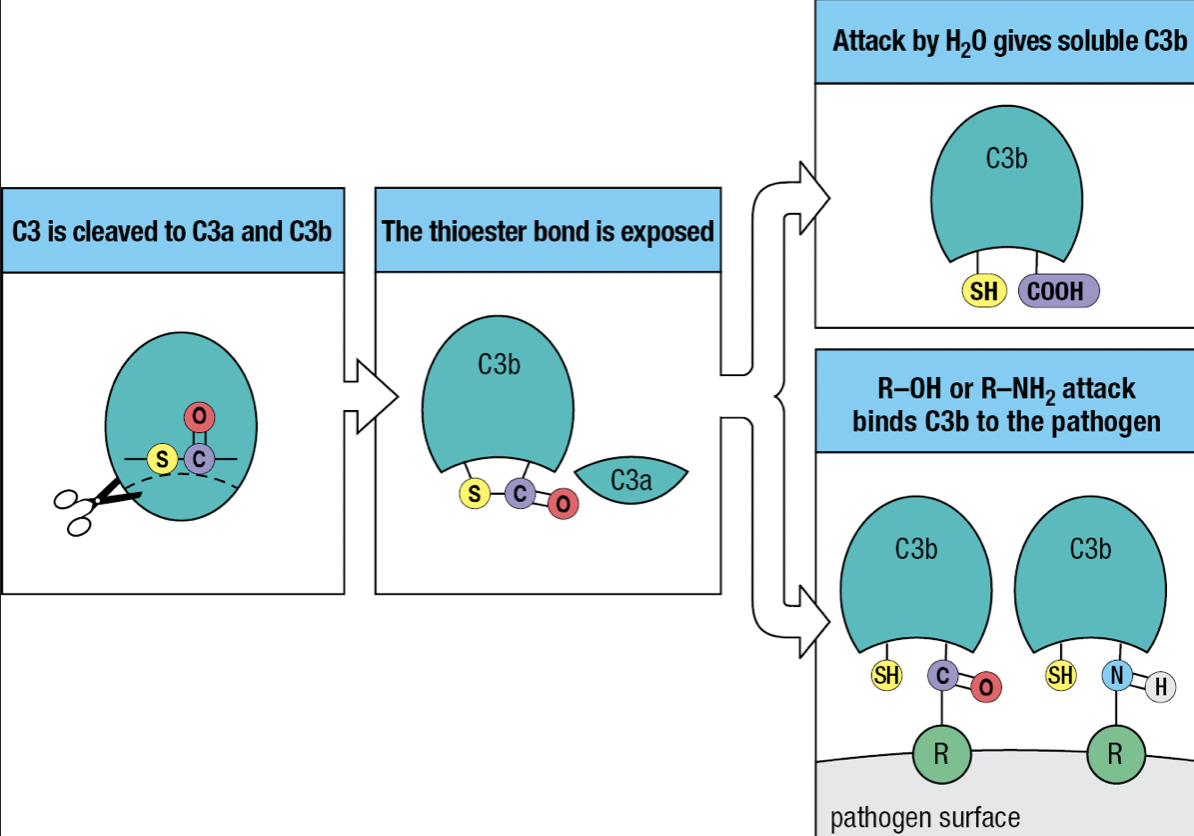
- Activation of the complement system by infection = cleavage of C3 into C3a and C3b
- C3b attaches to the pathogen’s surface = complement fixation
- Tags the pathogen for phagocyte-mediated destruction
- Organize the formation of protein complexes that damage the pathogen’s membrane
- C3a = acts as a chemoattractant to recruit phagocytes and other effector cells from the blood
- The thioester bond is exposed to the hydrophilic environment upon cleavage
- Most will hydrolyze but a minority will react with the hydroxyl or amino group on the pathogen’s surface
Pathways
- All lead to C3 activation, deposition of C3b and recruitment of effector mechanisms

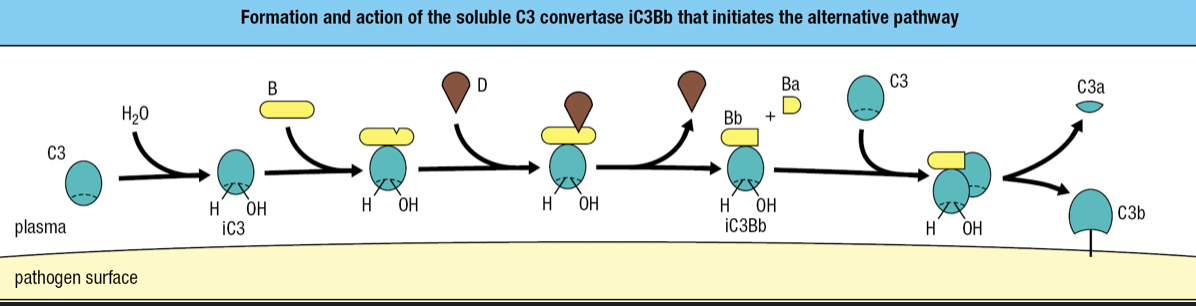
Alternative Pathway
- Start of infection
- Binding of C3 with water = iC3
- iC3 binds to factor B and is susceptible to cleavage by factor D
- fragment Ba is released while fragment Bb with protease activity remains bound to iC3 = iC3Bb complex
- iC3Bb cleaves C3 into C3a and C3b
- high concentration of C3 in blood = activation and cleavage of C3 in large quantity = some C3b will bind to pathogen’s surface
- C3 convertase = proteases that cleave and activate C3
- iC3Bb is an example
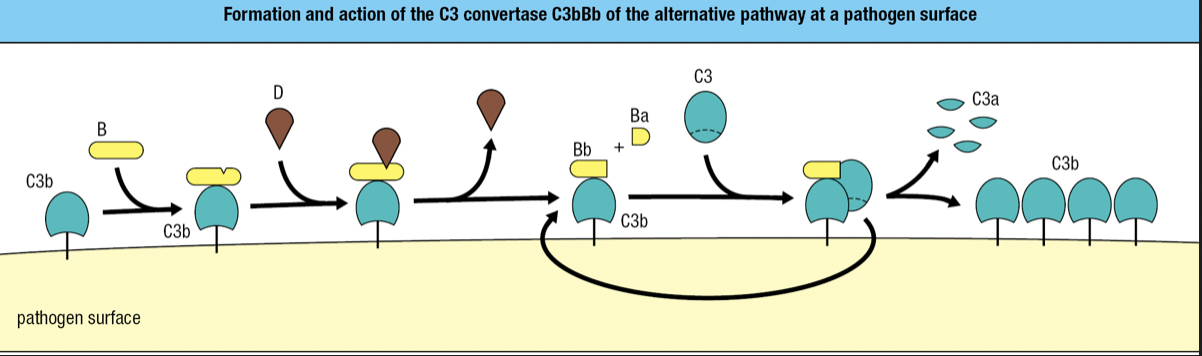
- C3b fragments bound to a pathogen also binds to factor B and cleaved by factor D
- Forms C3Bb complex
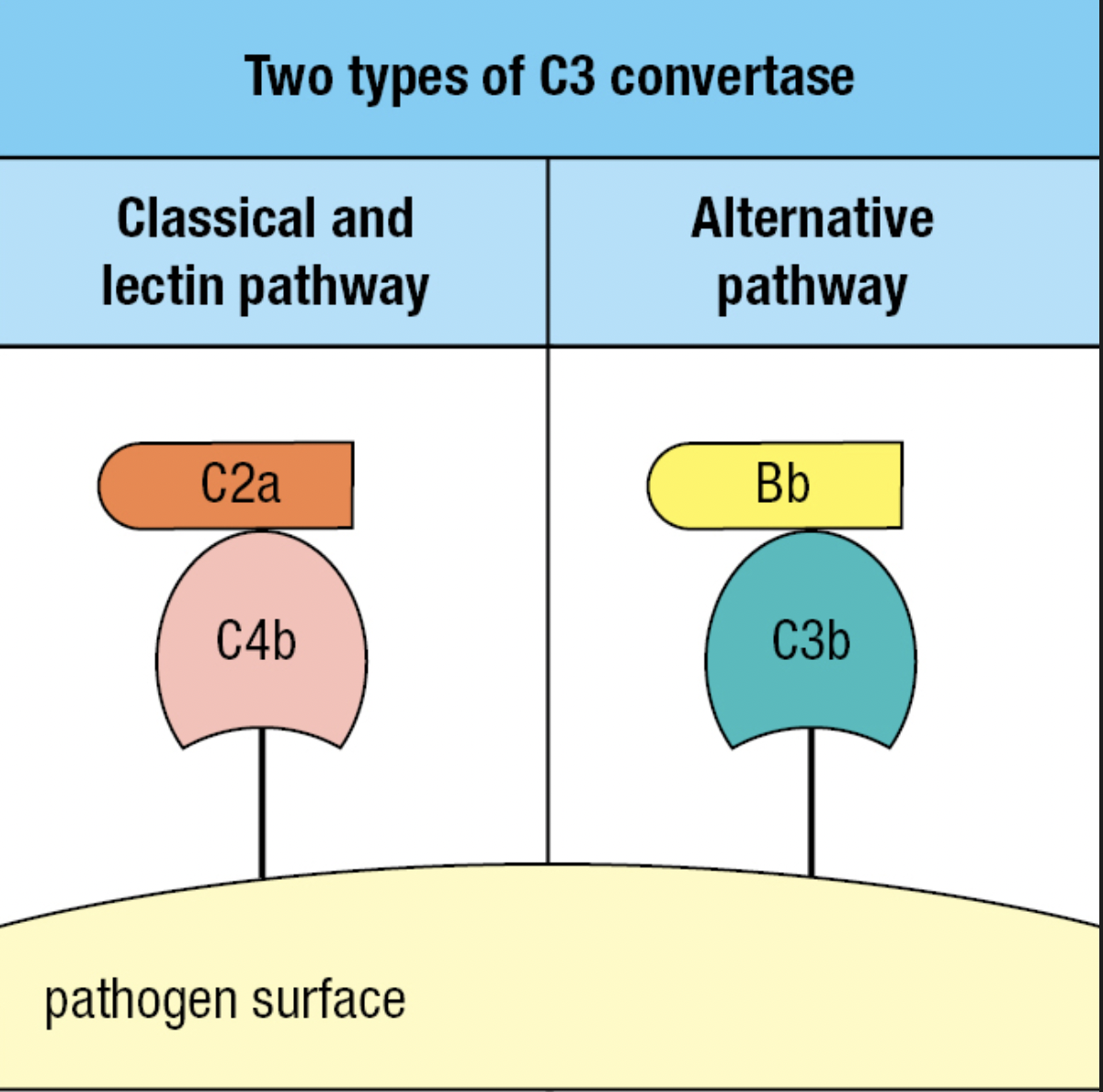
- The C3 convertase of the alternative pathway
- Works right at the pathogen surface
- Binds C3 and cleaves into C3a and C3b
- Because this convertase is present on the pathogen surface, it is more efficient in fixing C3b to the pathogen surface
- Rapid formation of additional C3Bb molecules
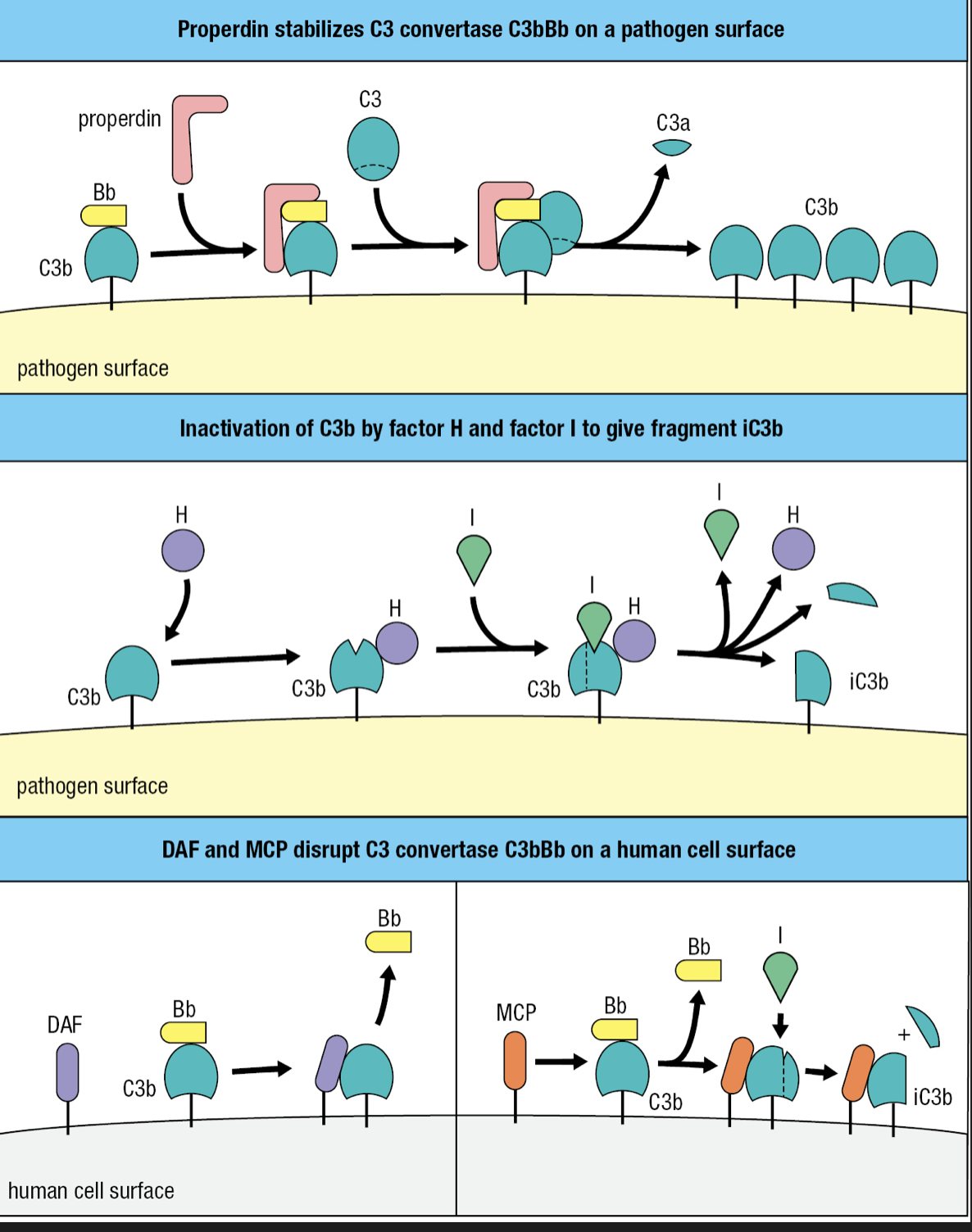
Regulatory Proteins
- Plasma protein properdin (factor P) increases complement activation
- Binds to C3Bb on pathogen surface and prevent its degradation by proteases
- Factor H plasma protein counters factor P
- Binds C3b and facilitates further cleavage to iC3b by factor I plasma serine protease
- Combined effects of factors H and I is to decrease the amount of C3 convertase on the pathogen surface
- Those who lack factor I = immunodeficiency
- Formation of C3Bb remains unregulated until the C3 reservoir in blood, extracellular fluid, and lymph is exhausted
- When faced with bacterial infections, individuals with factor I deficiency fix very little C3b to bacterial surfaces, causing inadequate clearance of bacteria by phagocytes
- More susceptible to ear infections and abscesses caused by encapsulated polysaccharide bacteria
- Decay-accelerating factor (DAF)
- Binds to C3b component of the alternative C3 convertase
- Causes its dissociation and inactivation
- membrane cofactor protein (MCP)
- Binding of MCP to C3b makes C3b susceptible to cleavage and inactivation by factor I
- Complement control protein (CCP) modules
- DAF, MCP, factor H
- Proteins made up of CCP modules = regulators of complement activation
- The combined effect of the reactions that promote and regulate C3 activation is to ensure that C3b is deposited only on pathogen surface and not on human cells
- Effective way of distinguishing human cells from microbial cells
Macrophage
- First effector cell to encounter invaded tissue
- Prevalent in connective tissues, linings of GI and respiratory tracts, liver (Kupffer cells), and alveoli of lungs
- Macrophage cell surface receptors enhances phagocytosis
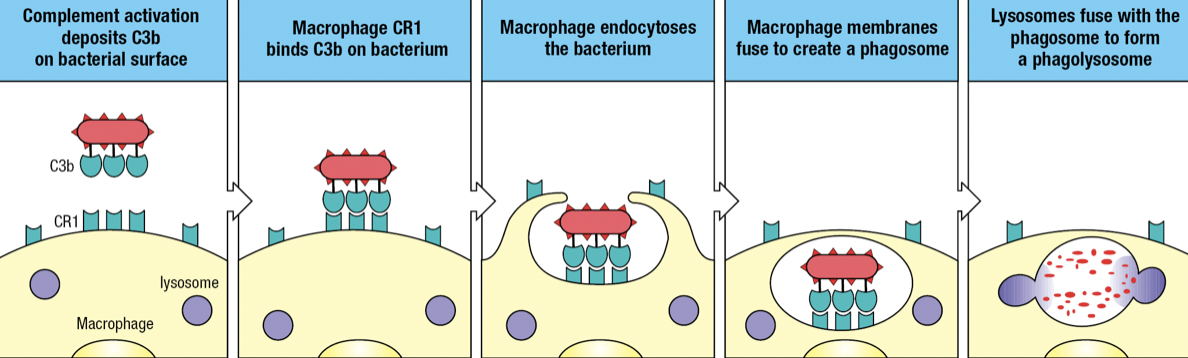
- Complement receptor 1 (CR1)
- Binds to C3b fragments on pathogen surface
- Facilitates engulfment of the pathogen into endosome or phagosome
- Fuses with lysosome which delivers toxins and enzymes to degrade the pathogen
- Opsonization
- Protects the surface of cells on which it is expressed
- Disrupts C3 convertase by making C3b susceptible to cleavage by factor I
- Made up of CCP modules
- CR3 and CR4
- Examples of integrins which contributes to adhesive interactions
- Bind iC3b fragments on microbial surfaces
- Although the iC3b fragment has no C3 convertase activity, it facilitates phagocytosis and pathogen destruction as a ligand for CR3 and CR4
- Combination of CR1, CR3 and CR4 > single complement receptor
Terminal Components
- C5, C6, C7, C8, C9 proteins
- Cooperate to form the membrane-attack complex (MAC)
- Large pore assembled in the pathogen membrane to disrupt its integrity
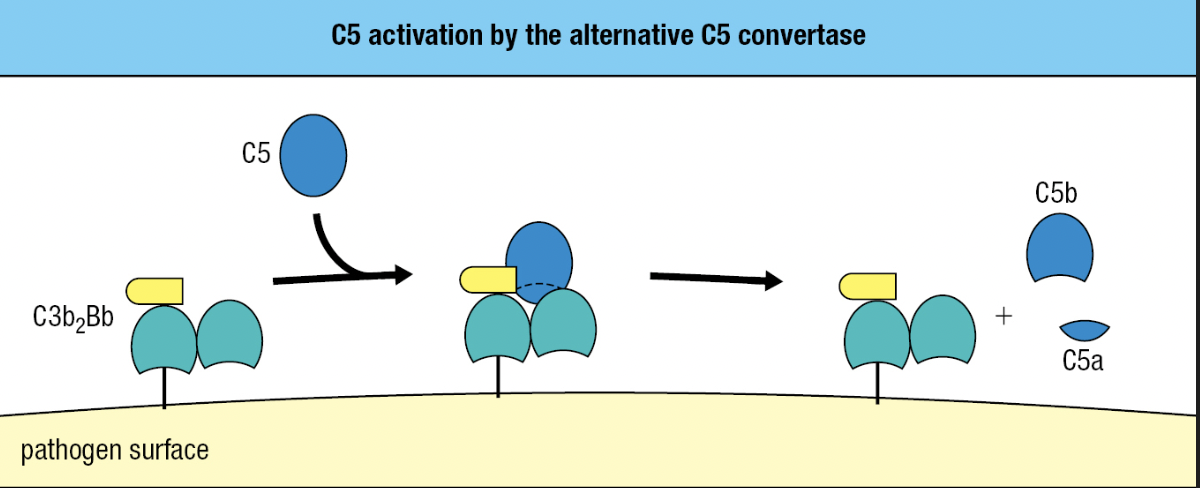
- C5
- Similar to C3 but lacks a thioester bond
- Activated by the alternative C5 convertase
- Has 2 fragments of C3b and one Bb fragment = C3b2Bb
- Cleaves C5 into C5a and C5b
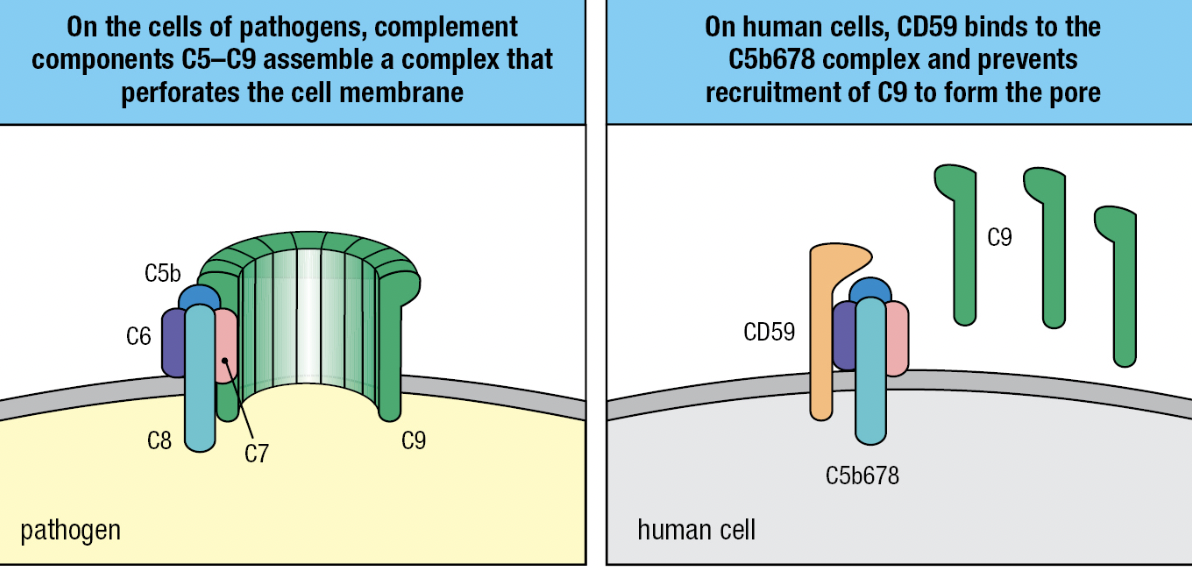
- C5b
- Initiates MAC formation
- C6 and C7 binds to C5b = forms complex to to expose hydrophobic region of C7
- C7 inserts into the lipid bilayer
- C8 joins complex
- C8 binds to C5b to expose its hydrophobic site and inserted into the membrane
- These events lead to the polymerization of 18 C9 molecules and MAC formation
- Final complex: one molecule each of C5b, C6, and C7, three molecules of C8, and 18 molecules of C9
- C5b

Soluble Proteins
- S protein, clusterin and factor J
- Prevent association of the C5b, C6 and C7 complex with the cell membranes
- Homologous restriction factor (HRF) and CD59 (protectin)
- Prevents C9 recruitment to the complex by binding to C5b678 complex
- Impaired synthesis of the glycosylphosphatidylinositol tail CD59 = human disease paroxysmal nocturnal hemoglobinuria (PNH) = complement-mediated lysis of RBCs which have no DAF, HRF, or CD59 = not protected from actions of the complement
- Treatment: monoclonal antibody specific for C5 and prevent its cleavage and activation by C5 convertase

Inflammatory Peptides
- C3a and C5a fragments are ligands for receptors on phagocytes, endothelial cells, and mast cells
- Increases inflammation at the site of complement activation
- Can induce anaphylactic shock = C3a and C5a are anaphylatoxins
- C5a more potent and stable than C3a
- Induces contraction of smooth muscle and degranulation of mast cells and basophils
- Release of histamine and other vasoactive substances that increase capillary permeability
- Increases exit of plasma and cells from the blood
- C5a induces neutrophils and monocytes to adhere to vessel walls and recruit phagocytes
- C5a increases CR1 and CR3 on surfaces
Plasma Proteins
- Vessels damaged by pathogens activate the coagulation system
- Enzymes in plasma that cooperates with platelets to form blood clots
- Pathogens are immobilized in the clots and cannot enter the blood and lymph
- During clotting, platelets degranulate and release active agents to recruit immune cells, antimicrobial defenses, and tissue repair
- Kinin system
- Enzymatic cascade of plasma proteins induced by damaged tissue
- Leads to the production of bradykinin
- Reduces hypertension and promotes vasodilation and smooth muscle relaxation
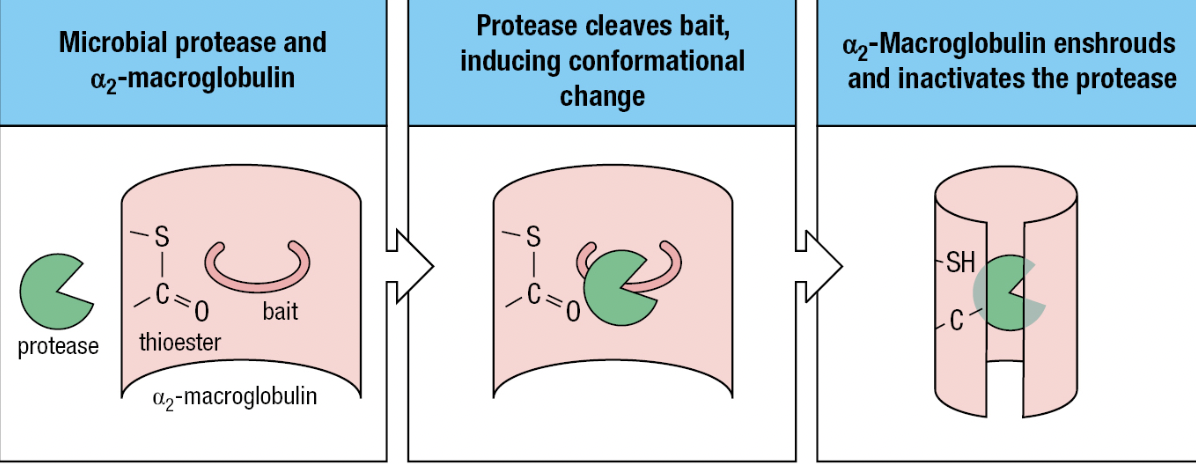
- Protease inhibitors
- Contained in human secretions and plasma
- For pathogens that have acquired proteases (i.e., plasmin) to hide from the immune system
- Ex: alpha2-macroglobulin
- Lures microbial protease with its “bait” region to be cleaved by the protease
- Activates the thioester in a2-macroglobulin = binds and envelops the protease
- Immediately bound by specific receptors on macrophages, hepatocytes, and fibroblasts
Defensins
- Antimicrobial peptide that can neutralize broad range of structurally diverse toxins
- Alpha-defensins and beta-defensins
- Amphipathic (has hydrophobic and hydrophilic regions to penetrate membranes) = pore formation
- When binding to microbial toxin, it promotes unfolding of the toxin = susceptible to proteases = anti-chaperones
Pentraxins
- Plasma proteins that bind microorganisms and deliver them to phagocytes
- Same role as antibodies
CHAPTER 3
INDUCED INNATE RESPONSES
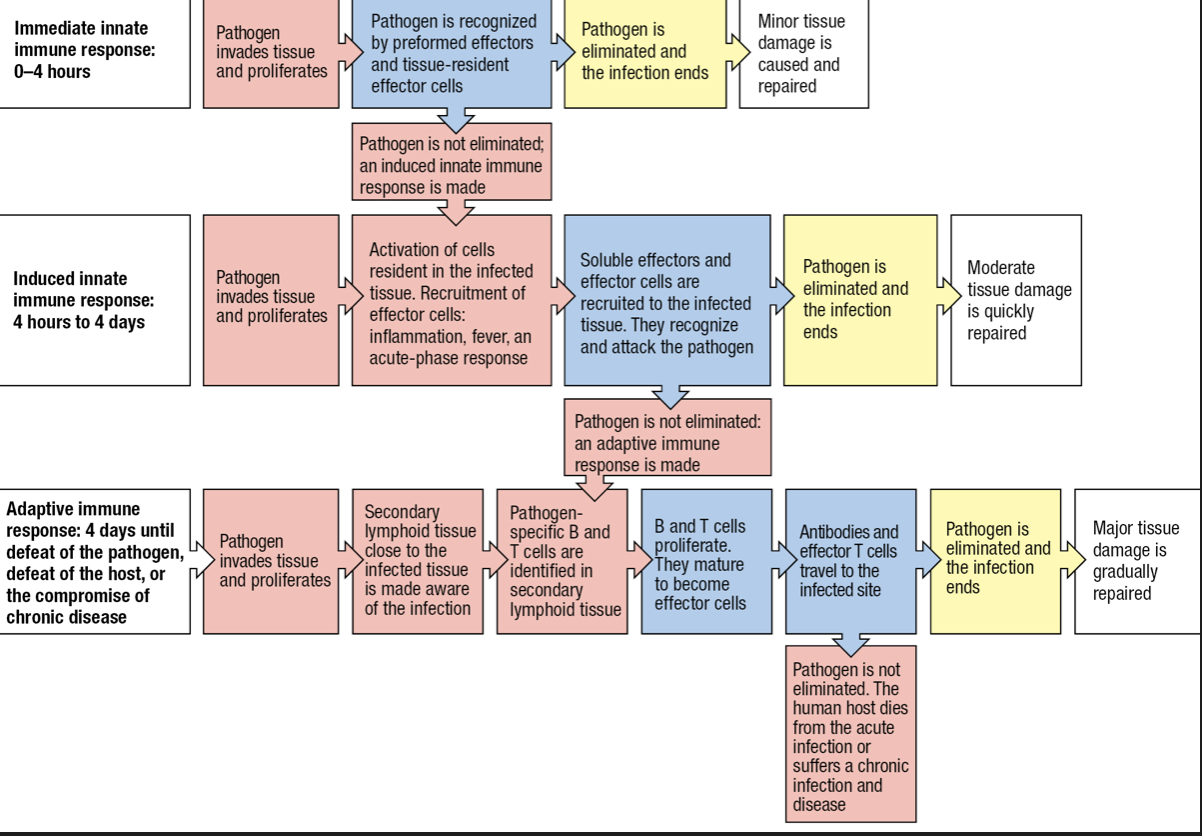
Inflammation
- Induced phase involves soluble and cellular receptors that detect the presence of a pathogen and recruit leukocytes to make an inflammatory response
- Inflammation will help induce a subsequent adaptive immune response
- Macrophages activated by the presence of an infection will release inflammatory cytokines
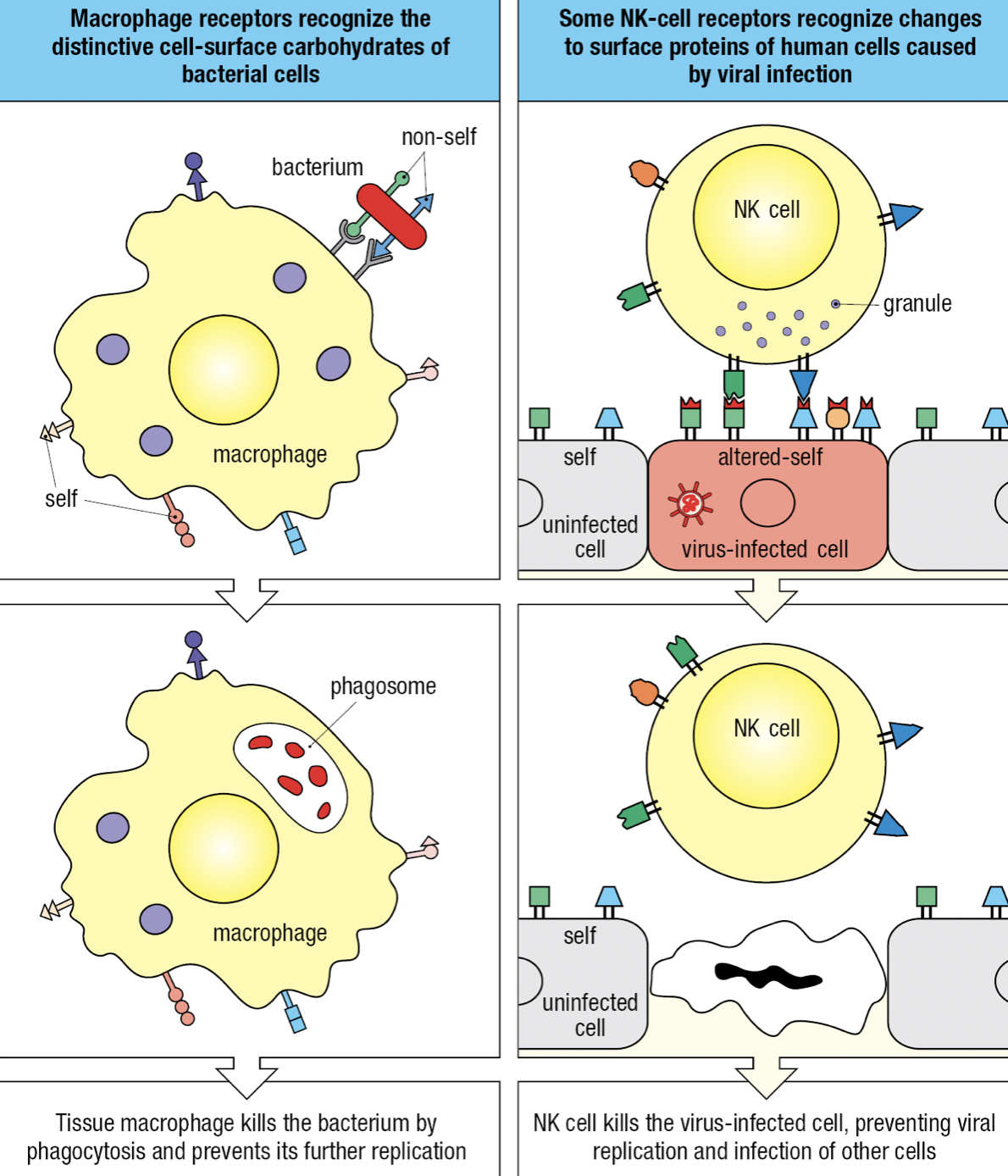
Self vs Non-Self vs Altered Self
- Receptors can recognize structural features that distinguish microbial macromolecules from human macromolecules
- Non-self: microbes
- Altered-self: infected cells and cancerous cells
- Individual cells express different combinations of receptors = functional diversity = increases likelihood that at least some of the cells will be able to mount an effective response to any given pathogen
Lectins
- Carbohydrate-specific receptors found in macrophages
Surface Receptors of Resident Macrophages
- Receptor-mediated endocytosis = macrophages surface receptors capture pathogens and deliver them to endosomes where the pathogen is killed and partly degraded
- Endosome fuses with lysosome where extreme acidity and concentration of degradative enzymes completely degrades the pathogen
- Pattern-recognition receptors (PRRs): receptors that recognize a structural feature common to many different types of pathogen
- Structural feature on microbe is called pathogen-associated molecular pattern (PAMP)
- Damage to cells or tissue that does not involve an infection: Damage-associated molecular pattern (DAMP)
- Scavenger receptors: most of the PRRs of macrophages
- Trigger phagocytosis, cell adhesion and intracellular signaling to identify microbes and molecules and cause their elimination
- SR classes A-L (11 classes)
- Bind to many different ligands including surface components of microbes
- In the absence of infection, SRs remove dead and dying cells and unwanted macromolecules and cells that have died by apoptosis (damaged or infected cells)
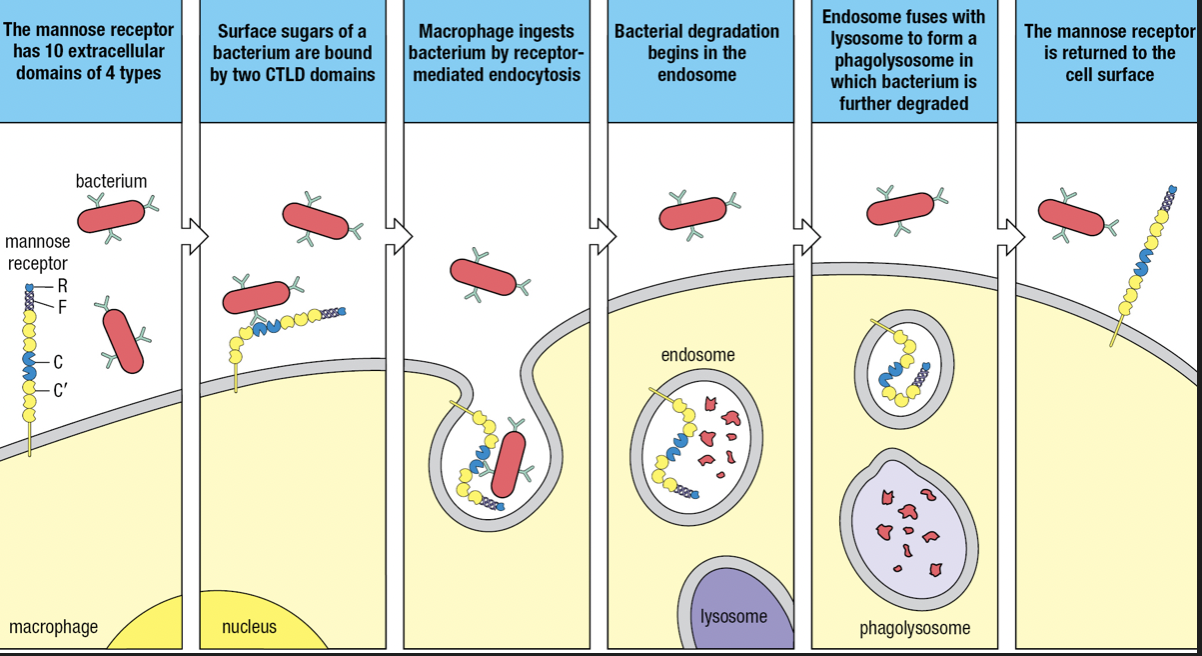
- Some SRs are lectins
- Mannose receptor (SR-E3) and Dectin-1 (SR-E2)
- MR recognizes mannose and other sugars present in the glycans of bacterial surfaces and internalized by receptor-mediated endocytosis
- Dectin recognizes beta-glucans (glucose polymers) on fungal and bacterial surfaces
- Belongs to C-type lectin domain (CTLD)
- Calcium ion coordinates the interaction of the carbohydrate ligand with the protein receptor
- Mannose receptor (SR-E3) and Dectin-1 (SR-E2)
- Some SRs are lectins
SR-A Class
- Macrophage receptor with collagenous structure (MARCO)
- Recognizes bacterial lipopolysaccharide (LPS) of the outer membrane of Gram-negative bacteria
- Also binds to Gram-positive bacteria
- SR-A1
- Recognizes LPS and lipoteichoic acid cell wall component of Gram-positive bacteria
- Additional ligands: hepatitis C virus, beta-amyloid, and heat shock proteins
CR3 and CR4
- Binds to iC3b
- Part of integrins that contribute to adhesive interactions between cells
- Recognizes LPS of E.coli, Salmonella, N. gonorrhoeae, and other Gram-negative bacteria
- Recognizes lipophosphoglycan of protozoan parasite
Toll-like Receptors
- Toll-like receptors (TLRs) = signaling receptors present on immune cells
- Responds to a range of microbial and viral products
- Main macrophage receptor for LPS = TLR4 which recognizes the lipid A component of LPS
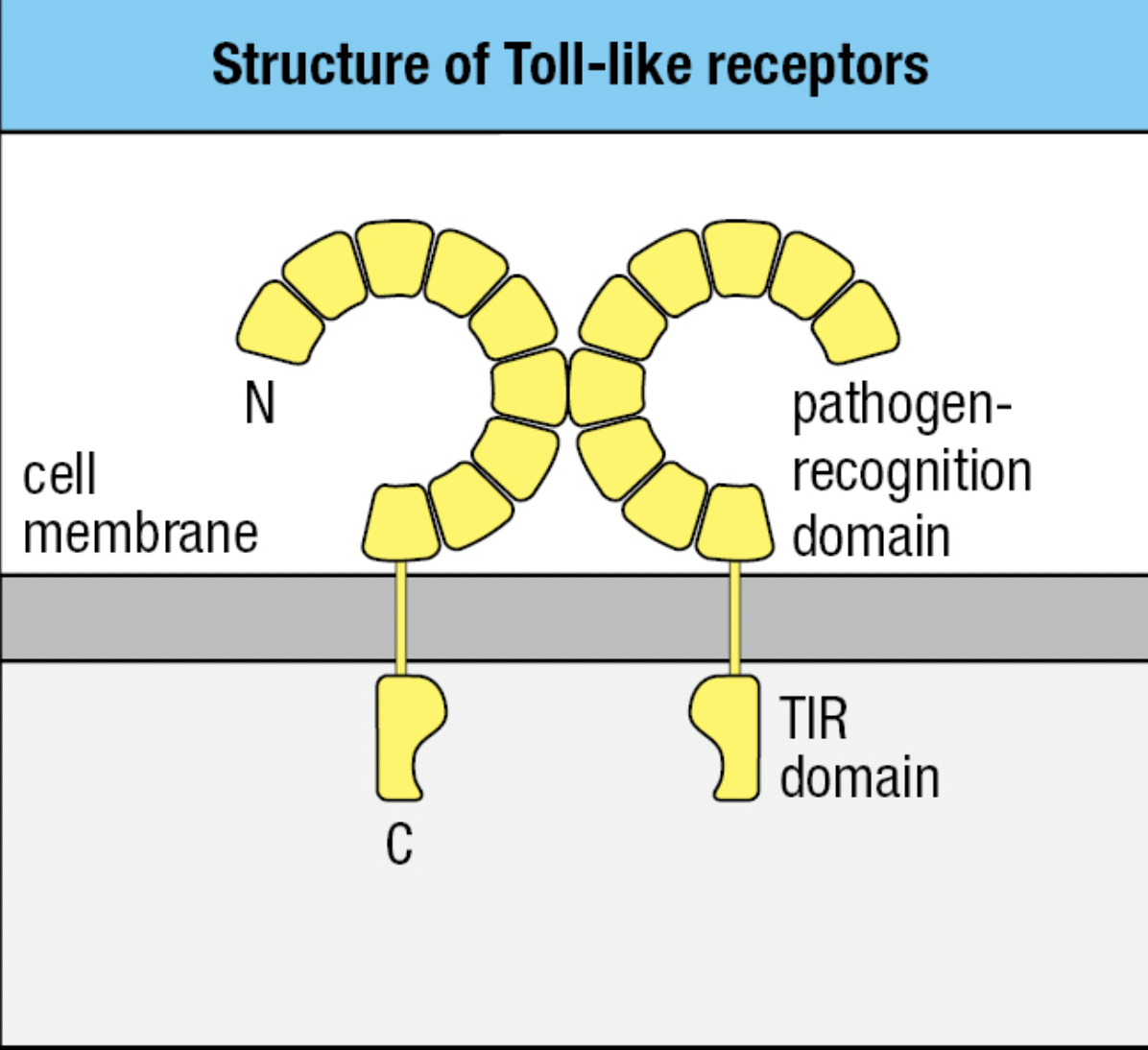
- TLR4
- Extracellular domain binds LPS
- Pathogen-recognition domain: has repeated leucine residues called leucine rich repeat region (LRR)
- Variation in the number of LRRs give each type of TLR a different ligand specificity
- Cytoplasmic domain signals macrophage to transcribe genes encoding proteins needed for innate immune response
- Signaling domain: Toll/interleukin-1 receptor (TIR) domain
- Can bind two molecules of LPS
- Extracellular domain binds LPS
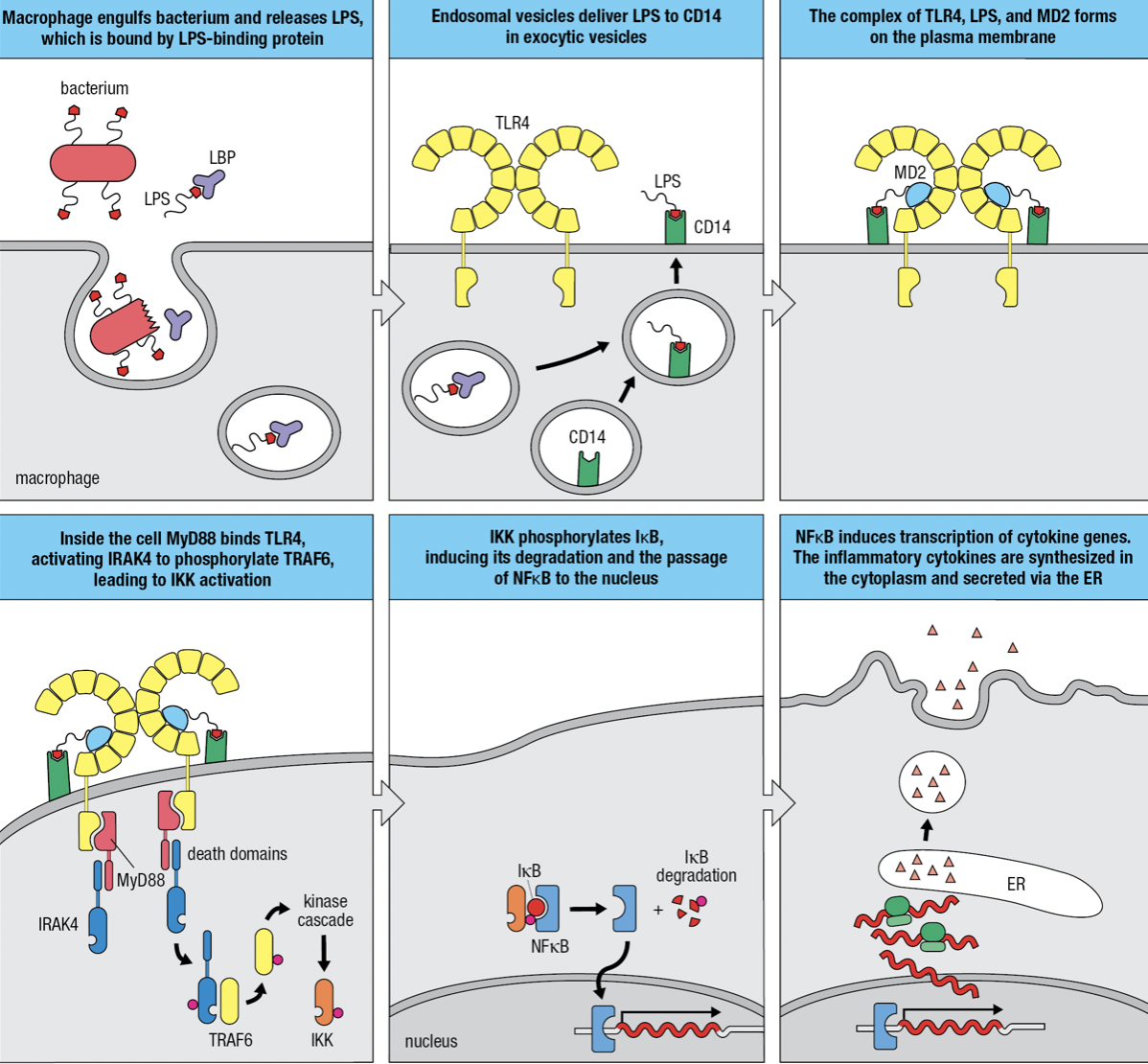
Gram-negative Infection
- Macrophages use mannose receptor to internalize and degrade bacteria and release LPS from the bacterial surface
- LPS is bound by LPS-binding protein and delivers it to CD14 at the macrophage surface
- TLR4 and myeloid differentiation factor 2 (MD2) forms a complex with CD14 and LPS
- MD2 only associates with extracellular domains of TLR4 but not other TLRs
- The TIR domain engages with the domain of MyD88 which acts as an adaptor protein
- The second domain of MyD88 engages with protein kinase IRAK4 (Interleukin-1 receptor-associated kinase 4)
- Induces IRAK4 to self-phosphorylate and dissociate from the complex and phosphorylate TRAF6 (tumor necrosis factor receptor-associated factor 6)
- Leads to the activation of inhibitor of kappa-B kinase (IKK) kinase complex
- IKK phosphorylates inhibitor of kappa-B (IkB)
- releases nuclear factor kappa-B (NFkB) to enter nucleus
- Initiates transcriptions of genes encoding cytokines and adhesion molecules
- When not needed, NFkB is held in the cytoplasm by IkB
- IkB degrades
NFkB Essential Modulator Deficiency (NEMO Deficiency)
- Loss of y subunit (NEMO) of IKK
- Impairs the activation of NFkB = impairs activation of macrophages by TLR4 signaling
- X-linked = males are more susceptible
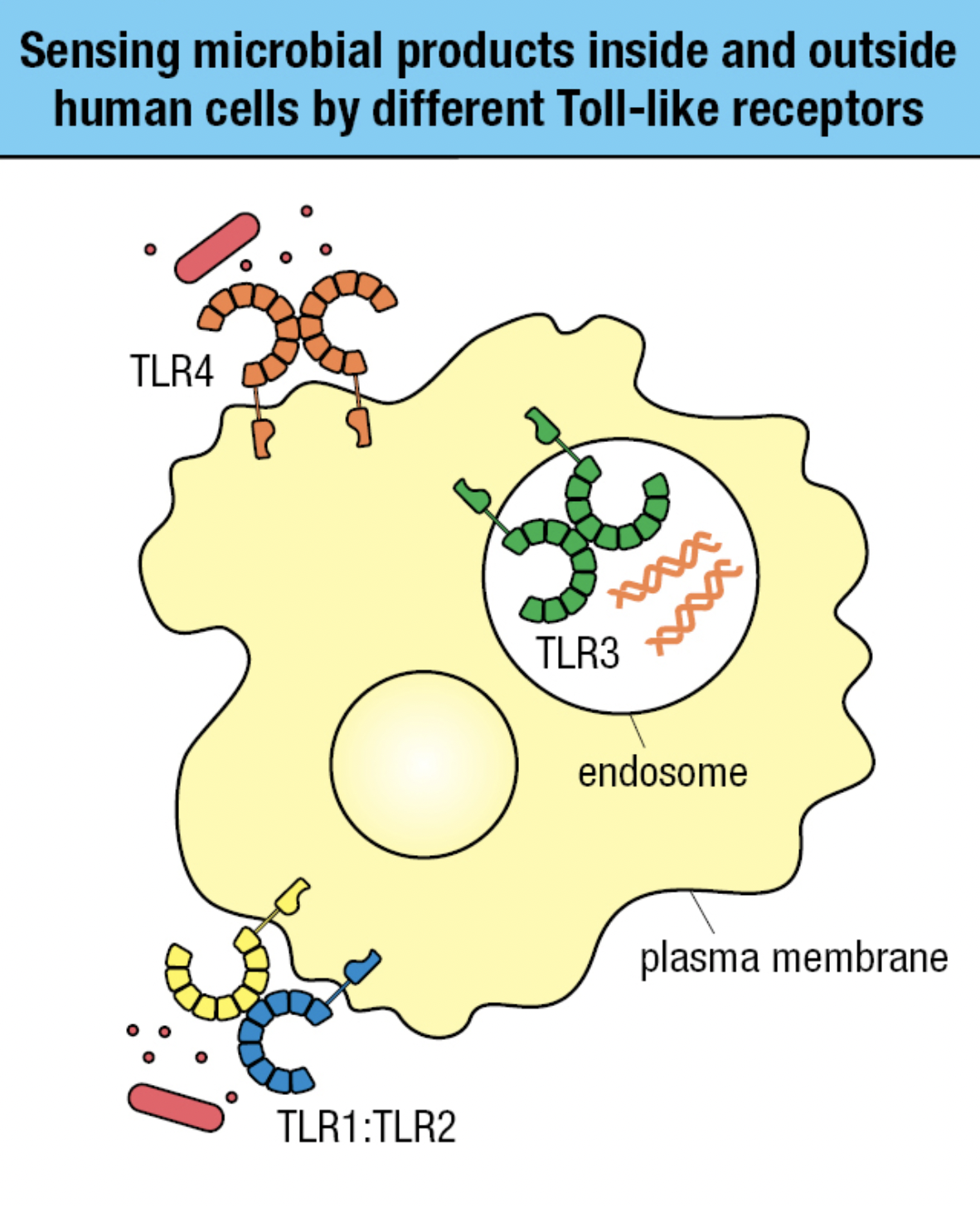
TLR Sensing
- Receptors on plasma membrane
- Recognizes carbs, lipid, and protein ligands on outer surface of pathogen
- Receptors within endosomal vesicle membranes
- Recognizes features of nuceic acids of pathogens and distinguish from human DNA and RNA
- TLR4 and TLR1:TLR2 sense bacterial infection
- TLR3 in endosomes sense viral infection
TLR4 Polymorphism
- Allotype: proteins encoded by different alleles of the same gene
- Causes genetic polymorphism
- More common in TLRs that recognize surface epitopes than nucleic acids due to greater diversity of carbs, lipids and proteins
- Septic shock
- Bacterial infection spreads from tissue to blood and becomes systemic
- Potent activation of the innate causes vasodilation and therefore leakage of fluid throughout the body
- Produces septic shock in which blood supply is perturbed and vital organs fail
- Caused by Gram-negative

NOD Proteins
- Intracellular
- Recognizes bacterial degradation products in cytoplasm
- Has three different regions
- Carboxy-terminal region = pathogen-recognition domain with binding site for degraded products
- Central region: nucleotide-binding oligomerization domain (NOD domain) which enables receptors to form oligomers
- Amino-terminal region = caspase-recruitment domain (CARD)
- contains binding site for RIPK2 (receptor-interacting serine-threonine protein kinase 2) that initiates signaling from NODs and acts as an adaptor molecule
- 1 in NOD1 and 2 in NOD2
- Mediates RIPK2 and NODs interaction
- Phosphorylation of RIPK2
- Activation of kinase TAK1
- Phosphorylation of IKK
- IKK activates NFkB
- Macrophage activation
- normally recruits caspases protease to protein complexes
Interferon Viral Response
- Cytoplasmic proteins that can detect viral nucleic acids and produce Type I Interferons
- Interferes with viral replication in the infected cell
- Instructs nearby uninfected cells to fight infection
- Alerts immune system that infection is present and causes virus-infected cells to be more vulnerable to NK cells
- In response to infection, one cell secretes cytokine that binds to a cytokine receptor on another cell
- Induces intracellular signals that change the behavior if second cell
- Makes contact with target cell before releasing cytokines to avoid unnecessary immune response damage to tissues
- When the cytokine and its receptor are both from same type of cell = give autocrine signals
- Cytokine and receptor are products of different cells: paracrine signals
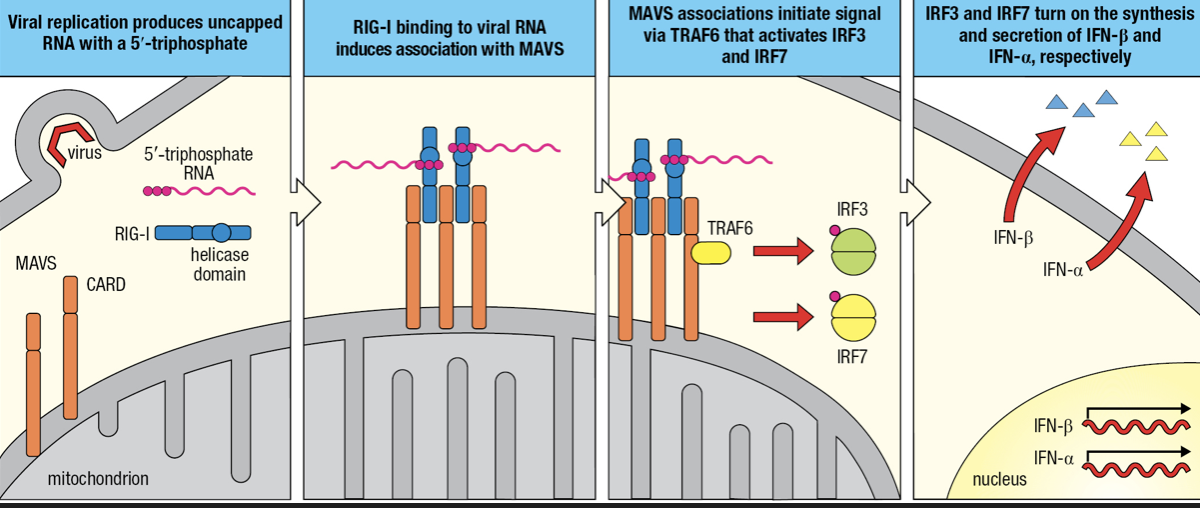
- RIG-I-like receptors (RLRs)
- Detects cytoplasmic viral RNAs
- Comprised of RIG-I and MDA-5
- Recognizes ds vRNA
- Two CARDs
- Interacts with mitochondrial antiviral signaling protein (MAVS)
- Forms a dimer on mitochondrial membrane
- MAVS engages TRAF6 which initiates signaling pathway to activation of IRF3 and IRF7
- IRF3 initiates IFN-beta gene granscription
- IRF7 initiates IFN-alpha gene transcription
- secretion of Type I interferons

- Secreted IFN-Beta maintains activation of infected cells (autocrine action) and binds to receptors on nearby uninfected cells (paracrine action)
- Infection induces phosphorylation of IRF3 to enter nucleus
- NFkB and IRF3 activates transcription of INF-beta gene followed by secretion of IFN-B
- Some IFN-B are bound by IFN-B receptors (autocrine)
- IRF7 is mobilized and induces production and secretion of IFN-a
- IFN-B receptors of uninfected cells bind to IFN-B secreted by infected cell (paracrine)
- Induces uninfected cell to secrete IFN-B and contribute to interferon response
Plasmacytoid Dendritic Cells
- Produce Type I IFNs
- Present in blood and lymph tissues
- Expresses TLR7
- Detects ss vRNA
- Expresses TLR9
- Detects presence of unmethylated CpG motifs in ds DNA
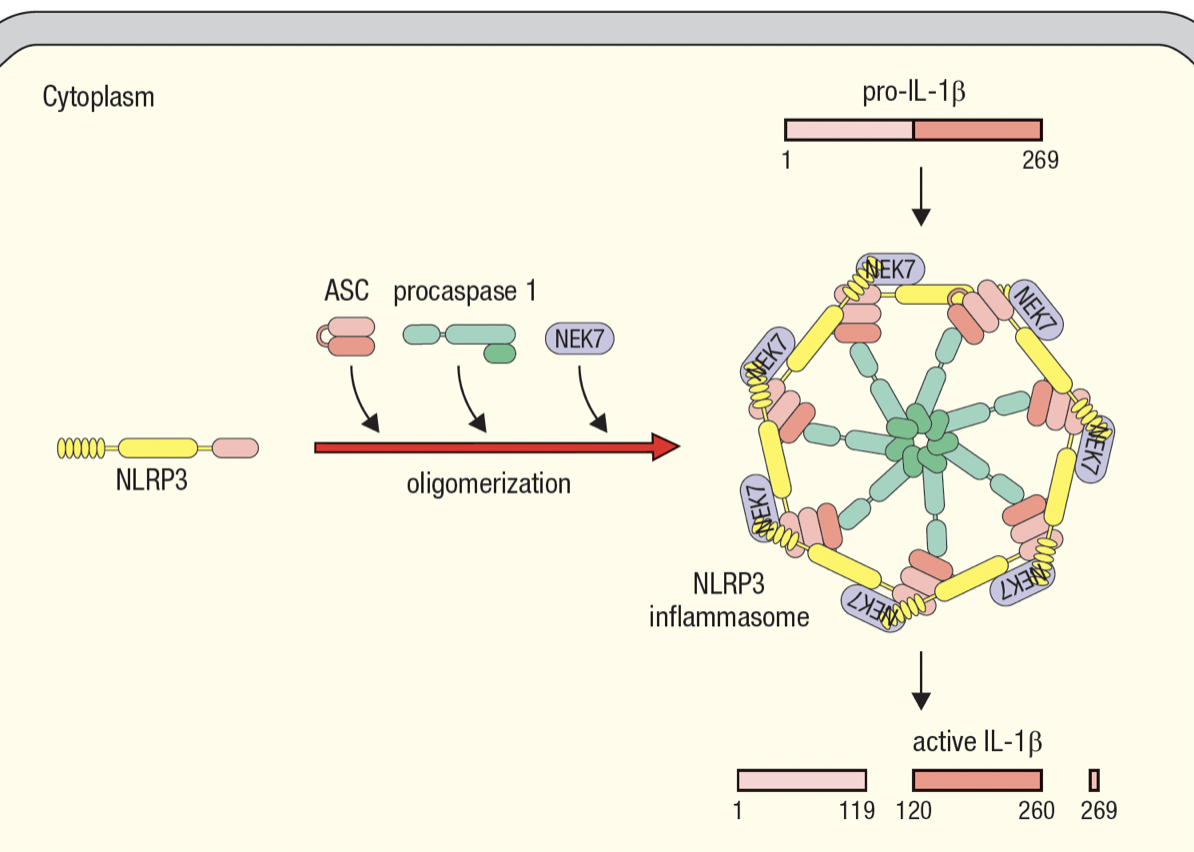
Inflammasomes
- Enable activated macrophages to release IL-1beta
- Interleukin-1B
- Master regulator of inflammation
- Inactive pro-form is stored in cytoplasm by macrophages
- Activated by cleavage and secreted in response to infection
- Macrophage assembles inflammasome to cleave pro-IL-1B into active IL-1B
- Inflammasome has NOD-like receptors (NLR) that detects infection
- NLR protein 3 basis for NLRP3 inflammasome
- same domains as NODs
- recruits molecules for oligomerization of inflammasome upon sensing infection
- procaspase 1 self cleaveage forms caspase 1 which cleaves pro-IL-1B
- Allows macrophage to respond to infection with large burst of IL-1B
- Gasdermin D
- To allow IL-1B to escape macrophage
- Upon sensing infection, and when pro-IL-1B is cleaved, caspase 4 cleaves gasdermin D
- Forms pores in PM where IL-1B leaves the dying macrophafe
- Pyroptosis = cytokine release, pore formation, death of macrophage
- IL-1a
- For homeostasis
- Always expressed by healthy cells
Autoinflammatory Diseases
- Chronic and recurrent bouts of systemic inflammation mediated by cells of innate immunity and do not involve adaptive immunity
Inflammation of Infected Tissue
- Attracts blood-borne immune effector cells
- Macrophage and neutrophils have distinct complementary properties
- Macrophage
- Long-lived
- Reside in tissues
- Work from the start of infection
- Raise the alarm
- Neutrophils
- Short-lived
- Dedicated killers that circulate in blood
- Awaiting call from macrophage and defend infected tissue
- First population of effector cells recruited to infected tissue

Release of IL-1B by Inflammasome
- Activates macrophages
- Two main effects
- Improve speed and efficiency in capturing and digesting pathogens
- Secretion of cytokines to recruit other effector cells
- Tumor necrosis factor-a (TNF-a) = vasodilation
- IL-6 = heat generation via metabolism of fat and muscle cells
- CXCL8 = chemokine to attract neutrophils
- CCL2 = chemokine to attract monocytes
- IL-12 = recruits and activates NK cells to secrete cytokines to enhance and maintain macrophage response

Recruitment of Neutrophils
- Macrophages release CXCL8 which guides neutrophils into site of infection
- Adhesion molecules on leukocyte surface and tissue-cell surface drives movement from blood and tissue
- In absence of infection, neutrophils move rapidly through capillaries and do not interact with vascular endothelium
- Within infected tissue, vasodilation and adhesion molecules of endothelial cells allow contact with endothelium
- Rolling adhesion
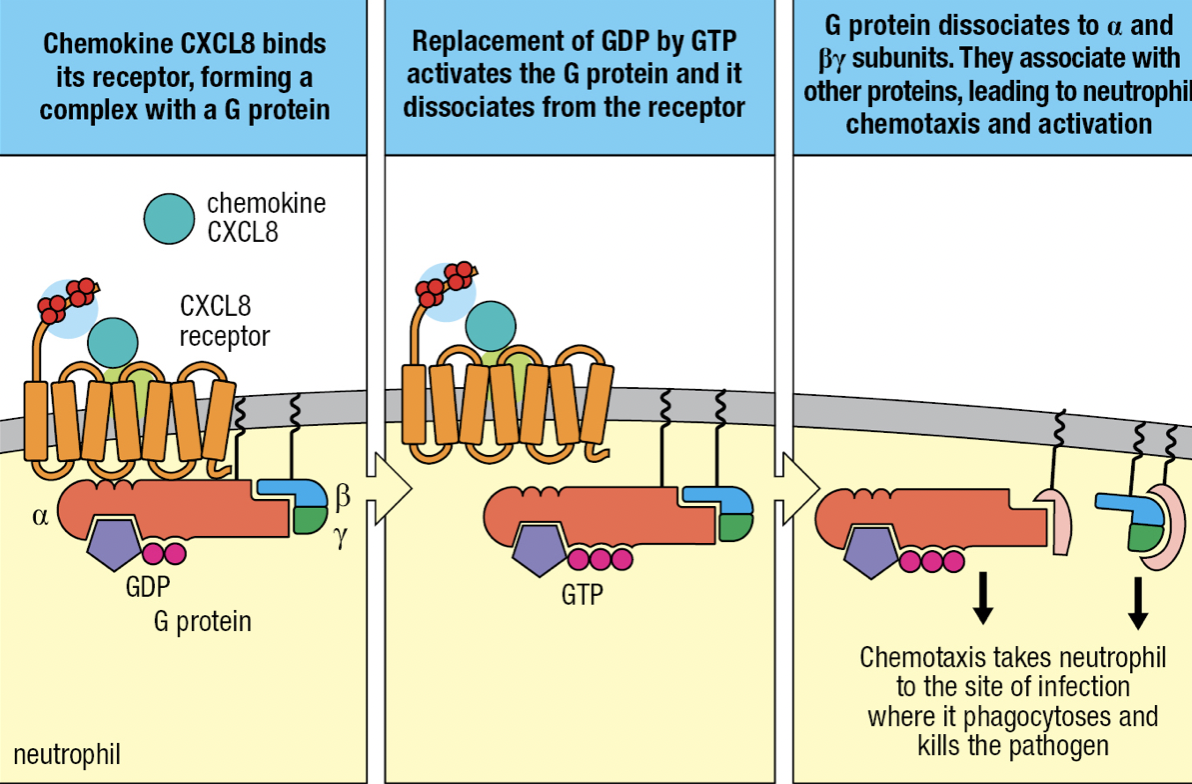
- TNF-a induces endothelium to express intercellular adhesion molecules 1 and 2 (ICAM-1 and ICAM-2) and neutrophils to express leukocyte function associated antigen-1 (LFA-1)
- ICAMs bind CR3 and LFA-1
- CXCL8 binds to neutrophil’s chemokine receptors CXCR1 or CXCR2
- Tight binding to immobilize neutrophil on endothelium
- CXCL8 binding with receptor
- Allows binding with G protein
- GDP is replaced by GTP
- Activates and releases G protein from receptor
- Neutrophil leaves blood by squeezing between cells = diapedesis
- Overall process of leaving the blood = extravasation
- Neutrophil migration is directed by gradient of CXCL8 bound to cell surface and ECM
- Neutrophil moves towards CXCL8 concentration at actuvated macrophage
- Pyogenic bacteria = pus-forming bacteria

Neutrophil Programmed Death
- They devote all their resources to storage and delivery of antimicrobial weaponry
- Has receptors for recognition: CR4 and CD14

Neutrophil Granules
- Order of loading of granules
- Azurophilic/ primary granules
- Specific/ secondary granules
- Gelatinase/ tertiary granules
- Secretory vesicles
- Order of degranulation/ releasing of granules is reversed
- Controlled by calcium concentration
- Low levels = sufficient for secretory vesicles
- Higher level in tissue = specific and gelatinase degranulation
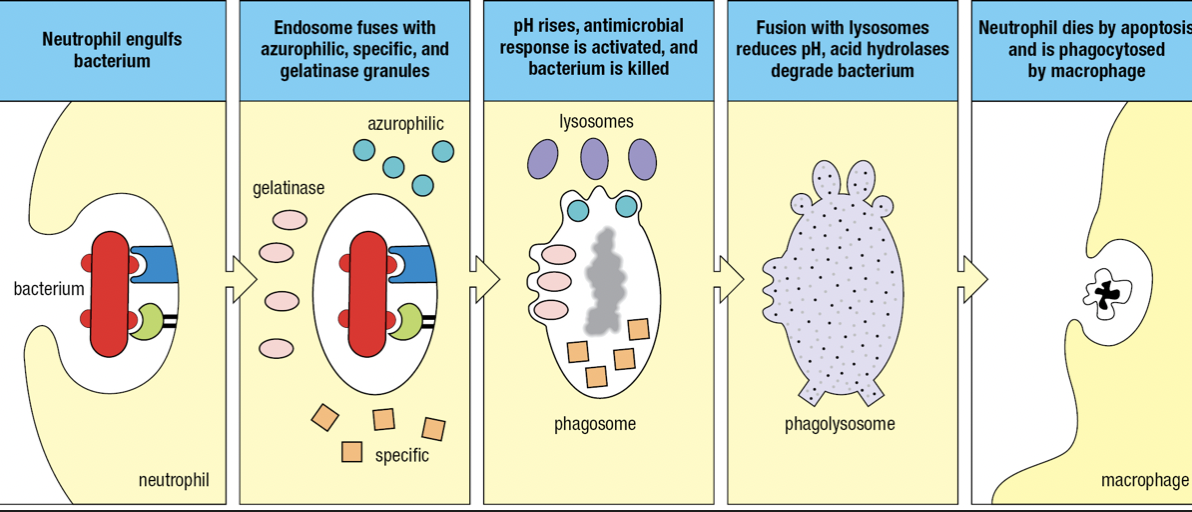
- Bacterium is engulfed to form endosome that fuses with azurophilic, specific and gelatinase granules
- Components of NADPH oxidase by specific granules facilitate a respiratory burst
- Raises pH = becomes phagosome and kills pathogen
- Fuses with lysosomes
- Lowers pH and activates hydrolases to degrade pathogen
- Respiratory burst
- Increase in oxygen consumption
- After neutrophils have deliverd all granules, they die
- Apoptosis
- NETosis which produces neutrophil extracellular traps (NETs) that capture and kill pathogens
- Nucleus swell and burst
Chronic Granulomatous Disease (CGD)
- Genetic syndrome caused by defective forms of genes encoding NADPH oxidase subunits
- Without functional NADPH oxidase = no respiratory burst
- Phagosome is too acidic to activate antimicrobial peptides
- Numbers of commensal cannot be controlled so they persist as chronic infections
- Infected macrophages are concentrated into nodules called granulomas which are unable to digest infected neutrophils
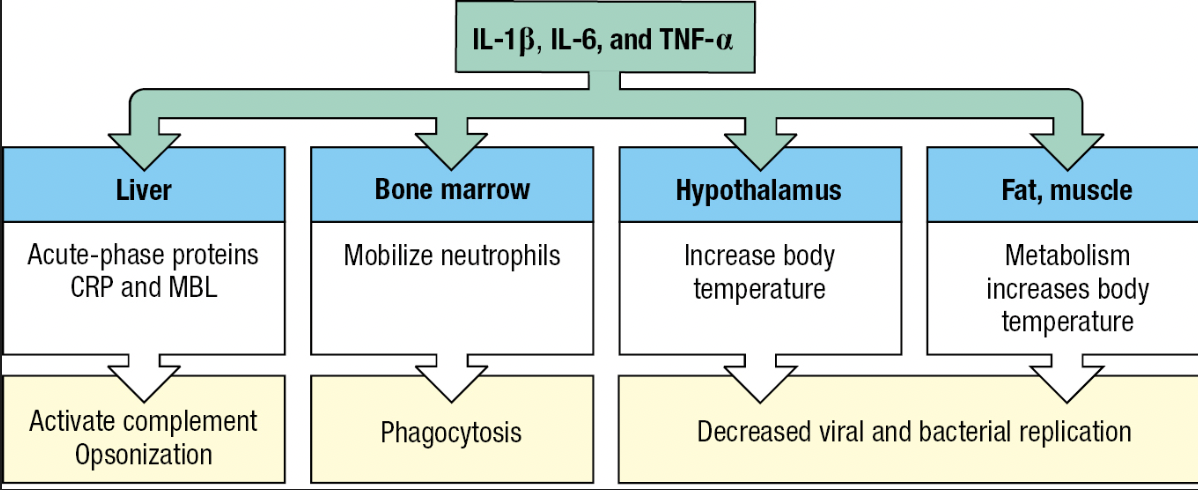
Fever
- Effect of cytokines IL-1B, IL-6 and TNF-a
- Called pyrogens
- Acts on temperature-control sites in hypothalamus and directly on muscle and fat cells = generate heat
- Inhibits the growth and replication of bacterial and viral pathogens
- Tissue cells become more resistant to damaging effects of TNF-a (capacity to kill tumor cells)
- Lethargy, anorexia = impedes use of energy to fight infection

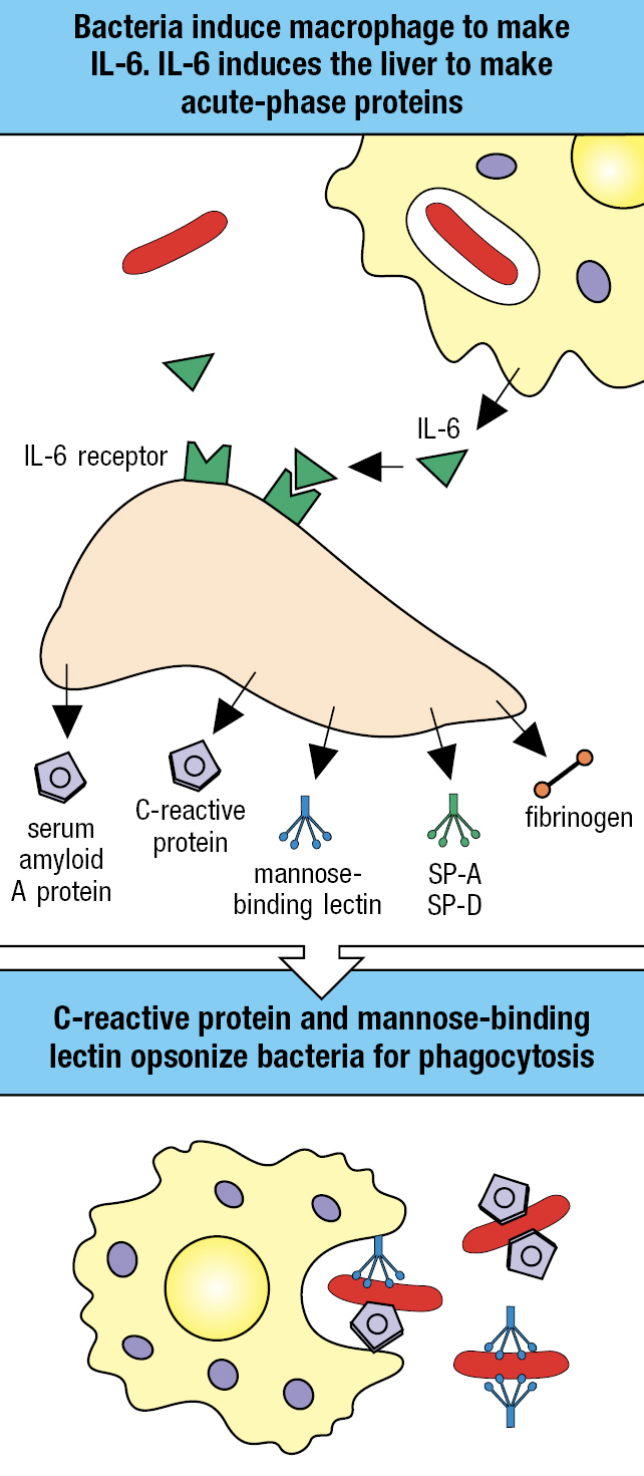
Acute-Phase Response
- Plasma proteins made by the liver are changed by IL-6
- Proteins that increase or decrease are called acute-phase proteins
- C-reactive protein (CRP) and serum amyloid A increases hundredfold
- CRP concentration – diagnostic test for infection and tissue damage
- CRP
- Targets phosphorylcholine of LPS
- Acts as opsonin and trigger classical pathway
- Serum Amyloid A
- Interacts with TLRs and SRs
- Activate cells to produce inflammatory cytokines
Lectin Pathway
- Induced by infection and requires time
- Contributes to innate immune response
- Initiated by acute-phase protein mannose-binding lectin (MBL) binding to pathogen surface
- Also an opsonin that facilitates uptake of pathogen by monocytes
- Circulates in plasma as complex
- MBL-associated serine protease (MASP)1 and 2
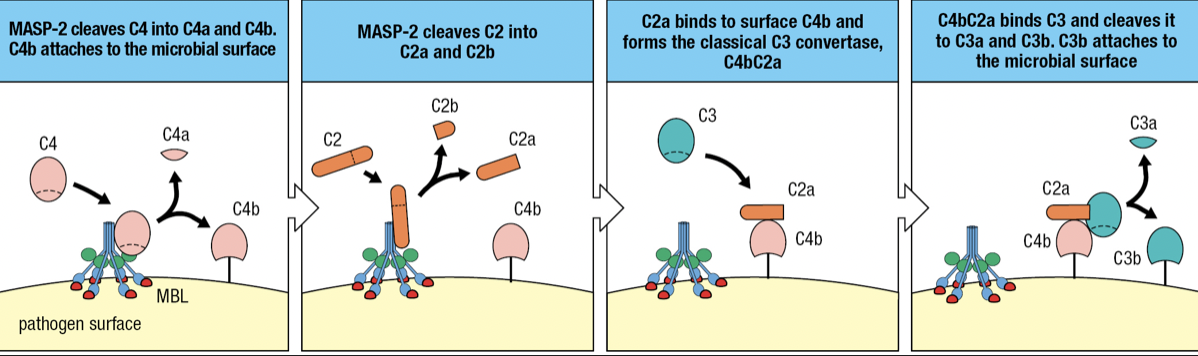
- when MBL binds to mannose-containing carbs, MASP-2 becomes active and cuts itself
- MASP-2 cuts C4 into C4a and C4b
- C4b attaches to pathogen surface
- C2 binds to MASP-2 and cleaves into C2a (larger) and C2b (smaller)
- C2a Forms complex with C4b = C4bC2a
- Forms Classical C3 convertase
- Cleaves C3 to assemble alternative C3 convertase
- when MBL binds to mannose-containing carbs, MASP-2 becomes active and cuts itself
Classical Pathway
- Contributes to innate and adaptive
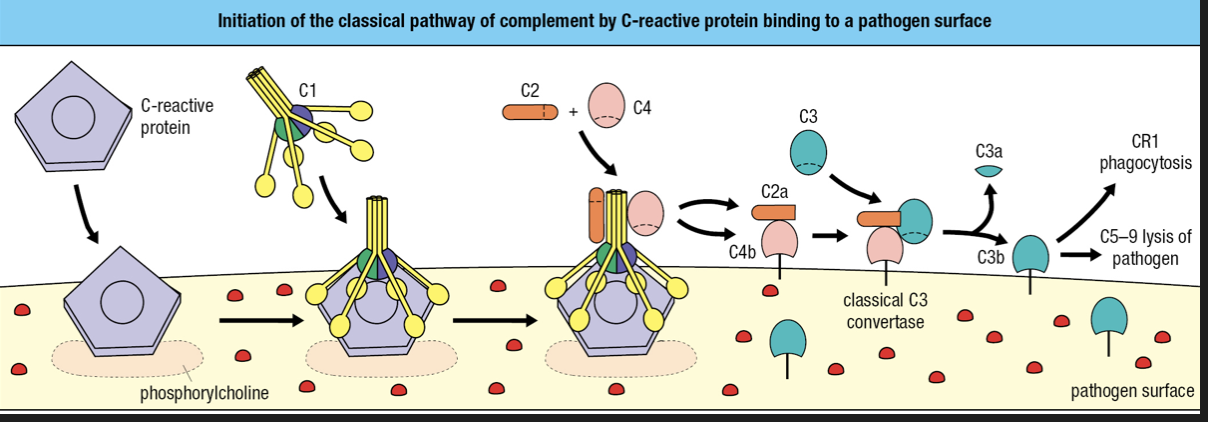
- Requires binding of antibody or C-reactive protein to pathogen surface
- CRP binds to phosphorylcholine on pathogen surface
- CRP binds with complement component 1 q (C1q) = cleavage and activation of C1r and C1s
- Activated C1s cleaves C4 and C2
- Forms classical C3 convertase
- C3b attaches to surface
- C3b is recognized by CR1 = engulfment
- C3b forms complex with C5-C9 = membrane destruction

Lymphoid Cells
- NK cells
- Cytotoxic innate lymphoid cells
- Circulate in blood and enter infected tissues
- Type I immunity
- Helper Innate Lymphoid cells (ILCs)
- Similar function to adaptive CD4 T cells
- Secretes cytokines to activate effector cells = macrophages and granulocytes
- Reside in tissues and make immediate response
- NK cells and Helper ILCs have no antigen receptor
- Instead expresses PRRs
- ILC1
- Intracellular (viral)
- Stops infection or limits spread until arrival of NK cells
- Type I immunity
- ILC2
- Mucosal surfaces
- Extracellular parasites
- Type 2 immunity
- ILC3
- Mucosal tissues
- Extracellular pathogens
- Participates in the containment of commensal microorganisms in the gut
- Tyupe 3 immunity
- Lymphoid-tissue inducer (LTi)
- Facilitates development of secondary lymphoid tissue
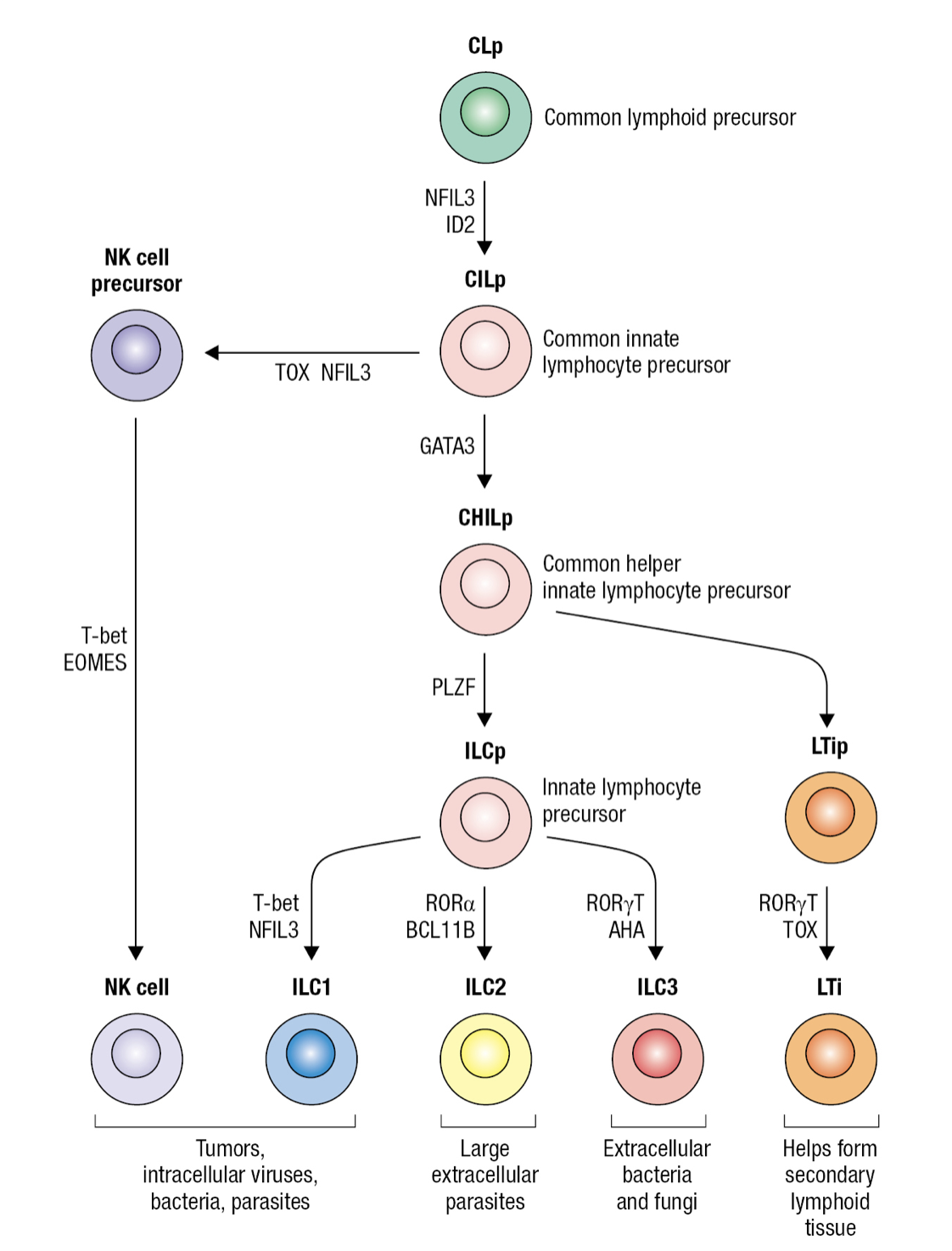
Innate Lymphocyte Precursor
- Common lymphocyte precursor cell (CLp)
- Gives rise to common precursor of all innate lymphocytes CILP
- Which then gives rise to precursor of NK cells NKp and precursor of all innate helper lymphocytes CHILp
- Which then gives rise to precursor of LTi cells LTIp and to common precursor of ILC1-3 ILCp
Circulating Lymphocytes
- NK = innate
- B and T cells = adaptive
- NK cells expresses CD56 but not CD3 that marks T cells
- No NK = persistent viral infections even with adaptive immune system
- NK importance in containing viral infections until cytotoxic T cells become effective
Subpopulation of NK cells
- CD56dim
- 90% of blood NK
- Less CD56 expression
- Differentiated cytotoxic cells
- CD56bright
- Gives rise to CD56dim during NK development
- Weak cytotoxic effector cells and more committed to making cytokines
- Most are taken up in tissues
- 80% of NK in lungs are CD56bright
- Uterine NK cells (uNK cells)
- Abundant in uterine
- Numbers fluctuate during menstrual cycle
- Contributes to embryo implantation and placenta formation
- Cannot kill cells or create inflammation
- Cooperates with fetal trophoblast nourishment to ensure mother can supply fetus with oxygen
NK Cytotoxicity
- Activated at viral infection sites
- To prevent NK from killing healthy human cells, cytotoxicity is regulated
- NK can only discharge weapons after contact
- Can only kill once cell at a time
- Decision to kill depends on sum of interactions between different NKRs and target cell ligands
- Inhibitory receptors prevent NK from killing healthy cells
- NK can only discharge weapons after contact
- NK Differentiation
- Infected cells secrete Type I interferons
- NK have IFN receptors that bind to IFN
- Aided by adhesion molecules CR3 and LFA-1
- Forms an immunological synapse that holds the cells together and forms channel for exchanging info = NK-cell synapse
- IFN activates NK to proliferate and differentiate into cytotoxic effector cells
- Effector NK kills infected cells by inducing apoptosis
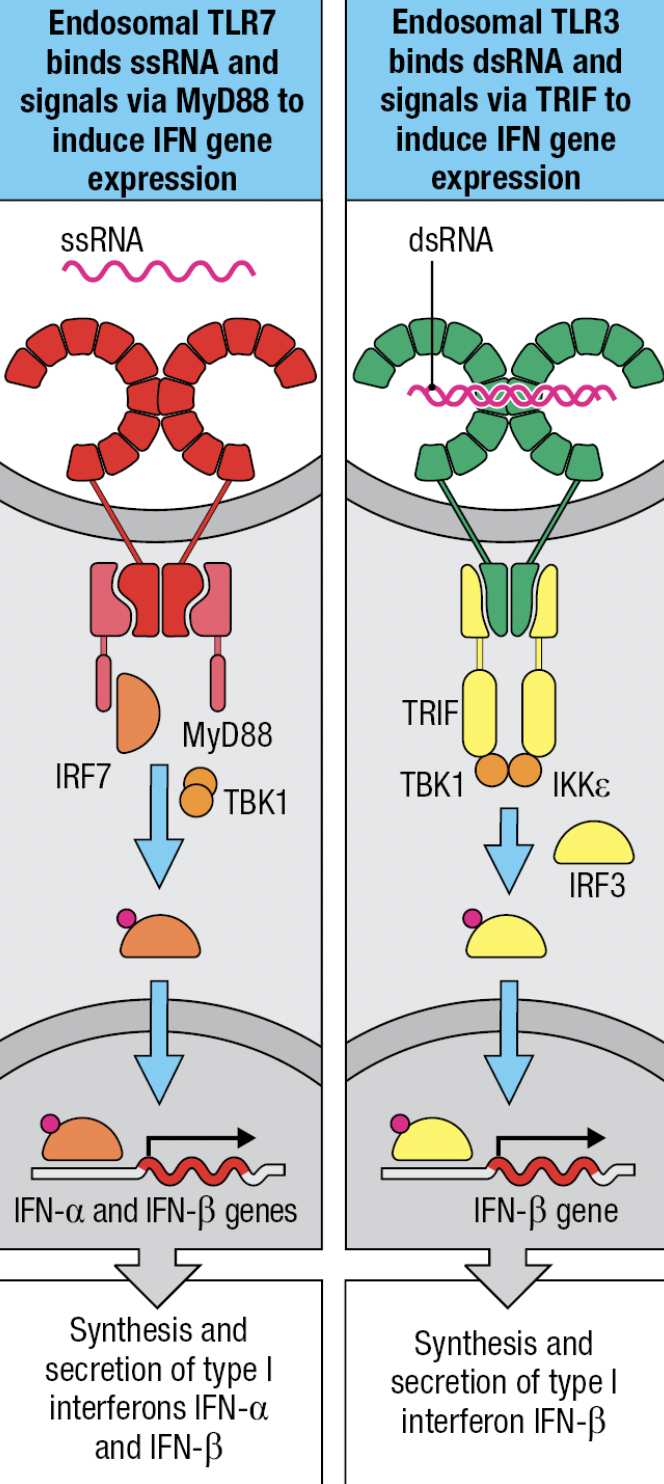
- NK expresses TLR3 (ds vRNA), TLR7 and 8 (ss vRNA)
- TLR7 and TLR8
- MyD88-dependent pathway that leads to IRF7 activation and production of IFN-a and IFN-B
- TLR3
- No MyD88 adaptor
- Signals by IRF3 pathway and leads to production of IFN-B
- Uses TRIF adaptor
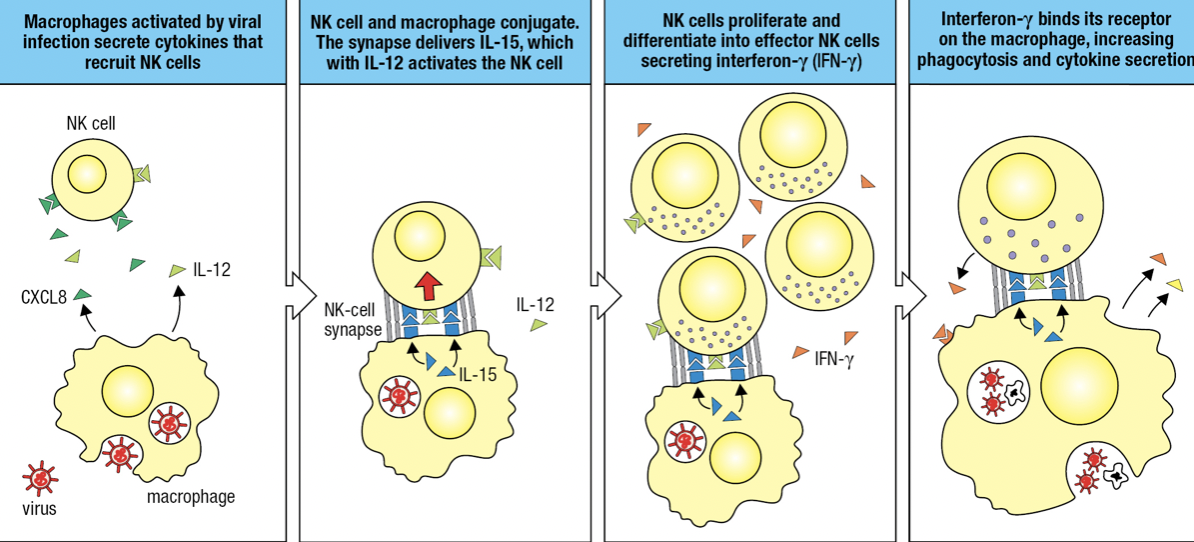
NK and Macrophages Activate Each Other
- Activated macrophages secrete CXCL8 and IL-12
- Activates and recruits NK
- NK and macrophage forms conjugate pair bound by NK-cell synapse
- Macrophage in synapse secretes IL-12 (for cytokine-secreting effector) and IL-15
- Activates NK
- NK proliferates and differentiates into effector NK secreting IFN-y
- IFN-y binds to macrophage and activates it
- Improved phagocytosis of virus particles and cells killed by NK
- IFN-y
- Potent inflammatory cytokine produced by NK
- Aka Type II IFN
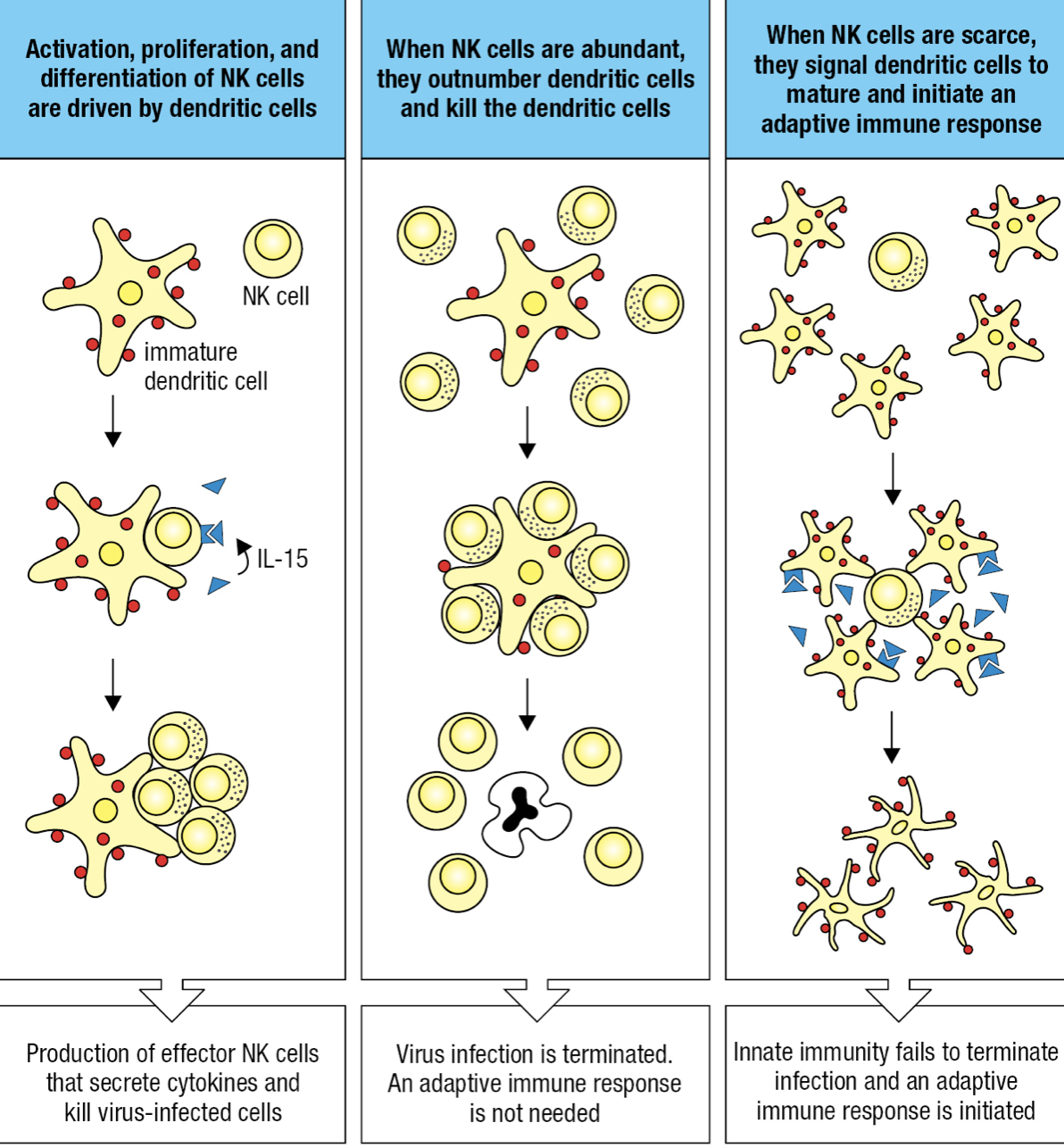
Dendritic Cells and NK cells
- DCs may take up pathogens or be infected by pathogens
- This causes changes on surface proteins monitored by NK receptors
- If infected, DC forms synapse with NK
- DC expresses IL-15 = cytotoxic NK proliferation
- If cytotoxic NK is high and innate immunity overcomes infection = NK kills DCs and prevent adaptive response
- If NK is low and innate cannot control infection = NK induces DCs to differentiate, migrate to lymphoid tissue and initiate adaptive response
NK Memory
- All nucleated cells express MHC class I = Human Leukocyte Antigen (HLA)
- Loss of HLA class I = marks disease
- Tumors and virus-infected cells escape T cell by reducing or losing HLA class I expression
- NK has receptors that detect low HLA class I and kills them
ANTIBODY FUNCTION
- Immunoglobulin
- General term for proteins produced by B cells that recognizes antigens (plasma cells)
- Attached or free-floating
- Antibody = free-floating
- Very specific
- Can recognize proteins and carbohydrates
- Their production is stimulated by vaccines or exposure to the antigen in question
- Antibody repertoire
- Mixing and matching of proteins to form immunoglobulins
- 10^16 but usually just 10^9
Antibodies have different functions:
- Neutralization
- Antibodies fully cover the antigen to recognize them
- Prevent adherence to cells
- Opsonization
- Antibodies fully cover antigen
- Makes it easier for phagocytes to engulf them
- Complement Action
- Attach to a certain antigen
- Activates a complement which enhances opsonization and lysis
Clonal Selection
- Transformation and proliferation of plasma cells that produce most specific antibodies from B cells
- Once a B cell recognizes an antigen, it will undergo further mutation of their antibodies
- Becomes more specific
ANTIBODY COMPONENTS
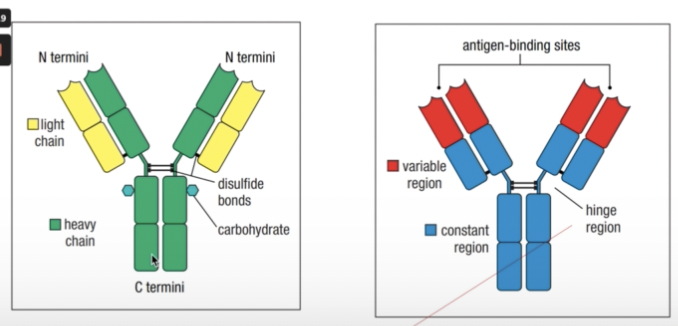
- Held together by a flexible hinge region composed of disulfide bonds
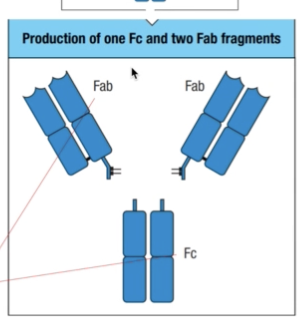
- Fab = fragment antigen binding
- Fc = fragment crystallizable
- Once it is done, they tend to precipitate and crystallize
ANTIBODY VARIABE REGION
- Epitope
- Antigens recognized by antibodies
- Antigenic determinant
- Can be linear or continuous depending on the structure
- Variable region light chain and variable region heavy chain
- Tend to vary to suit different shapes and sizes of all possible types of antigens

- Hinge region
- Can recognize different types of antigens no matter their positions
HYPERVARIABLE REGION
- Hypervariable region protein loops
- Found at the tip of both the light and heavy chain
- Aka complementary determining regions (CDR)
- CDR1, CDR2, CDR3
- Each variable region has three CDRs
- Framework regions
- Less variable
- Supports the CDR
- Affinity
- Binding strength of a single antigen binding site to the antigen
- Avidity
- Bing strength of multiple antigen binding sites to an antigen
- Binding to antigen depends on four covalent bonds
- Van der Waals forces
- Hydrophobic bonds
- Electrostatic bonds
- Hydrogen bonds
ANTIBODY ISOTYPES
- Isotypes
- Depends on structure of constant region
- Has diverse functions
- Named after the Greek letters they correspond to
- IgG = gamma
- IgM = mu
- IgD = delta
- IgA = alpha
- IgE = epsilon
GENETIC BASIS OF ANTIBODY DIVERSITY
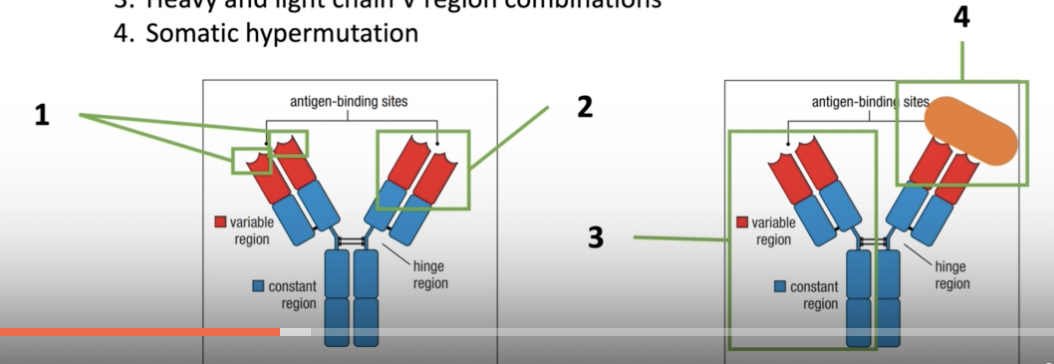
- Variable region gene segment combinations within heavy or light chain gene segments (combinatorial diversity)
- Junctional diversity between the gene segments
- Heavy and light chain V region combinations
- Somatic hypermutation
- Occurs after antibody recognizes an antigen
- Undergo further mutation/changes to become more specific
HEAVY AND LIGHT CHAIN DIVERISTY
- Recombination of antibody genes determine the specificity and diversity of the CDR and the antibody produced
- Light Chain
- Lambda locus gene segment from chromosome 22
- Kapa locus gene segment from chromosome 2
- Heavy chain
- Locus from chromosome 14
- Gene segments:
- V = variable
- L = Leader
- D = Diversity
- J = Junction
- C = Constant
ANTIBODY DIVERSITY

- Has a germline DNA origin
- From both sperm and egg
- Somatic recombination
- Antibody variable region diversity comes from the combination of the V, D, and J gene segments
- Light chain : V & J (variable) and C (constant)
- Heavy chain: V, D & J (variable) and 3 C gene segments (constant)
RSS
- Recombination signal sequences
- How a B cell know which g segments goes where
- Gene sequences that direct recombination
- Ensures gene segments are always in their proper orders
- V and J or V, D and J
- Have certain signals where the genes are supposed to join
- 12/23 Rule
- Segments with a 12 bp spacer will combine with a segment with a 23 bp space
RECOMBINATION
- Since the genes are in one linear chromosome, these gene segments are brought together closer first by your RAG complexes
- Recombination-activating genes (RAGs)
- RAG Complexes
- RAG complexes hold the RSS in place while the genes are recombined
- proteins coded by RAGs
- serve as a clip to place the V and J gene segments closer to each other before cleaving
- V(D)J Recombinase
- Set of enzymes needed to recombine the gene segments
JUNCTIONAL DIVERSITY
- Since these are always random gene segments, how can they combine next to each other when they don’t have complementary ends?
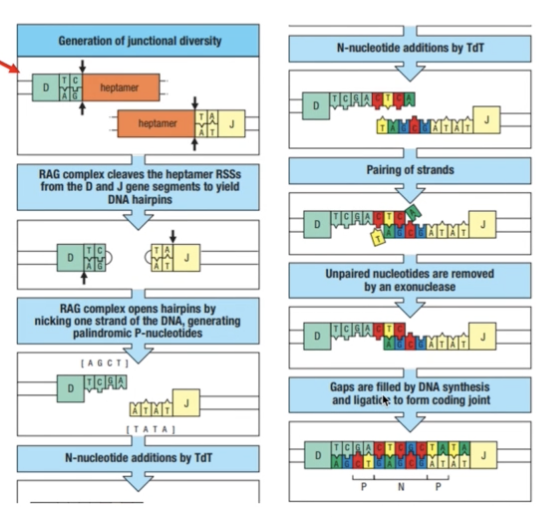
- RAG complex cleaves the gene segments to yield DNA hairpins
- RAG complex opens hairpins by nicking one strand of the DNA = palindromic P-nucleotides
- Terminal deoxynucleotidyl transferase (TdT) = adds random n- nucleotides
- Pairing of strands occur and non-complementary/unpaired parts are removed
- Gaps are filled by DNA synthesis and ligation to form coding joint
- This adds to the CDR3 diversity
WHICH GENE SEGMENTS CODE FOR WHICH CDR
- V segment = CDR 1 and CDR2
- V and J junction (and D) = CDR3
CONSTANT REGION DIVERSITY
- Free-floating = has different sets of proteins (hydrophilic) at the terminus end compared to transmembrane antibodies
- Transmembrane = protein ends help with hooking onto the surface of the B cell membrane
IgM STRUCTURE
- IgA and IgB accompany transmembrane IgM since these immunoglobulins do not have their own internal receptors
B CELLS TO PLASMA CELLS
- B Cells have their membrane-bound IgM
- Once it encounters an antigen, they will undergo somatic hypermutation to further produce more specialized antibodies that would better recognize antigens
- Eventually will transform into plasma cells which now solely produce free-floating antibodies specifically recognizing this specific antigen
- Allelic exclusion
- One allele is only expressed for producing antibodies while the other is silent
- Different alleles tend to code for antibody production but only one is expressed at a time
SOMATIC HYPERMUTATION
- After an antibody recognizes an antigen, the activation-induced cytidine deaminase (AID) will replace all the cytosine in the CDR3 with uracil
- Uracil-DNA glycosylase (UNG) will then remove the uracil and DNA polymerase will replace them with random nucleotides
ISOTYPE SWITCHING
- AID is also responsible for isotype switching
- It has a tendency to fold similar to Recombination of gene segments
- They put gene segments specific to an isotype closer to each other
- C mu and C delta gene segments are closest to the V, D, and J segments by default
- This is why IgM and IgD are produced first
- The rest of the C gene segments are next to each other
- Switch regions or switch genes are signals for isotype switching
- IgM antibodies exist as bulky pentamers
- Antigen activated T cells also mediate isotype switching
- Successive switching can occur (ex: IgM to IgG to IgA)
Ig SPECIALIZATION
IgM
- Produced in the spleen, bone marrow, and lymph nodes
- Low affinity
- Multiple binding sites of bulky pentamer free-floating form
IgD
- Mostly found in the respiratory tract
- Triggers local immune response when bound to basophils
IgA
- Monomeric form from spleen and bone marrow and circulates in blood
- Dimeric form from lymph nodes and circulate in lymph and other mucosal secretions (ex: tears, saliva, breast milk) and the digestive lumen
- May even target resident microbial microfauna to keep their populations in check
IgE
- Specialized toward recruiting effector functions of mast cells in epithelium, activated eosinophils in mucosal surfaces, and basophils in blood
- Targets parasites
- Triggers asthma and allergic reactions if parasites are not very common in the area
IgG
- Produced from the bone marrow, spleen, and lymph nodes
- Circulated blood and lymph
- Most commonly circulating antibody in bloodstream
- More flexible hinges compared to other antibodies for hard to reach antigens
- Also allows to bind to Fc receptors of phagocytes
- Different kinds based on flexibility, function, and targets
- Activates complements
IgG SPECIALIZATION
IgG1
- Most abundant and versatile
- Mostly binds to protein antigens
IgG2
- Second most abundant and less flexible
- Targets carbohydrates (bacterial capsules)
IgG3
- Most flexible with longest hinges
- Most susceptible to proteases
- Best in activating complement
IgG4
- Least abundant and does not activate complement unlike other 3
- Produced as monovalent together with IgE in case of allergic ractions
- Reduces allergic reaction by competing with IgE when binding
- Therapeutic uses
Caused by defects in immunoglobulin class switching and somatic hypermutation
Ig cannot adapt and patients become susceptible to infections
Defects in cd40 and cd40 ligand, AID, and UNG
Very low levels of other ig types but normal levels of igm
Cd40 ligand defects in t cells or cd40 defects in b cells
Cd40 activates isotype switching
AID mutation which disables somatic hypermutation and isotype switching
UNG mutation which disables somatic hypermutation
Effects:
Enlarged spleen or lymph nodes due to accumulation f immature b cells
Severe microbial infections and re-infections such as pneumocystis and cryptosporidium
Respiratory tract infections most common
Gastro are second most common
Includes entamoeba histolyca and salmonella enterica and giardia
Hindi naaalala ng immune system yung infections kaya nagkakaroon ng recurrent infections
Antibodies have Chlorophora or flurophore
Direct: recognize specific epitope
Indirect: gagawa muna ng serum or sample tapos sila marerecognize ng antibody
Polyclonal: antigen injected in animals tapos multiple antibodies are used targeting diff epitopes
Monoclonal: very specific using hybridoma and target the same epitope
- Antigen coat (glass slides
- Add blocking solution = hindi mag non-specific interactions with other artifcats
- Antibody incubation from human sample
- Fluorescent antibody incubation and visualization
Names of antibodies:
Source of the antbody, target antibody, conjygate chromo or fluorophore
Goat anti-human IgG alkaline phosphatase
Blocking solution: 1% BSA
For eliza chromophores
Horseradish peroxidase: brown or dark orange; stopping solution: hydrogen peroxide
Alkaline phosphatase: yellow to orange; no need for stopping solution
Immunofluorescent antibody technique
Fluorescein isothiocyanate: green
Western Blot
Transfer proteins from SDS PAGE gel to nitrocellulose membrane
Stain with antibody conjugated with horseradish peroxidase to determine the exact peptide detected by the antibodies
Flow cytometry
Monoclonal antibodies
Hybridoma grow and produce antibodies indefinitely
Separate hybridomas that produce certain antibodies specific for a protein
These are used in therapy as long as same source para marecognize as part of the body
Ex: Rituximab:
Binds to CD20 which is present in both malignant and normal B cells
Fc is the attachment for NK cells which cell-killing machinery
Plasma cells are still present to produce antibodies even if all B cells are being targeted
Non-hodgkin B cell lymphoma
LECTURE 5
CELL MEDIATED IMMUNE RESPONSE
Divided into two:
- MHC Class I = Cytotoxic or CD8 T Cells
- Deals with intracellular pathogens
- Requires CD8 receptors
- Found in all cells except RBCs
- Pair of protein segments
- MHC Class II = Helper or CD4 T Cells
- Deals with extracellular pathogens
- Requires CD4 receptors
- Found in macrophages, dendritic cells and B cells
- a chain of 4 protein segments
CMIR
- Uses both T Cell receptors and Major Histocompatibility Complex (MHC)
- unlike B Cells which produce antibodies that recognizes pathogens
T Cell Receptors
- attached to the cell membrane of T cells
- function like antibodies of B cells or immunoglobulins
- have alpha and beta chains = the light and heavy chains of antibodies
- they also have a variable and constant region
- Differences
- Can only recognize short peptides
- Unlike antibodies which can recognize larger proteins as well as carbohydrates
- Can only recognize short peptides
- short peptides are presented by an MHC
- no free-floating form
- no differentiation or somatic hypermutation after recognizing an epitope
T Cell Receptor Diversity
- Also dependent on the germline DNA
- Alpha chain: chromosome 14
- V and J
- Beta chain: chromosome 7
- V, D, and J
- Assembly of gene segments involve RAGs
- Relies on DNA recombination
- Only 1 constant alpha region gene and 2 constant beta region gene
- Both are functionally identical
Transposons
- Theorized origin of diversity
- Short gene segments that code for transposase
- Enzyme that cuts transposon and copy and paste it to the other parts of the DNA
- Recognizes the repetitive DNA sequences in the transposons
- Transposase and RAG recombinase are similar
- Led to the hypothesis that RAGs originated from transposons-transposase interaction
- A transposon was inserted into a gene encoding for a receptor of innate immunity
- The transposon separated the gene into two segments, each flanked by a piece of repetitive transposon DNA
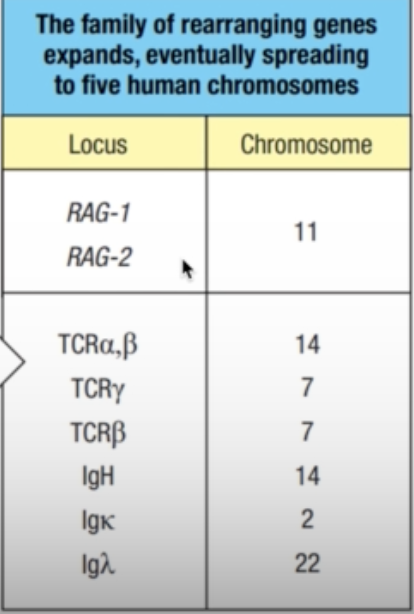
- Chromosomal rearrangements placed the transposase gene onto a different chromosome from the primordial rearranging gene
- The repetitive DNAs became the RSSs of a primordial rearranging gene and the transposase genes became the ancestral RAG-1 and RAG-2 genes (third panel).
- the family of rearranging genes expanded and eventually spread to five human chromosomes
- TCR: chromosomes 14 and 7
- Ig: chromosomes 14, 2, and 22

T Cell Receptor Complex
- TCRs do not penetrate through the transmembrane of the T cells
- Rely on support proteins and proteins that would serve as intracellular signal transmitter
- CD3 delta
- CD3 gamma
- CD3 epsilon
- Zeta chain functions in transducing signals

Two Classes of TCR
- Most studied are alpha and beta chains since these are similar in jawed vertebrates
- Meanwhile, gamma and delta chains are not similar in mice and humans
Overview of TCR and MHC Relationship
- Any pathogen that is engulf by the cell for the MHC II will have their proteins broken down
- presented by the MHC on the surface of that cell
- will be recognized by the respective T cell and T cell receptor

MHC I
- if a cell is infected by an intracellular pathogen, its proteins will be presented by the MHC I on the cell surface
- this will be identified by the CD8 T cells and CD8 TCRs
- signals apoptosis of the infected cell to prevent further infection of other cells
MHC II
- break down the segments of extracellular pathogen
- will be presented on the cell surface and be recognized by CD4 T cells and CD4 TCR
- Promotes production of cytokines or cell signals to
- activate macrophages to signal that there is an extracellular pathogen present
- signal B cells to transform into plasma cells and produce antibodies
Structure of MHC

- MHC I
- 3 alpha domains (alpha 1, alpha 2, alpha 3)
- Beta 2 macroglobulin
- Not encoded in MHC gene
- Has grooves and pockets in its structure
- Allows only small peptides (8-10 AA) to be presented
- MHC II
- Alpha 1, alpha 2
- Beta 1, beta 2
- No grooves or pockets
- Wider space for larger peptides (13-25 AA)
- MHCs exhibit promiscuous binding specificity
- Potential to bind to different peptides
- Not necessarily for a certain peptide
Self VS Non-Self Proteins
- Cells are programmed to distinguish their “self” proteins produced by the cell
- If they recognize them as “non-self”, they will be immediately presented by the MHCs
- MHC I
- foreign proteins are loaded in the ER
- If a pathogen enters the cell, the cell will trigger production of proteins for that pathogen
- These proteins will be recognized as “non self” by MHC I
- Triggers its presentation to CD8 T cells
- Proteosome breakdowns misfolded proteins and recycle the peptides
- Once they recognize non self proteins, interferon-gamma triggers their transformation into immunoproteosome and consequently, the breakdown of these cells
- In a normal setting, the broken down peptides pass through transporter associated proteins into the ER where they will be reused
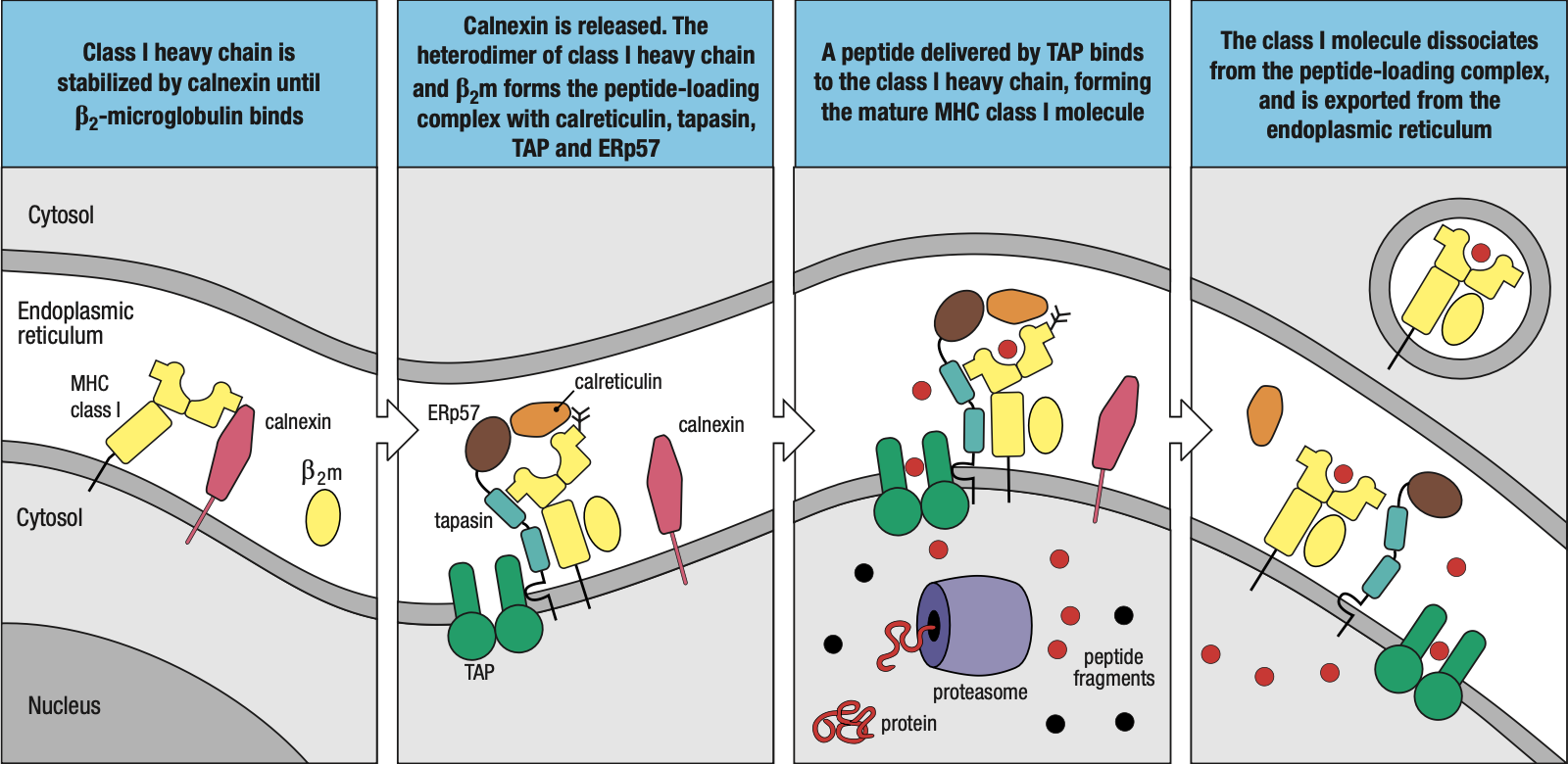
- In the case of foreign protein, the immunoproteosome will trigger the assembly of MHC I
- The chaperone protein calnexin attaches the beta-2 microglobulin to the 3 alpha domain proteins
- Forms the peptide-loading complex
- Tapasin: brings the MHC I closer to TAP = easier for foreign peptides to be loaded into MHC I
- ERp57 and Calreticulin: help in peptide bonding
- Foreign peptide is enclosed in a vacuole and transported to the surface of the cell
- Sometimes, protein segments are too long = ERAP (ER Aminopeptidase) protein
- Shortens the peptides to fit into MHC I
- Sometimes the peptide may not fit too well that it just falls off
- MHC I will then assist them back to the ER and try / load again
- MHC II
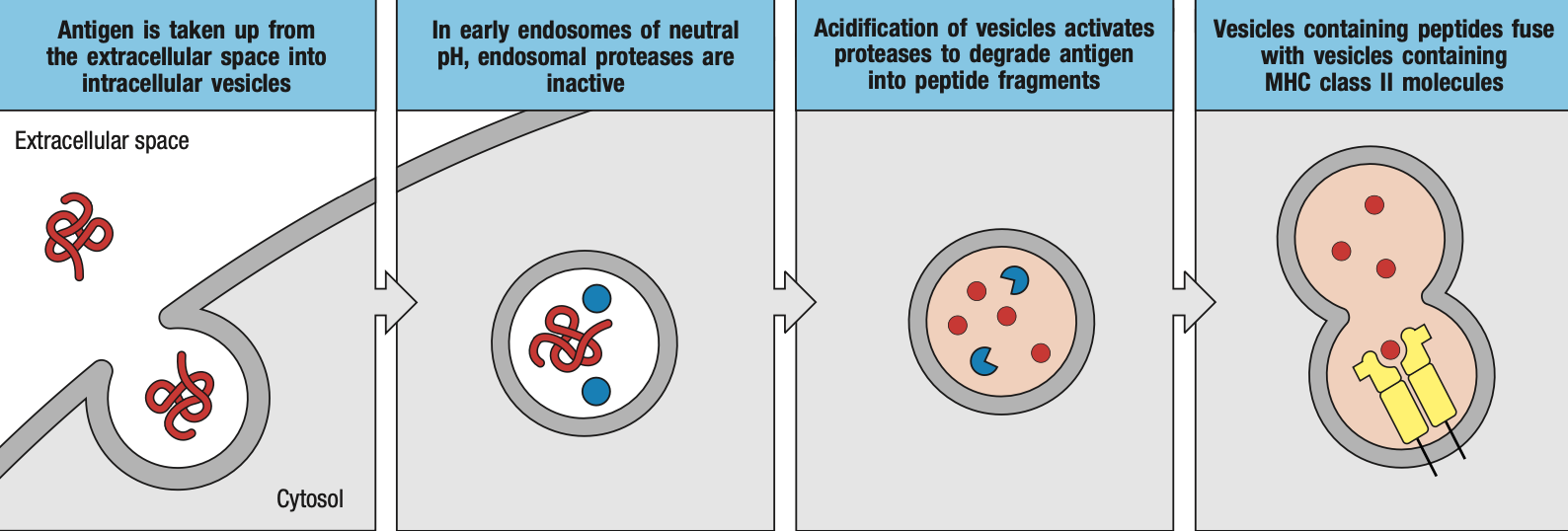
- foreign proteins are loaded in the endosomes
- extracellular pathogen will be engulfed by macrophages / dendritic cells and be enclosed in endosomes
- endosomes produce enzymes that will breakdown or inactivate proteins
- endosomes will then bind to specialized vacuoles containing MHC II
- the class II-associated invariant chain peptide (CLIP) will cover the groove or pocket on the MHC II while it has not encountered a foreign peptide yet
- for loading
- Human leukocyte antigen-DM (HLA-DM) removes the CLIP for proper loading of foreign peptide
- Human leukocyte antigen-DO (HLA-DO) retains the CLIP while there is no foreign peptide yet
Cross-Presentation
- Dendritic cells, B cells, and macrophages have MHC I and MHC II
- Sometimes, the peptides may enter through the MHC I pathway but end up being presented by MHC II or vice versa
MHC Diversity
- Human leukocyte antigen complex
- HLA I types: MHC I
- A, B, C, E, F, G
- HLA II types: MHC II
- DM, DO, DP, DQ, DR
- HLA I types: MHC I
- HLA diversity comes from
- MHC II alpha chains gene families
- MHC II beta chains gene families
- MHC I heavy chains gene families
- Genetic polymorphism
Genetics Review
- Gene = gene segments that codes for a certain protein
- In this case, HLA
- Alleles = variations (single or multiple nucleotide changes) that would alter the protein structure
- Each allele would code for a different allotype (of HLA)
- Can either be
- Monomorphic: there’s only 1 type
- Oligomorphic: dozen or less
- Polymorphic: more than a dozen
- From germ line
- Heterozygous: different allele from parents
- Homozygous: same allele from parents
- Haplotype: every set of different alleles represent a different kind of haplotype
MHC I and HLA-I Diversity
- HLA-A, -B, -C
- Highly polymorphic
- Present antigen to CD8 T Cells and ligands of natural killer cells
- HLA-E, -G
- Oligomorphic
- Present antigen to ligands of natural killer cells
- HLA-F
- Monomorphic
- Chaperone to MHC class I molecules that lose their antigen while travelling to cell membrane
MHC II and HLA-II Diversity
- HLA-DP, -DQ, -DR
- Highly polymorphic
- Present antigen to CD4 T Cells
- HLA-DM, -DO
- Oligomorphic
- Aids in loading antigen onto HLA-DP, -DQ, and -DR
Chromosome 6
- Class I Region for HLA-I
- Mostly MHC-related or antigen presentation-related genes
- Proteins of other functions
- Class II Region for HLA-II
- mostly MHC-related or antigen presentation-related genes
- TAPs
- Tapasins
- Related to immunoproteosomes
- Class III Region
- Proteins not related to MHC
HLA Gene Diversity
- MHC I
- Allotype diversity in both alpha-1 and alpha-2 domains
- Beta-2 immunoglobulin gene comes from another chromosome (chromosome 15)
- MHC II
- Allotype diversity in either alpha-1 or beta-1 domains
- The other domain is always non-functional
- Depends on MHC II isotype
MHC Activation
- Cell signals that activate MHC antigen presentation
- Interferon-alpha
- Interferon-beta
- Interferon-gamma
- Functions in both MHC I and II responses
- MHC I
- IFN-gamma activates proteosomes into immunoproteosomes
- For better breakdown of foreign peptides of intracellular pathogens
- MHC II
- IFN-gamma activates MHC class II transactivator (CIITA)
- Activates MHC II
MHC I and MHC II
- HLA-I most likely evolved first
- Most of HLA II genes code for proteins that are strictly for antigen presentation to T Cells while HLA-I genes contain proteins with other functions
- HLA I-related genes are found in other chromosomes
- HLA-II is more compact compared to HLA-I
- Some organisms such as Atlantic cod can survive with only HLA-I
MHC Restriction
- proper order of your MHC, TCR, and antigen for recognition
- different sides of the antigen bind to MHC and TCR
- peptide binding sites or set of anchor residues
- anchor residues or complementary pockets in the MHC binding sites
- TCR also recognize the residues on the surface of the alpha helices
- The part of the antigen recognized by HLA is different from that recognized by the TCR
- If same peptide but presented by a different HLA = no recognition
- If HLA carries a different antigen but same TCR = no recognition
Natural Selection
- MHC and TCR diversity are controlled by natural selection
- Balancing selection always ensure that highly polymorphic heterozygotes are always present
- Intermingling of populations = better immunity
- Ensure HLA are sufficiently heterozygous
- Ensures survival against new disease
- Meiotic recombination occurs at 2% frequency which increases with succeeding generation
- New alleles from point mutations and recombinations
- Interallelic conversion or segment exchange occurs when recombination combines segments with point mutations to homologous segments
GVHD occurs during hematopoietic stem cell transplants such as bone marrow
HSC ransplants are performed fo patients with damaged or malfunctioning immune system or bone arrow
Autologous = self MHC isoforms
Allogenic = non-self MHC
The donor tissue are recognized as allogeneic or different by the new T cells
Alloreactions
Alloreactive t cells will react against cells from another indiv
About 1-10% circulatnb T cells in an indiv are alloreactive
Alloantibodies of one indiv will atact the allotypic proteins of another
These mismatches can occur between mother and child
HLA types
- Combinations of HLA alleles in a person
- Should matcj in order for transplants to proceed
- Hla types testing by molecula methods
- Most likely matched at HLA-A B C and DRB1
Symptoms
Skin as the most likely first organ to be affected with presence of rashes
gI disease such as diarrhea
liver disease
chronic GVHD occurs after 100 days
acute GVHD within 100 days
organ transplant rejection
the recipient immune system attack the donor organ
all cels in immune cells possibly involved
can occur months after the transplant
Topic 6
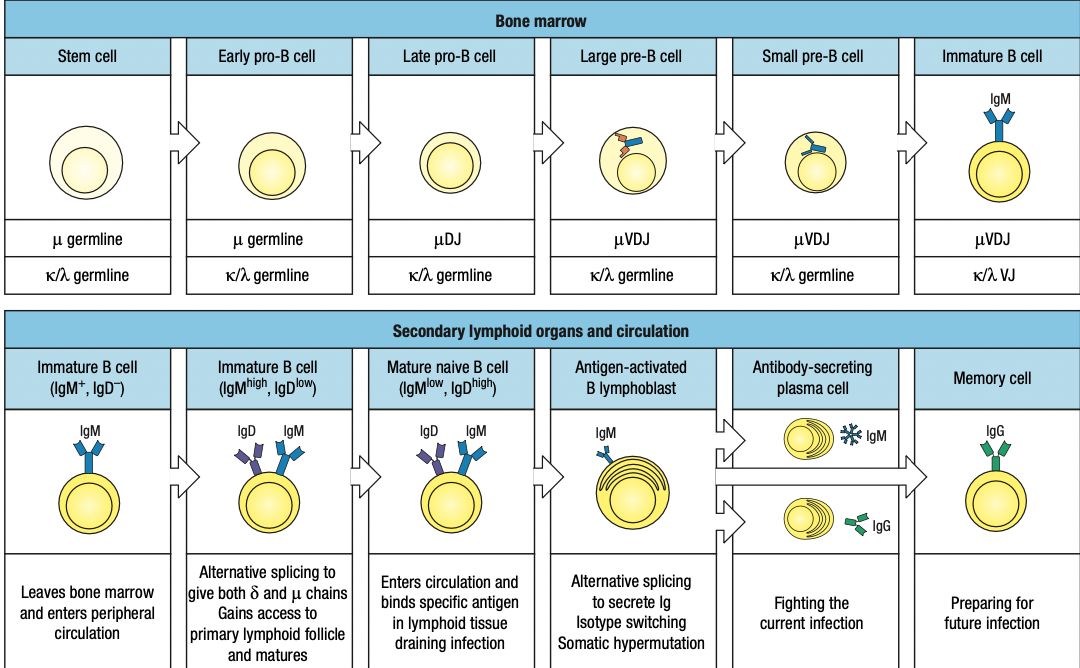
B Cell Development

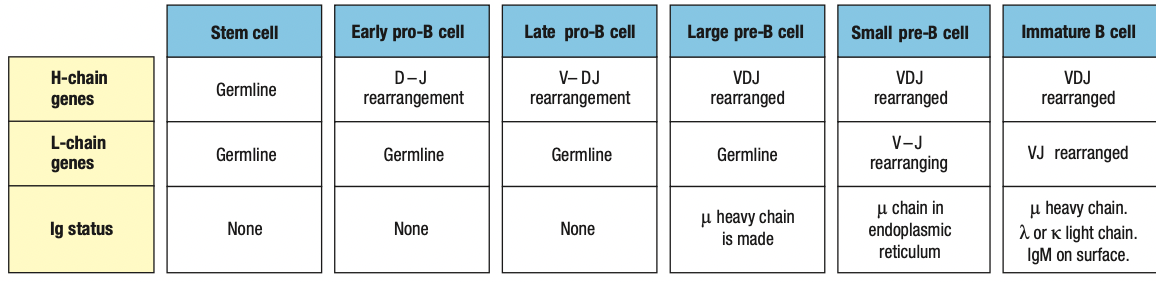
Pro-B cell
- main event in the pro-B-cell stage is the rearrangement of the heavy- chain genes
- Early: D and J segments
- Late: DJ and V segments
- The rearranged gene is transcribed through to the μ C-region gene, the nearest C gene to the rearranged V region
- The RNA transcript is spliced to produce mRNA for the μ heavy chain, the first type of immunoglobulin chain made by a developing B cell
Pre-B cell
- B cell expresses mu chain
- Large pre-B cell: less mature
- Heavy-chain rearrangement is done
- Makes mu heavy chain
- Small pre-B cell: mature
- Rearrangement of light chain gene
- First is kappa
- If successful, no lambda
- Light chain is synthesized and assembled sa ER
- Form IgM
- If fail, lambda
- Same as above
- If fail both = apoptosis
Immature B cell
- Heavy and light done rearranging
Stromal
- Environmental cells sa marrow for maturation
- Interacts as adhesion molecules to ligands
- Has growth factors that attach to b cells
- SCF = Kit receptor on maturing B cell
- IL-7 acts on late pro and pre
- Moves from subendosteum to marrow cavity
- Later stages are less dependent on stromal cells = leaves BM
Pro-B Heavy Rearrangement
- Inefficient and imprecise = due to addition random N and P nucleotides in V, D, J segments
- Limited material tapos random
- Parang sandwich na sakto yung parts
- One in three chances of maintaining correct frame
- Nonproductive rearrangements = not useful
- Productive = useful
- B cell has two copies of heavy chain locus (one from each parent)
- Can still produce heavy chain if one chromosome locus is productive
- Express RAG1 and RAG2 = rearrangement starts
- Activated by E2A and EBF which expresses Pax5
- Apoptosis is default unless there are survival signals
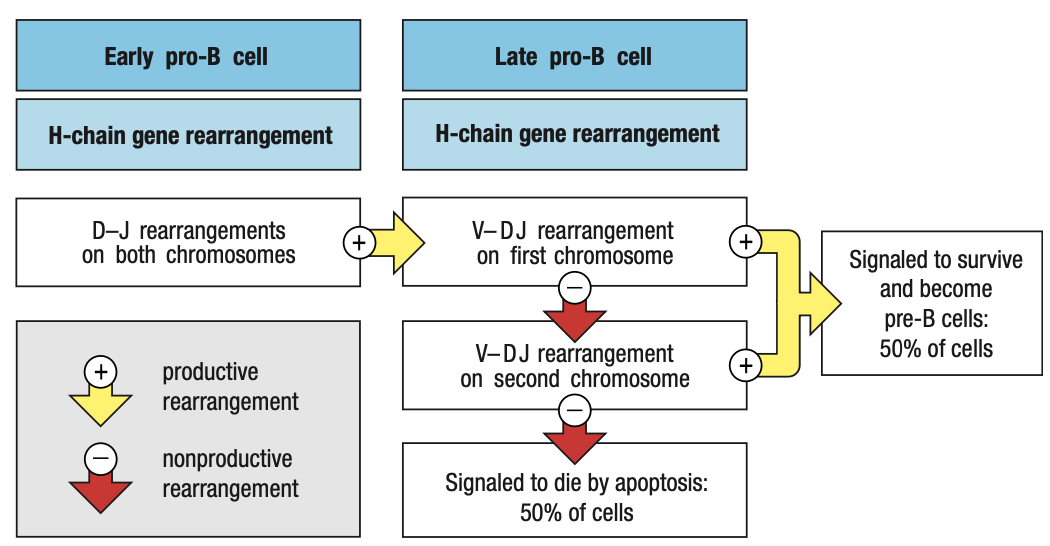
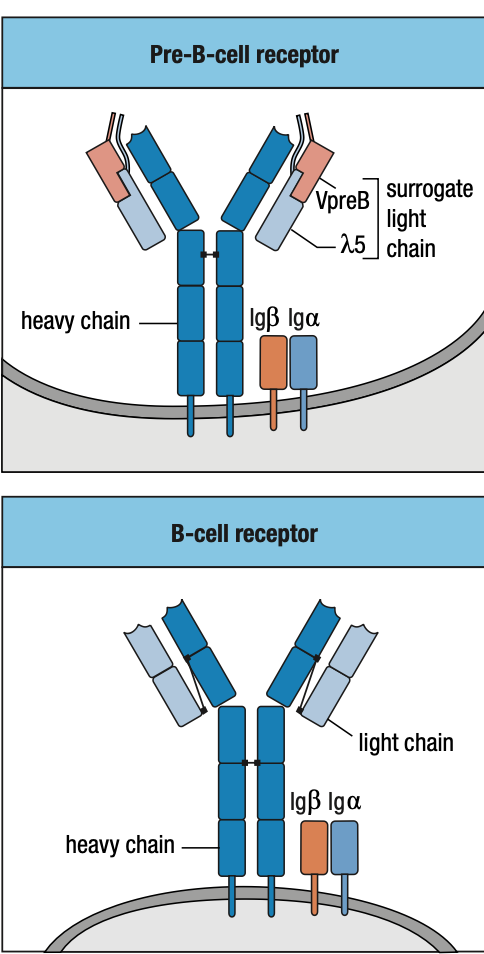
Pre-B Cell Receptors
- Pro-B needs to make mu chain to survive and mu chain must be able to combine with Ig light chain
- E2A and EBF roduce surrogate light chain
- To test kung functional heavy chain, surrogate light chain ay parang substitute since light chain wont be formed unless functional ang heavy chain
- Synthesizes VpreB (mock variable region) and lambda5 (mock constant region)
- In the ER of pro-B, mu chain binds with surrogate and IgB = pre-B cell receptor = sends signals okay/functional ang heavy chain = interacts with BM ligands = stop RAG = pro-B cell div = large pre-B
- Pre-B cell interacts with BM ligands heparin sulphate and galectin-1 to stop RAG
- Unlike the B-cell receptor (IgM), the pre-B-cell receptor does not have an antigen-binding site, nor is it well represented at the cell surface
Allelic Exclusion
- Pre-B receptor
- Eliminates nonfunctional mu chains
- Prevents making more than one functional mu chain
- In a pro-B
- rearrangement of the first immunoglobu- lin locus succeeds
- synthesis of μ chain
- assembly of the pre-B-cell receptor
- stop RAG transcription
- Alleles from each parent, As long as one is working, ignore the other
SMALL Pre-B Light Chain Rearrangement
- Large pre-B undergoes several rounds of division = CLONING
- Yields resting pre-B cells that express identical μ chains
- no longer make the surrogate light chain and the pre-B-cell receptor
- RAG is reactivated = light chain rearrangement
- One locus at a time: kappa locus first before lambda
- One recombination event V and J: several attempts can be made in kappa before lambda rearrangement
- The organization of the light-chain loci allows initial nonproductive rearrangements at one locus to be followed by further rearrangements of that same locus that can lead to the production of a functional light chain
- If IgM is self-reactive
- Receptor editing = mature B cells (for light chain lang)
- Repeated fail = apoptosis
- Bind to monovalent self-antigens = anergy
- Arrest of B cells
- Prduce IgM and IgD but IGD allowed to develop
- Lifespan is 1-5 days
- Bawal na mag-undergo ng receptor editing pag nasa bloodstream na
- Peripheral tolerance = apoptosis or anergic lang
- Central tolerance = receptor editing, apoptosis or anergic

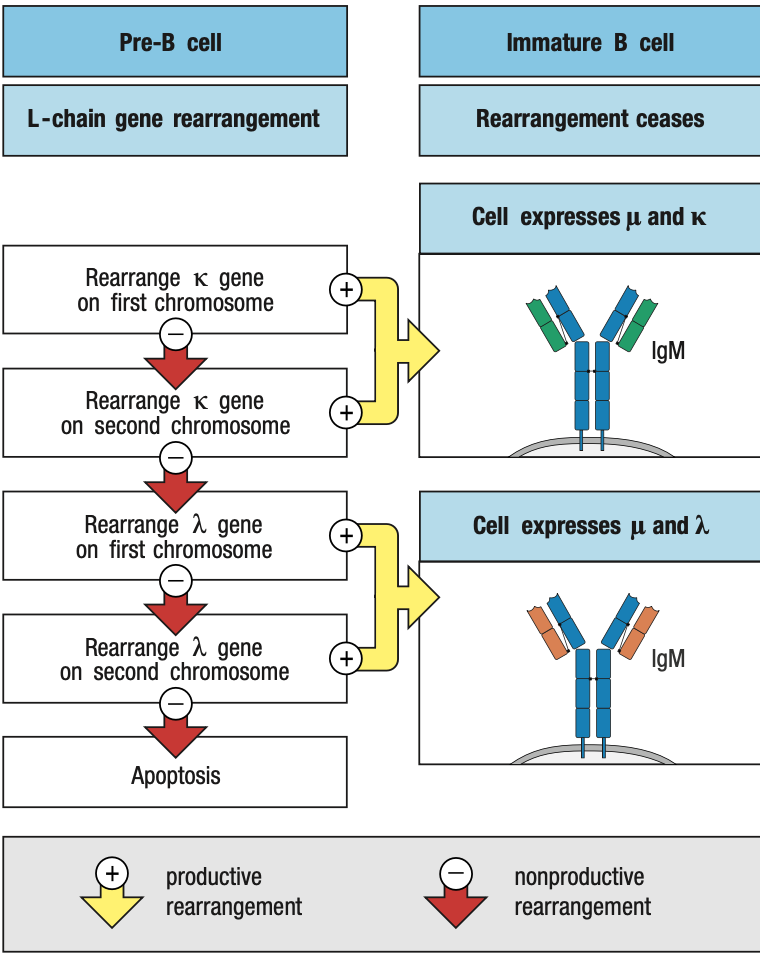
- Functional light chain + mu chain in ER = IgM
- IgM + IgA + IgB = B-cell receptor in surface = stop light chain rearrangement = immature B cell
Two Check Points in Bone Marrow
- First: late pro-B if may pre-B receptor or wala
- Second: small pre-B kung may B-cell receptor or wala
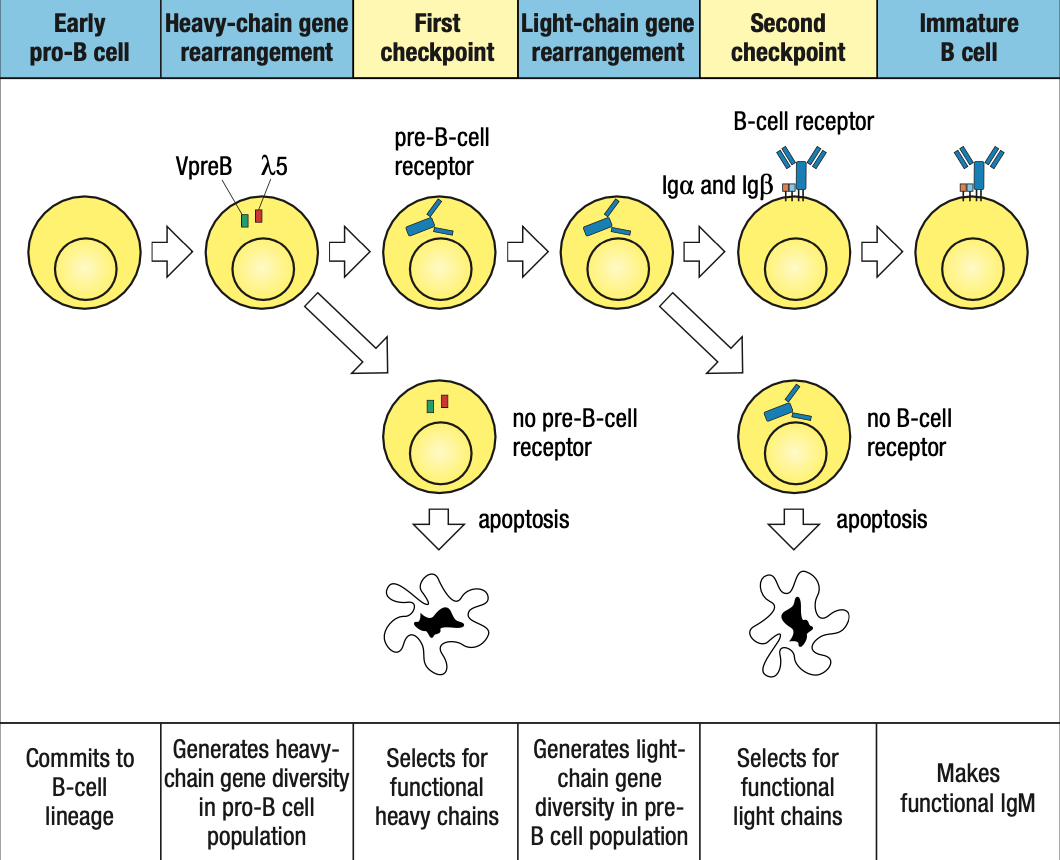
Protein Expression
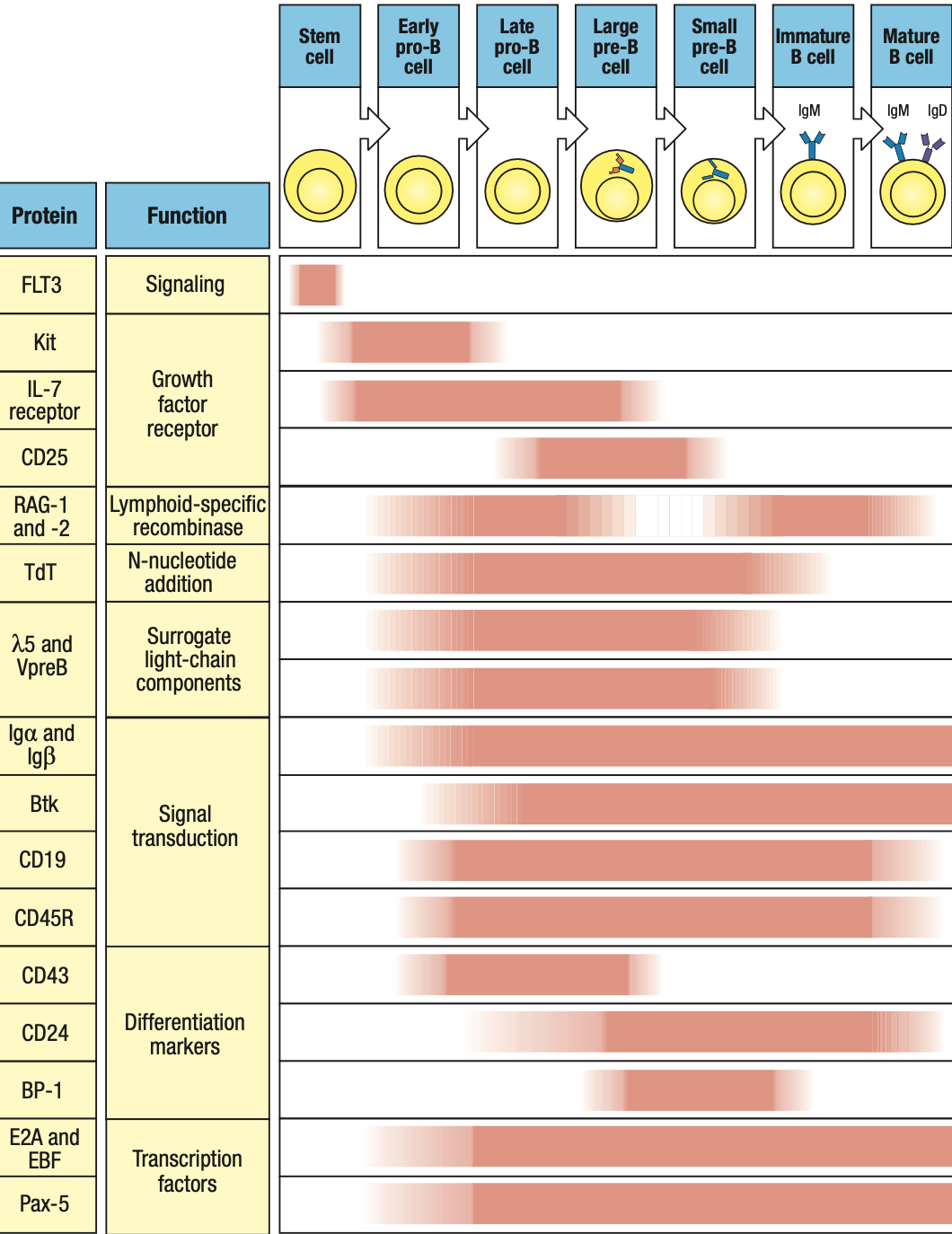
- RAG1 and RAG2: on during heavy/light rearrangement, off during checkpoints
- TdT: on during heavy rearrangement, off during light
- IgA and IgB: forms receptors so on from pro-B stage until end
- Pax5: on early pro-B
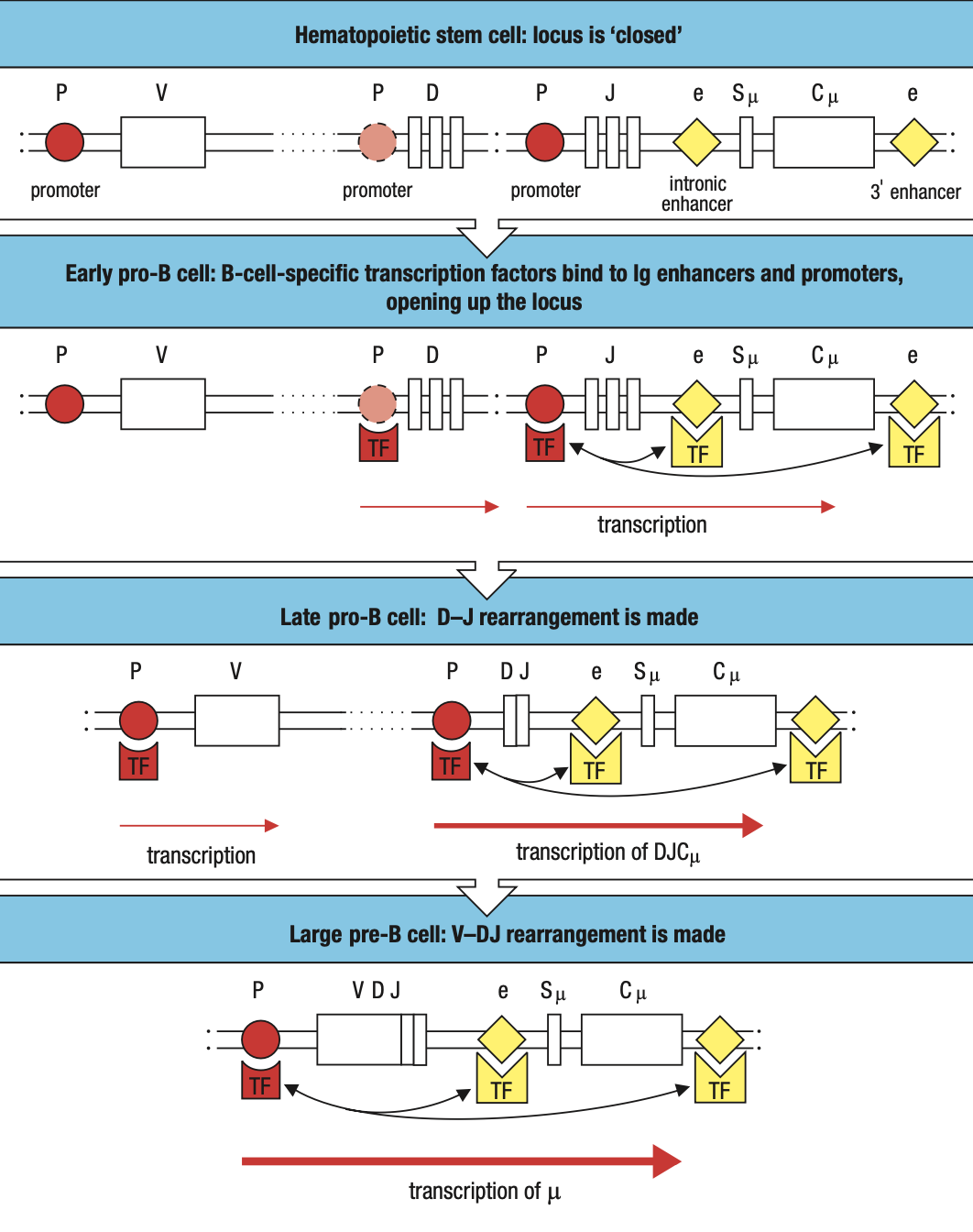
B-Cell Translocation
- Genes that cause cancer when their function or expression is perturbed are collectively called proto-oncogenes
B Cell CD5 Expression
- B1/CD5 B Cells: little to no IgD
- Produced pre-natally
- Lack N nucleotides cuz no TdT during prenatal
- Polyspecificity = binds to many diff antigens
Summary
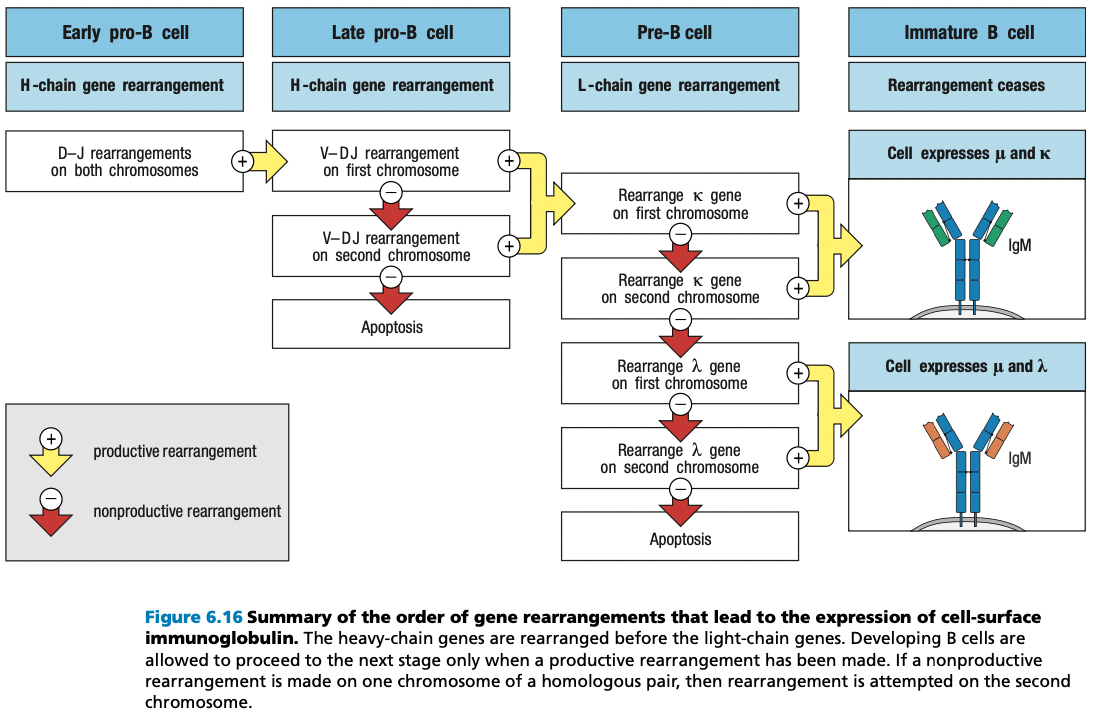
self-reactive immature B cells that encounter their self antigens, either in the bone mar- row or in the peripheral circulation, are prevented from advancing from the immature B-cell stage to the mature B-cell stage.
This repertoire of immature B-cell receptors includes many with affinity for self antigens that are components of healthy human tissue.
Activation of mature B cells carrying such receptors by these antigens would produce self-reactive antibodies
To prevent this from happening, the B-cell receptors of immature B cells are wired to generate negative intracellular signals when they bind to antigen. These signals cause the B cell either to die by apoptosis or to become inactivated.
In this way, the immature B-cell population is subject to a process of negative selection that prevents the maturation of self-reactive B cells. As a consequence, the resulting population of mature B cells in a healthy individual does not respond to self antigens and is said to be self-tolerant.
Bone marrow = blood stream = lymph organs = plasma cell to BM or glial cells remain in lymph organs
Stromal cells = stem cell development into B cells
Pro BCell
Early: DJ
Late: VDJ
CCL21 CCL19 = from lymph cortec xcells
CXCL13 = from follicular DC
Lymphotoxin preserve integridity of FDC
B cell activating factor BAFF to promote B cell survival
Mature Naïve B Cells
Have IgM not self reactive but yet to biun to antigen
B cells pass thru T cell zone into lymphoid follicles
Anergic B cells pass thru lymphoid follicles but stay outside T cell zone and later die by apoptosis
Plasma Cells
Activated by antigen and CD4 T cells
Terminal differentiation = stop proliferation and differntioation
Return to bone marrow to produce more antibodies = meaning effective na
Memory B cells
Resting quiescent b cells
Reserve in case similar infection occurs
Lasts for a lifetime and more quickly activated
CHAPTER 7
T-CELL DEVELOPMENT
- Origin: bone marrow
- Maturation: thymus
- Before they can undergo gene rearrangements
- “Thymus-dependent lymphocytes”
- Positive Selection
- T cells that can recognize self MHC class I and II isoforms survive
- Negative Selection
- T cells that bind too strongly to self MHC proteins (autoreactive) die by apoptosis
- a:B Lineage is more dominant than y:d Lineage
- a:B has two sublineages distinguished by CD4 and CD8 receptors
- can recognize MHC class II and I respectively
DEVELOPMENT IN THE THYMUS
- Thymocytes: immature T cells
- Embedded in epithelial network thymic stroma
- Cortex: outer, close-packed, ectodermal
- Medulla: inner, less dense, endodermal
- Thymus is a primary lymphoid organ
- Only concerned with production of useful lymphocytes
- Not concerned with application to problems with infection
- Not involved in lymphocyte recirculation
- Does not receive lymph from tissues
- Involution of thymus
- Human thymus is fully developed at birth but degenerates one year after birth
- gradually replaced by fatty tissue
- does not impair T-cell immunity
- same case with thymectomy (removal of thymus)
- T cell repertoire are either long-lived, self-renewing, or both
- Thymic anlage: rudimentary thymus of cortex + medulla cells
- Colonized by progenitor cells
- Gives rise to thymocytes and DCs
- DCs and macrophages populate the medulla
- T cell progenitors enter the thymus at the junction between cortex and medulla: cortico-medullary junction
- Thymocytes move out through the cortex > subcapsular region > outer cortex > inner cortex > medulla
- Hassall’s corpuscles: sites of cell destruction
- DiGeorge Syndrome
- Thymus fails to develop = T cells are absent
- Suffers from opportunistic infections
- No adaptive immune system
COMMITMENT TO T-CELL LINEAGE
- Progenitor cells are not committed to the T-cell lineage when they enter the thymus
- Expresses stem cell markers CD34
- CD3: generic marker of T cells
- CD19: generic marker of B cells
- CD56: NK cells
- Interacts with thymic stromal cells = signals to divide and differentiate
- lose stem cell markers = become thymocytes committed to T-cell lineage
- expresses CD2 adhesion molecule and CD5
- double-negative thymocytes (DN thymocytes)
- Thymocytes still lack T-cell receptor complex but begins to rearrange T-cell receptor genes
- Express neither CD4 or CD8
- Critical cytokines
- IL-7
- Secreted by stromal cells and binds to receptors on CD34-expressing progenitor cells
- Defective IL-7 receptors = T cells absent
- Notch1
- Cell-surface receptor on thymocytes
- That’s why they need to go to the thymus because andoon ang Notch ligand transcription factor that it needs to interact with
- Binds to transmembrane ligands on thymic epithelial cells
- Drives cells along the pathway of T-cell differentiation and away from B-cell differentiation
- Parallels that of Pax5 in B cell development
- IL-7

- Notch1 Protein
- Has intracellular and extracellular domains
- Extracellular domain binds to ligand on thymic epithelium = cleavage of the two domains
- Intracellular domain translocate to nucleus
- Initiates gene transcription
COMMON THYMOCYTE PROGENITOR
- Both lineages derive from DN thymocyte precursors
- Where T-cell receptor gene rearrangement is initiated
- Rearrangement of y, d, and B loci occur at the same time
- If thymocyte makes a functional y:d receptor before a functional B = committed y:d T cell
- If thymocyte makes a functional B chain first = incorporated into pre-T cell receptor
- Still not committed to a:B lineage
- double-positive thymocytes (DP thymocytes)
- gene arrangement stops when functional B chain is incorporated into pre-T cell receptor
- cell proliferates and expresses CD4 and CD8 co-receptors
- allows a-chain rearrangement
- continues y and d-chain rearrangement
- If functional a:B receptor first: committed a:B lineage
- If functional y:d receptor first: committed y:d lineage
DN THYMOCYTE GENE REARRANGEMENT
- B and d chain : V+(D+J) segments
- a and y chain: V+J segments
- Functional y:d receptor gene arrangement is made first
- y:d heterodimer is formed
- assembles CD3 signaling complex and moves to surface
- signals to stop B chain rearrangement
- y:d T cell receptors are not subjected to positive and negative selection
- Functional B chain gene arrangement is made first
- More frequent outcome
- Translocate to ER
- Binds to surrogate a chain pTa to test capacity
- Pre-T cell Receptor
- If functional B chain successfully binds to pTa
- Forms heterodimer with CD3 complex and ζ chain
- pTA interacts with C and V regions of the B chain
- generates signals through the CD3 complex and Lck cytoplasmic protein
- Assembly of pre-T cell receptor signals to stop rearrangement of B, y, and d chain genes
- First checkpoint to ensure B chain can bind to a chain
- Cells become pre-T cells
- If functional B chain successfully binds to pTa
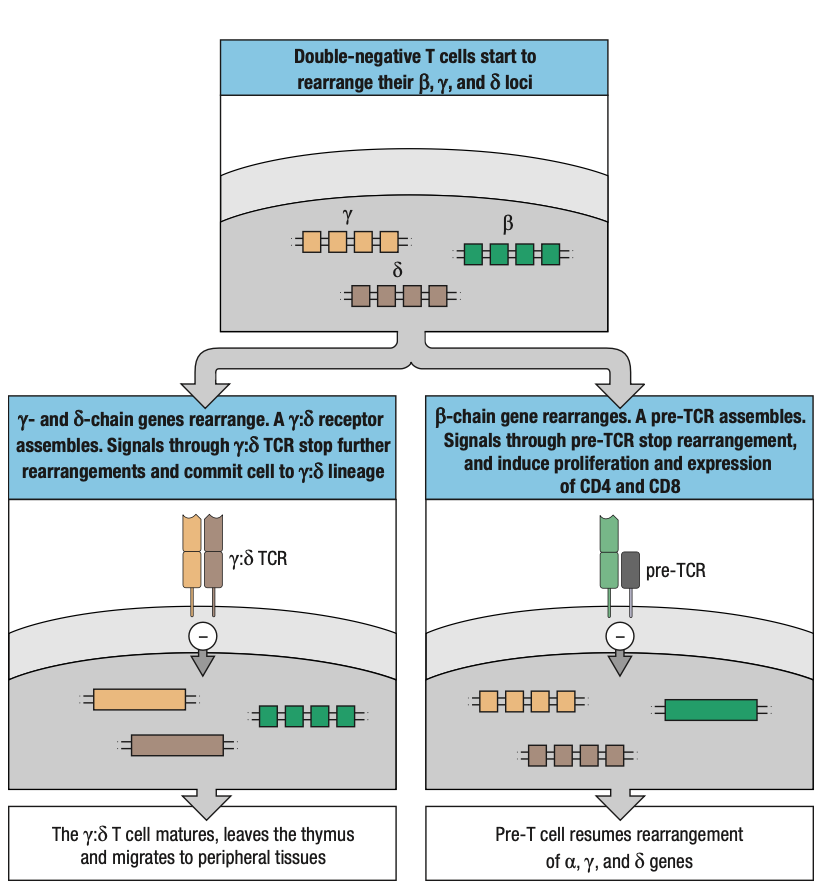
B CHAIN REARRANGEMENT
- a:B lineage is favored because it only requires one productive gene rearrangement while y:d requires at least two
- this increases the potential to rescue nonproductive B chain rearrangements
- two C genes and their associated D and J segments
- if rearrangement is nonproductive: thymocyte can attempt rearrangement at the other chromosome or same locus
- Four attempts to rearrange B chain gene
- 80% of T cells make productive B chains
A CHAIN REARRANGEMENT
- Successful rearrangement of B chain signals via pre-T cell receptors to stop gene rearrangement
- Express RAG1 and RAG2 genes
- Ensures that only one productive B chain is expressed = allelic exclusion
- Pre-T cell is induced to proliferate and create clones expressing the same B chain
- Accompanied by expression of CD4 first and followed by CD8 = DP thymocytes
- Constitute majority of thymocytes and are found in the inner cortex
- Interacts with epithelial cells
- Once Pre-T cells stop proliferating, they become small DP thymocytes
- Reactivation of recombination machinery and targeted to the a, y, and d loci
- T-cell receptor a chain genes
- Similar to immunoglobulin kappa and lambda light chain genes
- No D segments
- Rearranged only after B chain has been expressed
- Repeated rearranging attempts possible
- Multiple V segments and J segments
- Continues until productive rearrangement occurs or until the supply of V and J segments is exhausted
- Rearrangement of a chain
- d locus within the a locus will be deleted irrespective of whether the rearrangement is productive or not
- reduces the probability that committed a:B lineage expresses y:d receptors and a:B receptors
- d locus within the a locus will be deleted irrespective of whether the rearrangement is productive or not
- when DP thymocytes make a productive a chain rearrangement
- gene transcription starts to form a chain
- translocate to ER
- tested to bind to B chain and form T-cell receptor
- second checkpoint
- if successful: proceed to positive selection


GENE EXPRESSION CHANGES DURING DEVELOPMENT
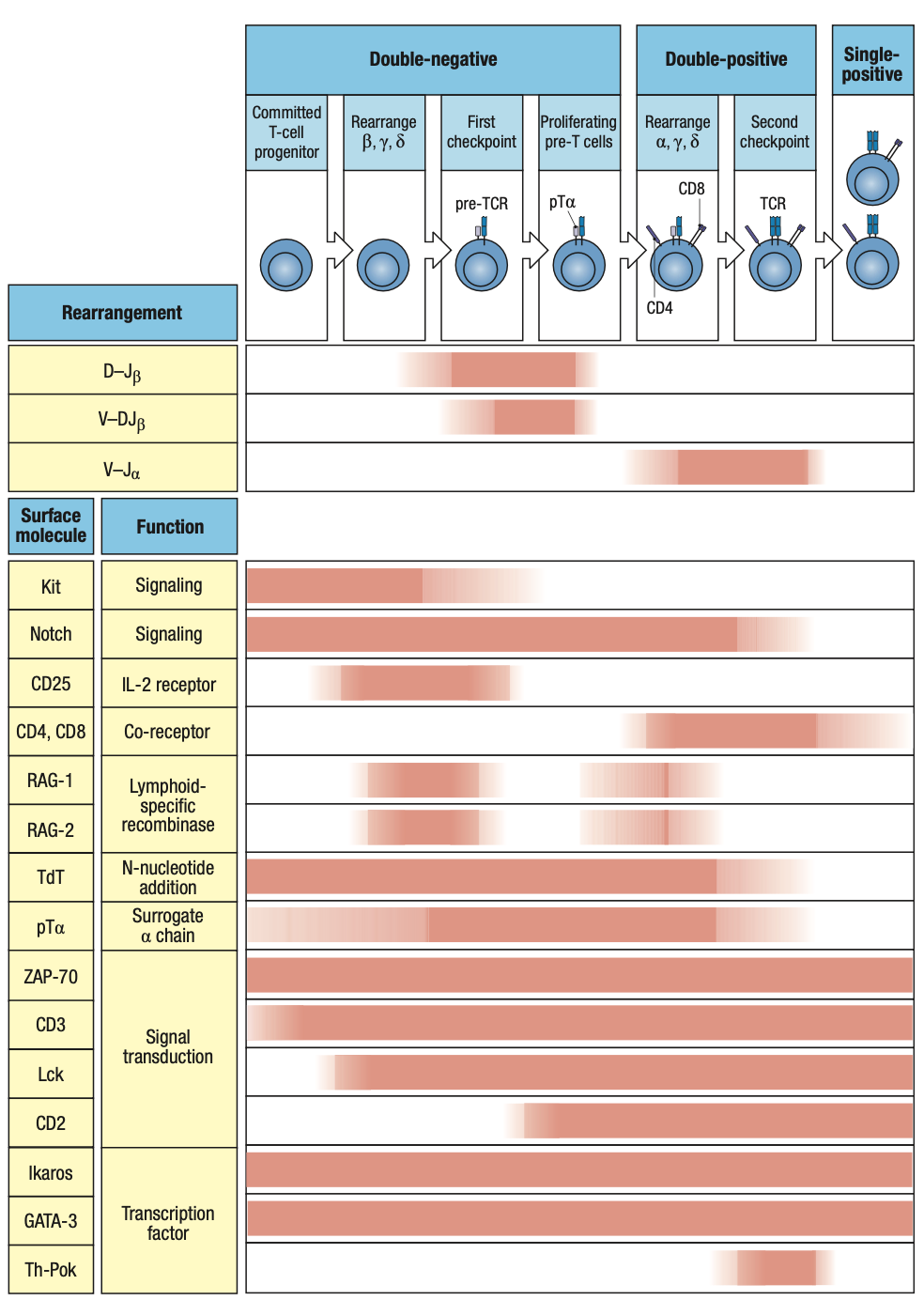
Notch transcription factor
RAG1, RAG2: may stop bc of allelic exclusion (checkpoint testing)
TdT: junctional diversity
pTa: surrogate alpha chain
- RAG1, RAG2: expressed at the two stages where B and a gene rearrangements are made respectively
- TdT, pTa: also expressed throughout this phase of development
- pTa: expressed throughout the period when rearrangements occur
- CD4, CD8: relay signals from pre-T cell receptors
- Tyrosine kinase Lck: signaling from pre-T cell receptor and co-receptor
- CD2: adhesion molecule on T cells that interact with cell-surface CD58 on other cells
- Ikaros, GATA-3: transcription factors in early progenitors
- Th-POK: expressed late in development and is for development of CD4 T cells from DP thymocytes

POSITIVE SELECTION
- First phase of T cell dev: produce T cells irrespective of their antigenic specificity
- Second phase: examination of receptors produced and selection of those that can work effectively with self MHC molecules
- Only involves a:B T cells
- y:d cells do not depend on MHC molecules for detecting infection
- primary T-cell receptor repertoire has a bias towards MHC molecules but T cell receptors are not tailored specifically
- only 2% of total can interact with MHC I or II
- Positive Selection
- Small population that can interact with MHC molecules are signaled to mature further
- Takes place in cortex of thymus where thymic cortical epithelium expresses both MHC I and II molecules
- Forms a web of cell processes that envelop and make contact with DP thymocytes
- Potential interactions are tested
- If peptide:MHC is bound within 3-4 days after the thymocyte first expresses a functional receptor = further maturation

CONTINUATION OF GENE REARRANGEMENT
- If first a chain rearrangement of pre-T cell is productive = positive selection occurs within a few hours
- Turns off recombination machinery
- Degrades RAG proteins
- Stops transcription of RAG genes
- Proliferates
- If not = rearrangement continues
- Can continue throughout the 3-4 days of positive selection
- After successful B chain = developing T cell turns off recombination machinery = allelic exclusion
- a chain is not subject to allelic exclusion
- rearrangements can occur at both copies of a chain locus
- DP thymocytes can express 2 a chains = 2 T cell receptors
- Receptor editing
- Trying out different a chains in order to become reactive with self MHC molecules
- Opposite to B cells = receptor editing is to get rid of reactivity
CD4 CD8 DETERMINATION
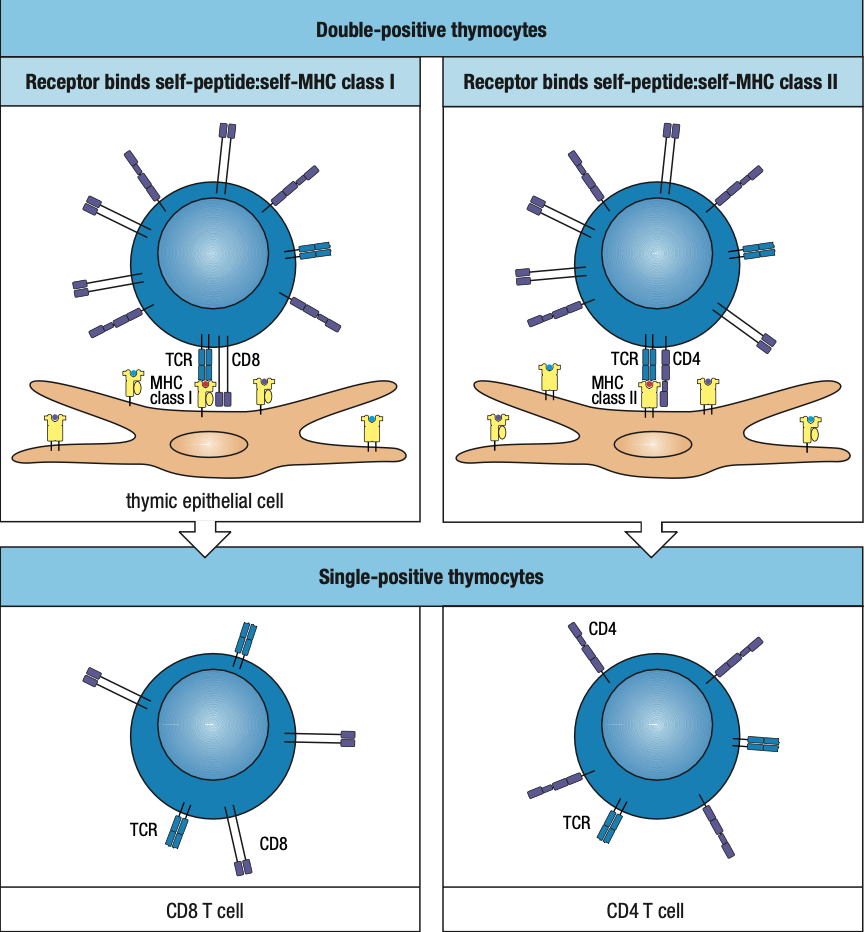
- Single-positive thymocytes (SP thymocytes)
- result of positive selection
- DP thymocytes mature into SP thymocytes
- If peptide interacts with MHC I = CD8 is recruited
- Any cell that is nucleated should be able to present it via MHC I in case infected
- If peptide interacts with MHC II = CD4 is recruited
- CD4 coordinating cell in innate and adaptive immune system
- HIV kills CD4 = acquired immue deficiency syndrome
- ACPs magnify signals of infection and is needed sa MHC II since CD4 is what coordinates everything
- Bare lymphocyte syndromes
- Lack of expression of either MHC I or II molecules by lymphocytes and thymic epithelial cells
- Patients who lack MHC I have CD4 cells but no CD8 ; vice versa
NEGATIVE SELECTION
- T cells whose antigen receptors bind too strongly to self-MHC molecules ( potential autoreactive) are deleted
- To avoid tissue damage and autoimmune diseases
- Mediators
- Positive selection: epithelial cells
- Negative selection: dendritic cells, macrophages, thymocytes
- T cells that bind too strongly to MHC I molecules on DCs or macrophages are signaled to die
- To extend negative selection to proteins that are specific to one or a few cell types, such as the insulin made only by Beta cells of the Langerhans cell of the pancreas, transcription factor autoimmune regulator (AIRE) causes several hundred of these tissue-specific genes to be transcribed by a subpopulation of thymic epithelial cells
- Activation of AIRE allows thymus to express those specific proteins na di makikita sa iba, para ma-practice ng T-cell kung madedetect and kung magiging autoreactive ba o hindi
- Peptides derived from these proteins can be bound by MHC class I molecules to form complexes that participate in negative selection
- Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) or autoimmune polyglandular syndrome type 1
- T cells specific for tissue-specific antigens are not eliminated by negative selection and thus attack variety of tissues
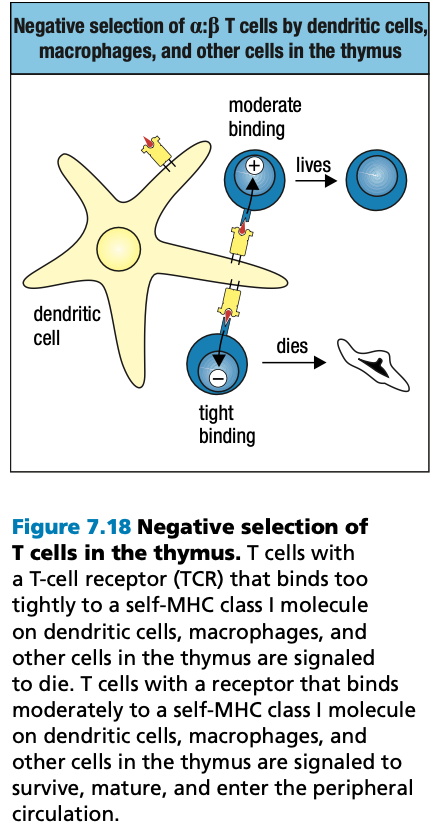
REGULATORY CD4 T CELLS
- Functions of CD4 T cells
- Helper T cells: activates macrophages and B cells
- Regulatory T cells
- Suppress response of self-reactive CD4 to their specific antigens
- Despite AIRE activity, self-reactive CD4 T cells are still present in healthy people
- Have T cell receptors specific for self-antigens
- distinguished from other CD4 T cells by expression of CD25 on cell surface and transcriptional repressor protein FoxP3
- On contacting self-antigens presented by MHC II
- Treg do not proliferate
- Suppress the proliferation of naïve T cells responding to self-antigens presented on the same antigen-presenting cell

FURTHER DIFFERENTIATION IN SECONDARY LYMPHOID TISSUES
- Only a small fraction of a:B T cells survive positive and negative selection and leave the thymus as mature naïve T cells
- Secondary lymphoid tissues provide specialized sites where naïve T cells are activated by their specific antigens
- Encounter with antigen provokes the final phases of T cell development and differentiation: the mature T cells divide and differentiate into effector T cells
- Some stay in lymphoid tissues while others migrate to sites of infection
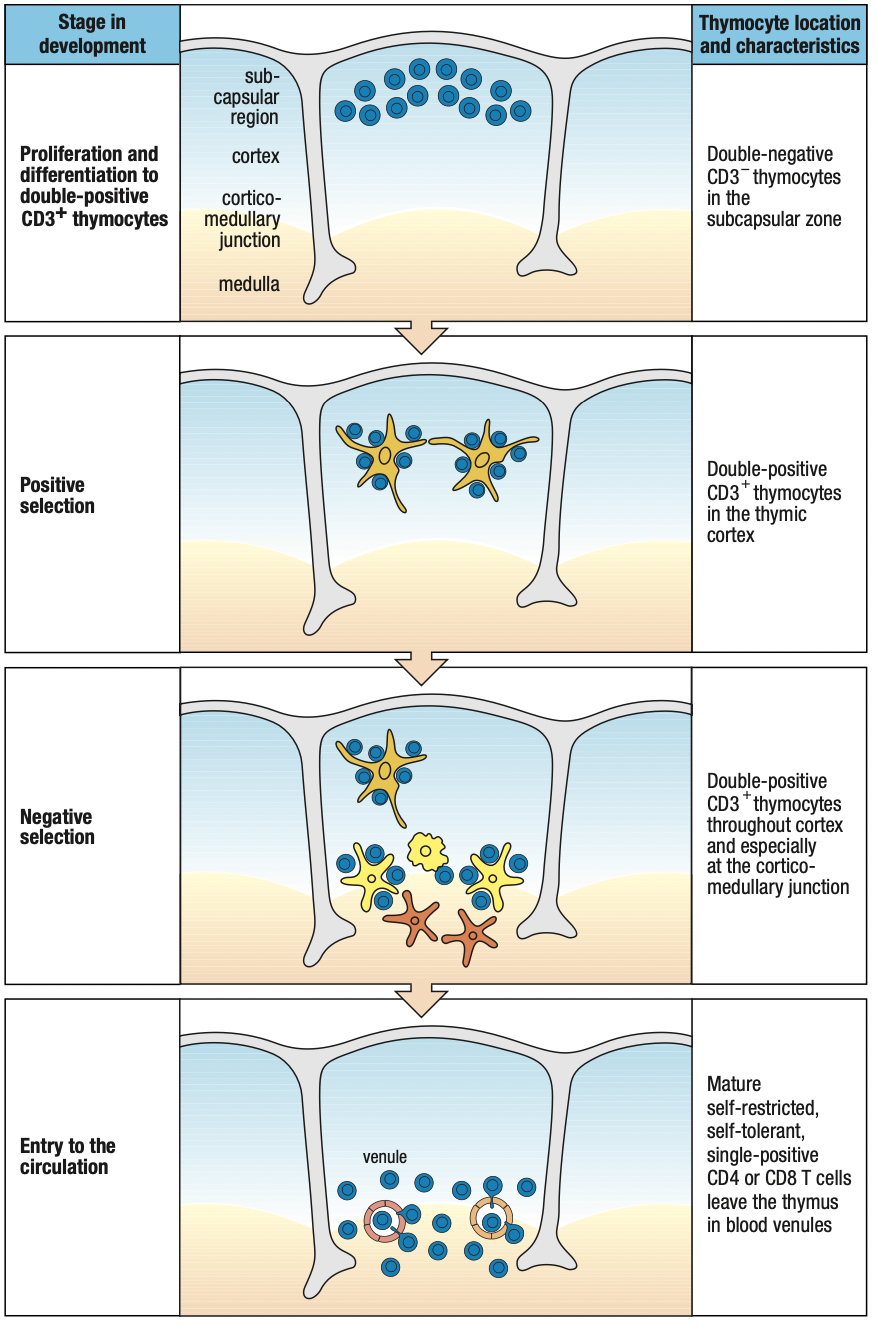
Discussion Notes
B cells (Humoral)
- have role in viruses
- Already circulating antibodies can neutralization before virus can enter cells
T cells (Cell-Mediated)
- Antigen presentation by MHC before T cells can act
- Necrosis: cell death due to external factors
- MHC I: all nucleated cells (kaya hindi kasama RBCs)
- MHC II: professional antigen-presenting cells
- Dendritic
- Macrophage: recruitment
- Mature B cells
Similarity: Somatic recombination (gene rearrangement)
- Diversity of receptors
- B cell receptors are identical
- Allelic exclusion: ano na-arrange, ayun lang ie-express nila
- More effective as an immune cell kasi isang target lang ang need i-focus
- The more diverse MHC = the more diverse receptors you need to have
- B cells need input from cell-mediated immunity before activation
- Mas derived ang humoral since it cannot function without pre-existing xdsfunction of cell-mediated
Rubor: redness : increased blood flow to recruit more immune cells
Tumor: inflammation due to vasodilation which looses tight junctions
Dolor: pain because of triggering pain receptors
Calor: heat to impede the propagation of pathogens
B cells leave bone marrow as immature B cells
They mature upon meeting follicular dendritic cell (different from APC dendritic cell)
“follicular” = lymph node
Will give B cell maturation factor
Then start looking for infection to complete its maturation
Kaya called lymphocytes kasi paikot ikot sa lymph nodes
B cells only recognize antigen
For t cells, they need to recognize both MHC and antigen
After being able to attach to MHC molecules (positive selection), they must be able to let go para iwas autoreactivity (negative selection)
TLRs= receptors of the innate system
PAMPs= pathogen associated molecular patterns
NK cells are non specific but fast acting
Pero since nonspecific, if a pathogen dominates, di na kaya labanan kaya saka need ng adaptive
Cytokine storm = nagpapanic kasi di macontrol ang infection/ mapatay ang pathogen so release lang ng release ng cytokines = over inflammation
Immune system is aware that inflammation can go overboard
T regulatory = anti-inflammatory cytokines to regulate/
IgG4 is anti-inflammatory sa innate and acts as regulator
CHAPTER 8
T-CELL MEDIATED IMMUNITY
Remember: Adaptive immunity is initiated through interactions of NK cells and dendritic cells (refer to ch. 3)
8-1: DENDRITIC CELLS CARRY ANTIGENS FROM SITES OF INFECTION TO SECONDARY LYMPHOID TISSUES
- Adaptive immune responses are initiated by capturing some of the pathogen and sequestering them in the secondary lymphoid tissue
- Myeloid dendritic cells capture antigens and present them to naïve T cells
- T-cell response to infections is made in the draining lymph node
- Blood infections: spleen
- Mucosal tissues: mucosal secondary lymphoid organs
Dendritic Cells
- Immature DCs
- In the skin and peripheral tissues
- Active in capture, uptake, and processing of antigens
- Mature/activated DCs
- No longer active in capture, uptake, and processing of antigens
- Upon moving to secondary lymphoid tissues
- Gains capacity to activate naïve T cells
- Confined to the outermost part of the cortex where T cells congregate
Macrophage
- Present in the cortex and medulla
- Removes pathogens and their breakdown products from the afferent lymph that arrives from the site of infection
- Macrophage-mediated lymph filtration
- Prevents infectious microorganisms from passing through the node and gaining access to the blood via efferent lymph
- Prevents systemic infections
- Eliminate lymphocytes that are signaled to die by apoptosis
8-2 DCs PROCESS ANTIGENS
- DCs present peptides on MHC II molecules to activate naïve T cells
- DCs have various endocytic and signaling receptors
- Receptor-mediated endocytosis
- Capture bacteria/virus particles from EC fluid and process in lysosomes
- Aka micropinocytosis
- Micropinocytosis involves nonspecific ingestion of larger volumes of ECF and is used to capture pathogens not recognized by any endocytic receptor
- DCs can become infected and make viral proteins that enter the endosomal ER and presented on MHC I molecules
- If infection not lethal, DCs carry the virus to the draining lymph node and activate naïve CD8 T cells
- If infection is lethal, DCs reach secondary lymphoid tissue but too sick to activate naïve T cells
- The virus released by dying DCs can infect healthy DCs which then present the antigens on their MHC I molecules and activate CD8
- If virus do not infect DCs, cross-presentation is used to stimulate a CD8 response
- DCs take up antigens by the endocytic pathway
- Transfer the virus degraded peptides to the exocytic pathway for cross-presentation by MHC I

- DCs carry all TLRs except TLR9
- Highly sensitive to the presence of all manner of pathogens
- Signals from TLRs lead to DC activation
- Increase efficiency in uptake, processing, and presenting
- Appearance of CCR7 on the cell surface which is the receptor for the CCL21 chemokine in the secondary lymphoid tissue
- Leave lymph and enter the tissue of the draining node
- Maturation of DCs to focus on presentation
8-3 NAÏVE T CELLS FIRST ENCOUNTER WITH PEPTIDE:MHC COMPLEX
- T cells bind to the endothelial cells of the thin-walled high endothelial venules (HEV)
- They squeeze through the vessel wall and enter the T-cell area
- They encounter mature DCs
- T-cell receptors examine the peptide:MHC complex on DC surfaces
- If it binds successfully, T cell is activated and retained in LN
- T cells can arrive via the afferent lymph
- Entered an “upstream” LN from the blood but did not encounter specific antigen, leave the LN via the efferent lymph and carried “downstream”
- T cells arrive at the node in a different afferent vessel from that carrying pathogens and antigens and DCs from the infected tissue

- If T cell does not find its specific antigen, it leaves the medulla in the efferent lymph to continue recirculation
- If T cell is activated, it takes several days for the cell to proliferate and differentiate into effector cells
- Accounts for much of the delay between the onset of an infection and the appearance of the adaptive immune response
8-4 HOMING OF NAÏVE T CELLS
- Homing: naïve T cells leave bloodstream and enter T-cell area of LN
- Guided by CCL21 and CCL19 chemokines
- Secreted by stromal cells and DCs in T cell area
- Establish a concentration gradient along the endothelial surface
- Naïve T cells express CCR7 chemokine receptor which binds to CCL21 and CCL19
- Guides T cells up the chemokine gradient
- Exit route: cortex > medulla > efferent lymph
- Controlled by T cell receptor that can recognize sphingosine 1-phosphate (S1P) that establishes gradient on the LN
- Lowest in T-cell area and increases in the direction toward the efferent
- For T-cells that recognized antigen on DCs
- Express CD69
- Moves S1P receptors inside the cell to prevent external sensing
- Prevents leaving LN during nurturing
- Upon maturation into effector T cells, they stop expressing CD69
- Express CD69
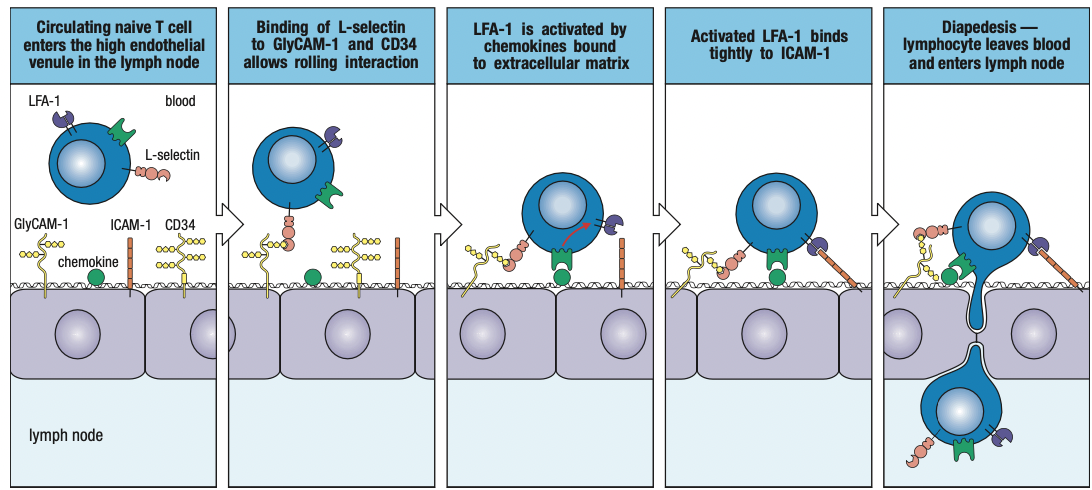
Adhesion Molecules
- L-selectin
- On the T cell surface
- Binds to CD34 and GlyCAM-1 on endothelium
- Causes naïve T cell to slow down and attach to endothelial surface
- LFA-1
- On T cell surface
- Binds with ICAM-1 and ICAM-2 on endothelium
- Activated by chemokines secreted by LN
- Strengthens hold on the ICAM
- Transient binding with DCs
- Upon encountering specific peptide:MHC complex, LFA-1 molecules increases affinity for ICAMs = conjugate
- ICAM-3
- On T cell surface
- Binds to LFA-1 and DC-SIGN (unique to activated DCs)
- Transient binding with DCs
- CD2
- On T cell surface
- Binds with LFA-3
- Strengthens adhesion
8-5 ACTIVATION OF NAÏVE T-CELLS
- Co-stimulatory signal
- Required because signals generated by T cell receptor and co-receptor (CD4/8) with peptde:MHC complex is not enough for activation
- Without this, T cells cannot divide nor survive
- CD28 T cell co-stimulatory receptor which binds to B7 co-stimulatory molecule on DCs
- Only professional APCs express B7 in the presence of infection through presence of inflammation and not constitutively
- When DCs take up pathogens/antigens, signals are generated to induce B7 expression
- DCs arrive at the draining lymph node already expressing B7
- Inflammation is needed or else anergy = pag isang e.coli lang, hindi dapat ma-activate adaptive immune system
- Hindi enough ang pathogen population to induce inflammation = not a concern = anergy
- Effector T cells activates macrophage and naïve pathogen-specific B cells to express co-stimulatory molecules
- Once T cells are activated, they express an additional complementary B7 receptor CTLA4
- Binds B7 twentyfold
- Functions as brake on CD28 to inhibit activation and proliferation of T cells
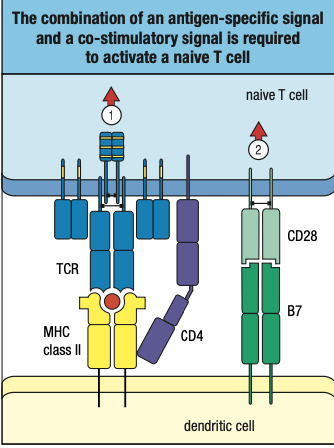
8-6 RECEPTOR SIGNALS FOR ACTIVATION
- Steroid signals can pass through to bilayer and into the nucleus kasi andoon ang receptor
- Protein signals cant pass through the bilayer kaya receptor niya ay membrane-bound to start signal cascade towards nucleus
- T-cell synapse: region of contact and communication between TCs and DCs
- Central supramolecular activation complex (c-SMAC): TC receptors, co-receptors, co-stimulatory receptors, and CD2 adhesion molecule
- Peripheral supramolecular activation complex (p-SMAC): LFA-1, ICAM-1, and talin which forms a tight sea, around the SMAC
- Interactions of MHC ligands with TC receptors activate cytoplasmic protein tyrosine kinases
- Phosphorylate tyrosine residues (immunoreceptor tyrosine-based activation motifs or ITAMs) in the CD3 tails and the associated zeta chain CD247
- Extracellular binding of antigen to TC receptor initiates pathways of intracellular signaling that leads to TC differentiation
- Cytoplasmic tails of co-receptors associate with Lck
- Phosphorylate the CD3 ITAMS and ZAP-70 upon synapse formation
- Binds to the phosphorylated tyrosines of the zeta chain
- Activated ZAP-70
- Leads to activation of nuclear factor of activated T cells (NFAT)
- Leads to activation of protein kinase C-theta which induces NFkB
- Leads to activation of nuclear protein Fos, one of the component of transcription factor AP-1
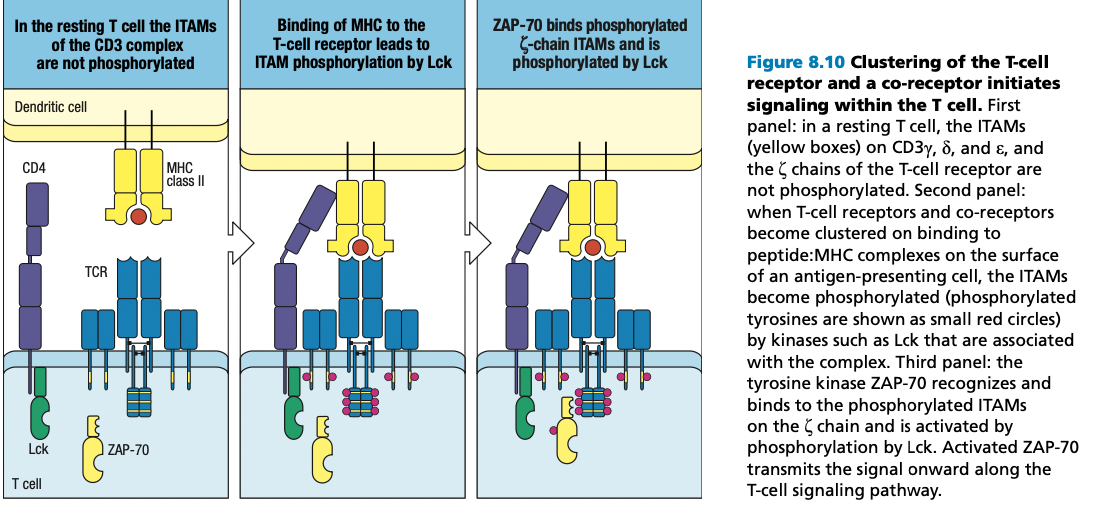
8-7 IL-2 FOR PROLIFERATION AND DIFFERENTIATION
- Synthesized and secreted by activated T cells
- Autocrine action
- Paracrine action: cytokine acts on a different type of cell from the one that made it
- Requires signals from TC receptor:co-receptor complex and the co-stimulatory signal delivered by CD28 and activation of NFAT
- Co-stimulatory signal stabilizes cytokine mRNA to increase TC production of IL-2 and rate of transcription from the IL-2 gene
- Naïve TC has B and Y chain of IL-2 receptor
- Activated TCs: alpha chain is synthesized
- Triggers TCs to progress through cell division
8-8 T CELL ANERGY
- Some T cells with specificity for self-antigens escape negative selection in the thymus because the self-antigens they recognize are not expressed in the thymus
- These cells will not express B7
- Absence of B7 and co-stimulation, engagement of peptide:MHC complex by TC receptor and co-receptor = TC anergy
- State in which it cannot respond to any external signal
- Cannot be revived
- Fails to make IL-2
- Irreversible mechanism of self-tolerance to render harmless those self-reactive T cells
8-9-10 CD4 T CELL POPULATIONS
- CD4 TCs do not directly attack the pathogens
- Form a cognate pair with target cell

TH1
- Help macrophages
- For intracellular bacterial and viral infections
- Stimulated by IL-12 (by DCs) and IFN-y (by NK cells)
- Controlled by T-bet transcription factor
- Causes TC’s own IFN-y gene to be turned on
- Local production and secretion of IFN-y in the secondary lymphoid tissue
- Increases inflammation
TH2
- Help eosinophils, basophils, mast cells, and B cells
- For parasites
- Do not generate inflammation but promote repair and recovery of damaged tissues
- Favors production of pathogen-specific IgE antibodies to eliminate parasites
- Stimulated by IL-4 which binding induces GATA3 to commit to the TH2 pathway
- Turns on IL-4 and IL-5 which are secreted by TH2
- Histamines = expulsion of allergen
- Sneeze, diarrhea, scratching
- IgE is found on surface of eosinophil and mast cells
TH17
- Secrete IL-17 and CXCL8
- Recruits neutrophils to infected tissues
- Stimulated by IL-16 and TGF-B
- Induces TC to make IL-21 which activates STAT3
- Controlled by RORyT which turns on IL-17
TFH
- Cooperate with naïve B cells
- Initiate antibody response
- Induced by IL-6 and controlled by Bcl6
- Co-stimualtion involves the Inducible TC co-stimulator (ICOS)
- Bcl6 express CXCR5 the receptor for CXCL13 produced by stromal cells of B cell follicles and causes exit from T cell area into follicles of the B cell areas
- Guides switching of immunoglobulin isotypes in B cells
Treg
- Keeps immune response under control
- Limits tissue damage and facilitate healing
- Reduce chance of secondary infections
- Natural regulatory cells: commit to regulatory function during thymic development
- Induced Treg: emerge in the course of the immune response to infection
- activated by TGF-B but in the absence of IL-6 and other pro-inflammatory cytokines
- both expresses FoxP3 and CD4 and CD25
- Induced Treg produces TGF-B and IL-10 to inhibit inflammation
8-11 POSITIVE FEEDBACK
- For both TH1 and TH2 cells, the cytokine that drives their differentiation is central to their effector function and secreted in quantity
- TH1: IFN-y
- TH2: IL-4
- Positive feedback: in which the functioning effector TCs drive further differentiation
- Polarized: if population is dominated by either TH1 or TH2
- Cell-mediated immunity
- Polarized TH1
- Humoral immunity
- Polarized TH2
- Tuberculoid Leprosy
- TH1 bias
- Help macrophages to suppress growth and dissemination of bacteria
- Chronic inflammation damages skin and peripheral nerves
- Disease progress slowly
- Lepromatous Leprosy
- TH2 bias
- Large pathogen-antibodies are made but ineffective against bacteria hidden inside macrophages
- Bacteria become disseminated to other sites in body
- Causes gross tissue destruction which is fatal
8-12 CD8 TCs REQUIRE STRONGER ACTIVATION
- CD8 TCs are functionally homogenous but must interact with a wide range of target cells
- For intracellular infections, mostly viral
- Recognize antigens presented on MHC I molecules of DCs
- For some viral infections, the interaction of a naïve CD8 with DC that presents antigen via MHC I is enough for activation and differentiation
- Synthesize IL-2 and receptor
- Induce CD8 TCs to proliferate
- For other viral infections, DCs solicit help from virus-specific effector CD4
- Gives IL-2 to jump-start activation
- Forms viral peptide:MHC I complex and viral-peptide:MHC II complex
- The effector CD4 is activated to make and secrete IL-2 which then binds to the receptor on CD8
- Drives the naïve CD8 to proliferate and differentiate
- CD8 are only activated when the evidence of infection is unambiguous
- Cytotoxic TCs inflict tissue damage

8-13 CD8 TCs and Effector CD4 TCs
- CD8 cytotoxic TCs and CD4 TH1, TH2, and TH17 TCs travel to infected tissue after differentiation
- Adhesion molecules on effector T cells allow them to leave the secondary lymphoid organ
- Differentiates them from naïve TCs
- L-selectin is replaced by VLA-4
- Binds to adhesion molecule VCAM-1 on activated endothelial cells at inflammation sites
- Halts passing effector TCs and direct them to enter the infected tissue
- Effector TCs express more CD2 and LFA-1 than naïve TCs
- Makes them more sensitive to ICAM-1 and LFA-3 than APCs
- Signals from TC receptor and co-receptor are sufficient to activate effector TCs
- Whereas induces anergy in naïve TCs
- Co-stimulation through CD28 and B7 is not needed for activation
- Allows effector TCs to kill any type of cell infected with virus
- CD4 effector TCs are activated by antigen recognition on all cells that express MHC II
- Including vascular endothelial cells induced to express MHC II by IFN-y secreted by NK cells and effector TCs at sites of infection
- Naïve TCs are activated by antigen recognition of DCs only
- Co-stimulation restricts initiation of TC responses
- Relaxing this requirement allows effector TCs to work quickly and efficiently
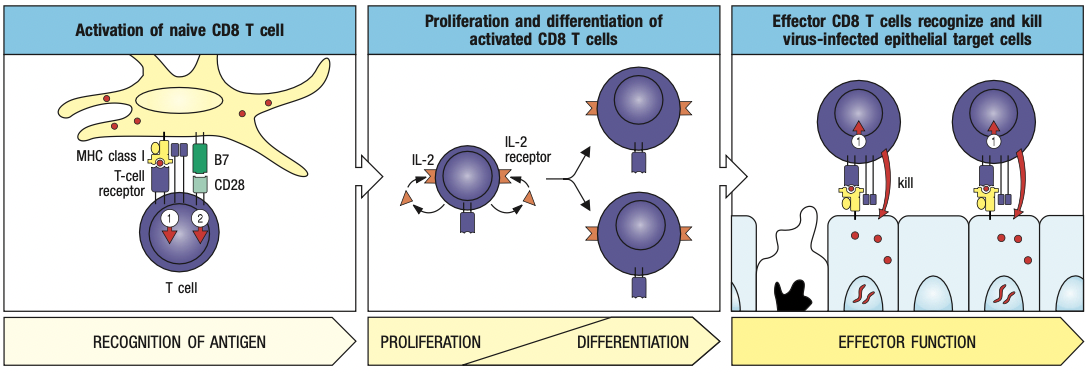
8-14 CYTOKINES AND CYTOTOXINS
- Cytokines: alter behavior of target cells
- Interleukins by TCs
- Made only after the effector TC has formed a conjugate
- Never made and stored in CD4 TCs for future use
- Cytotoxins: kill target cells
- Fewer than cytokines
- Effector CD8 manufacture and store them in lytic granules before an encounter with a target cell
- Comprised of granzymes, serglycin, and granulysin
8-15 CYTOKINES
- Janus kinases (JAKs)
- Dimerization of receptor polypeptides = activates JAKs into enzymes
- Phosphorylates STATs (signal tranducers and activators of transcription)
- Phosphorylated STAT dimerize = allows them to move from cytoplasm to nucleus
- Change gene expression in target cell
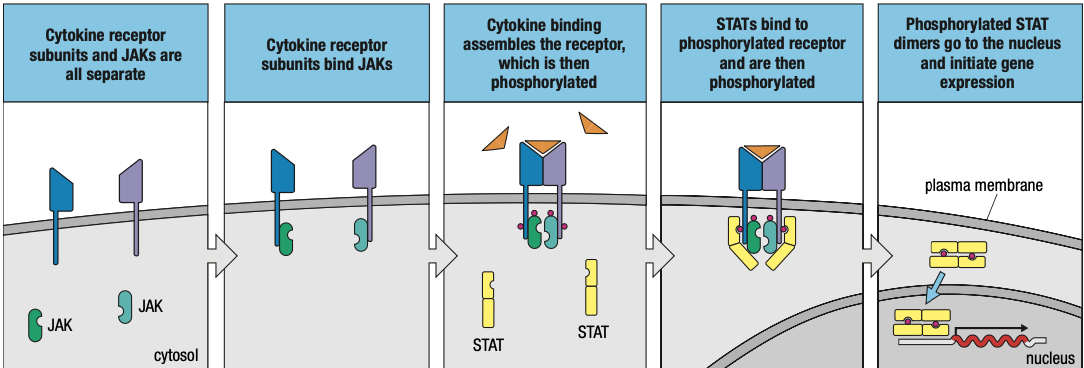
- Effects of cytokines can be turned off
- Phosphatases remove phosphate groups from JAKs, STATs, and cytokine receptors
- Suppressors of cytokine signaling (SOCS) proteins inhibits signaling pathway
8-16 SELECTIVE CYTOTOXINS
- CD8 synthesizes cytotoxins in inactive forms and package them into lytic granules
- Antigen specificity allows TCs to pick out only infected cells
- Cytoskeleton directs them to go towards T cell receptor with antigen kaya specific
- TC focuses granule secretion at the small, localized area of the synapse
- Granules only kill the infected cell
- CD8 also secretes cytokines
- IFN-y to inhibit viral replication in infected cells, increase viral antigen presentation, and activate macrophages

- One cytotoxic TC can kill many infected cells
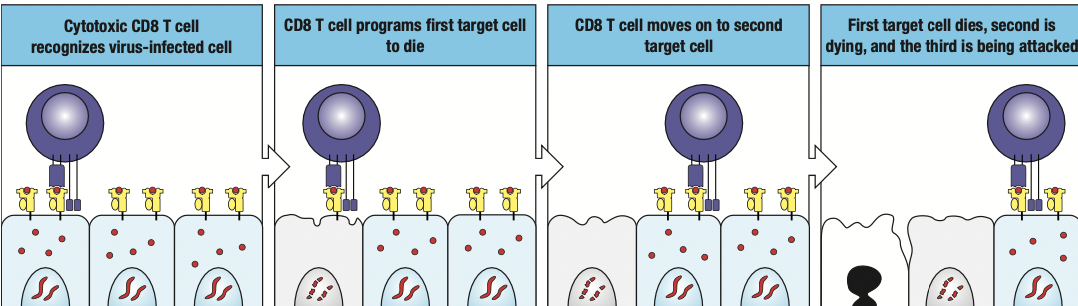
8-17 APOPTOSIS
- Cytotoxic CD8 kill cells by apoptosis aka programmed cell death
- prevents pathogen replication
- prevents release of infectious pathogen particles from infected cell
- cytotoxins make pores in target cell membrane
- phosphatidyl serine = marker of apoptosis because PS is normally in the intracellular side of the cell membrane
- if induced to undergo apoptosis, it will flip and move to the extracellular side
- anexinfide stains PS
8-18 MACROPHAGE ACTIVATION BY TH1 CD4
- TH1 CD4 help macrophages at infection sites to be more proficient in killing pathogens by secreting cytokines
- Macrophages containing captured pathogens fuse more efficiently with lysosomes
- Increased synthesis of potent microbicidal agents such as nitric oxide, oxygen radicals and proteases
- Overall enhancement of macrophage function by effector TCs is called macrophage activation
- Macrophages require two signals for activation from TH1
- Delivered by IFN-y and CD40 ligand
- Induces changes in gene expression for activation
- After forming conjugate with macrophage, TH1 takes several hours to initiate cytokine gene transcription
- Cytokines are translocated to the ER of TH1
- Delivered by secretory vesicles to synapse
- Only cytokine receptors on the macrophage become loaded with cytokines and subject to activation
- activation is antigen-specific
- only macrophages that present pathogen-derived antigens are selected for activation
- prevents unnecessary damage to healthy tissue
- regulatory mechanism: TGF-B, IL-4, IL-10, IL-13
- inhibit macrophage activation
- counter effects of TH1 cells
- evident in lepromatous leprosy whose macrophages become overwhelmed by proliferating bacteria

8-19 TFH CELLS AND NAÏVE B CELLS
- TFH cells help B cells make antibodies
- TFH cells move from the T cell area of secondary lymphoid organ to near B cell area
- Naïve B cells enter lymph node from blood because of chemokines CCL21 and CCL19
- Allows for interaction of TFH with naïve B cells
- once TFH and B cell forms synapse, TFH synthesizes CD40 ligand
- binds with CD40 on B cell
- causes proliferation, differentiation, and maturation of B cell into plasma cell
- Linked recognition
- Bounded B cells and T cells are specific for epitopes of the same antigen that was first bound and internalized by B cell receptor

8-19 REGULATORY TCs
- Treg expresses high levels of CD25
- Treg make immunosuppressive and anti-inflammatory cytokines
- IL-4, IL-10, TGF-B
- Suppression depends on physical contact between Treg and target cell
- Treg interacts with DCs to prevent them from interacting and activating naïve T cells
- Treg may also interact directly with effector TCs
- Human Treg has FoxP3
- Infants with no functional FoxP3 have no Treg
- fatal because it permits immune responses that are directed at self-antigens
NK cells similar to CD8 cytotoxic
No specific receptor for antigen
Nonspecific PAMPs triggers TLRs of NK cells
Activated dendritic (pag na-outnumber niya ang NK cells) detach from tissue and get washed by lymphatic fluid
MHC I magaling sa intracellular bc proteosome ang naglload ng peptide sa MHC I
Mas efficient ang cross presentation kasi it triggers both CD8 and CD4 (so cytotoxic and b cells are activated)
Kaya sa sars-cov-2 antibodies are needed kasi they play a role both in innate and adaptive
Cytokines that induces inflammation can loosen the tight junctions = recruitment of both neutrophils and T cells
Chemokines parang bread crumbs to direct t cells where to go
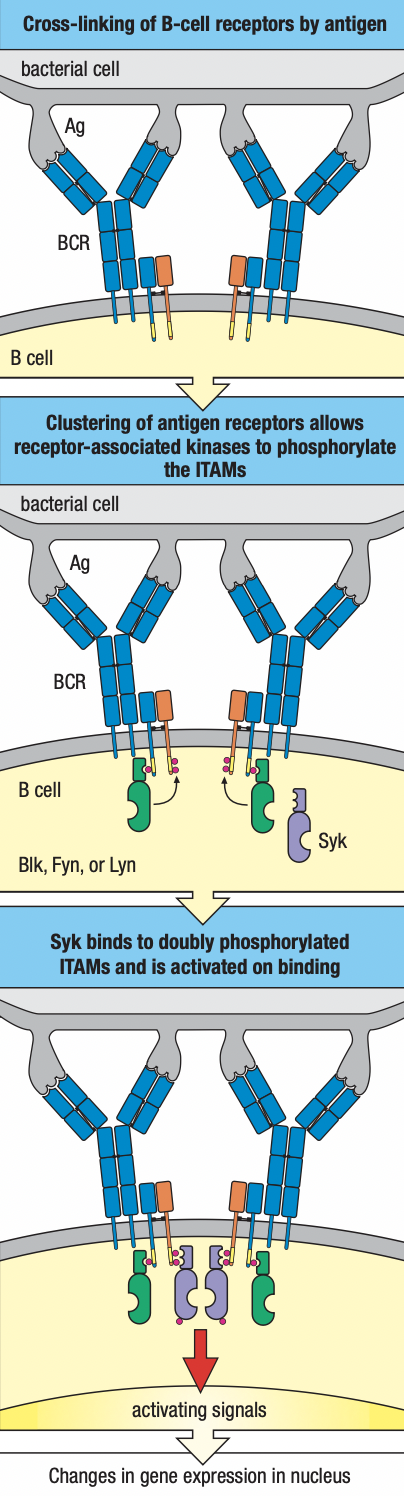
Chapter 9
Humoral Immunity
- TFH stays in the lymph node to activate B-cell arm of adaptive immunity
- Mature B cells has IgM molecules that become cross-linked by the antigen
- Sends signal from receptors to inside of cell
- Interaction of antigen and immunoglobulin is communicated by IgA and IgB (B cell receptor together with IgM)
- Not enough to activate naïve B cell

- Additional signals: B cell receptor:co-receptor
- CR2/CD21: recognizes iC3b and C3d derivatives deposited on the pathogen
- CD19: signaling chain
- CD81: binds to CD19 to bring it to the surface
- Generation of the iC3b and C3d ligands
- CR1 + C3b with ligand
- Becomes susceptible to cleavage Factor I
- Gives iC3b fragment and C3d fragment
- CR1 increases abundance of ligands for B cell co-receptor on pathogen surface
- CR2 component of co-receptor
- Upon binding to antigen, binds to C3d
- Causes kinase cascade
- Simultaneous ligation of the B cell receptor:co-receptor increases the B cell sensitivity to antigen
- TFH recognizes peptides presented by B cell MHC II
- Sends cytokines and signals to induce division and differentiation
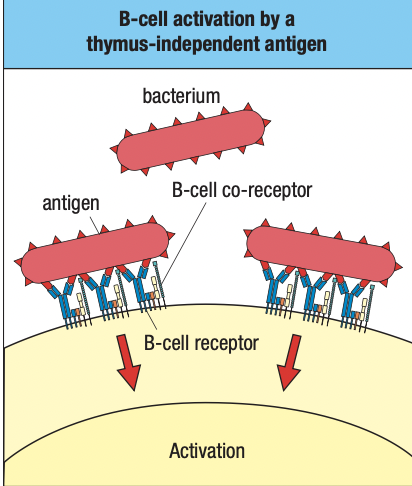
- DiGeorge Syndrome
- No thymus
- Little to no T cells
- Normal B cell levels
- B1 cells produce low-affinity IgM
- Does not require T cell activation
- Polyspecific and no isotype switching
- Carb or protein epitopes can cross-link B cell receptors and co-receptors and sufficient to activate B cell even without additional signals
- Aka thymus-independent antigens (TI antigens)
- Highlights importance of B2 cell and T cell activation
Follicular DCs
- FDCs provide signals to allow B cells to mature and survive
- Diff from myeloid DCs that present antigens to T cells
- Diff from plasmatycoid DCs that make type I IFN
- FDCs serve as a depository of intact antigens
- Available for interaction with antigen receptors of B cells
- They lack phagocytic activity = preserves antigens intact
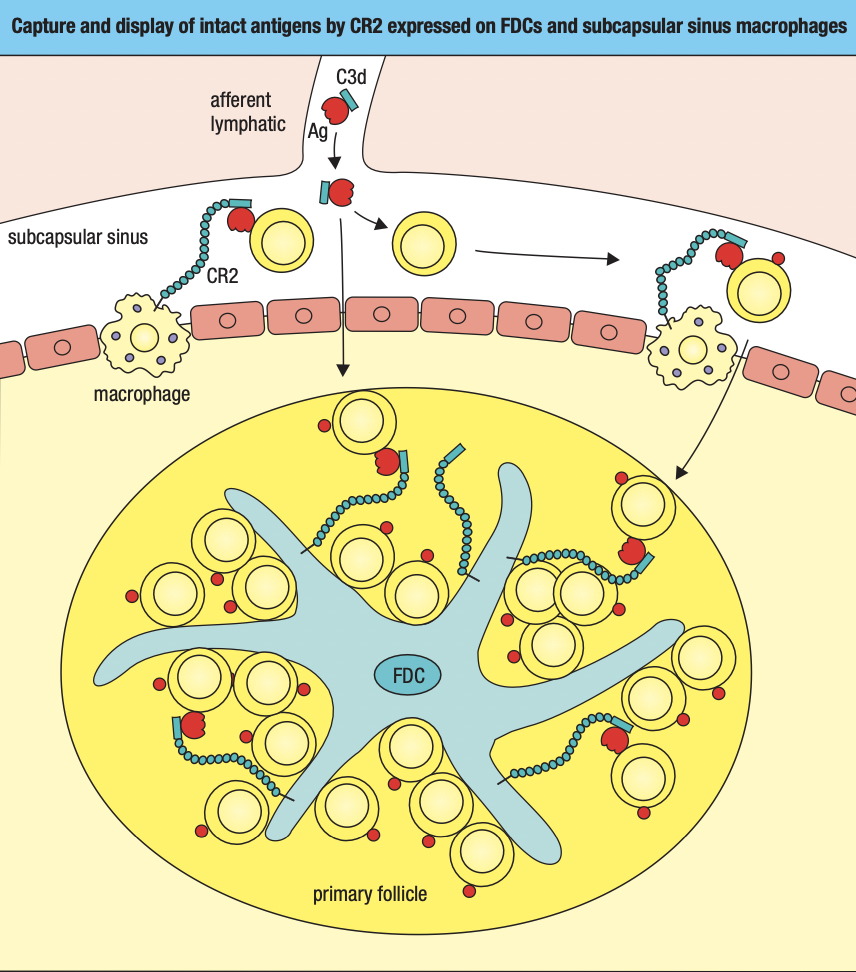
- FCs take up antigens from lymph
- C3b and C3d fragment binds to pathogen surface
- Those tagged with C3b and C3d are taken up by FCs
- Subcapsular sinus macrophage
- Little phagocytic activity
- Has CR1 and CR2 to take up antigens tagged with C3d or C3b
- Holds them on surface to be screened by naïve B cells
- Medullary sinus macrophage
- Highly phagocytic
- Filters lymph before leaving node
B cell and T cell interaction
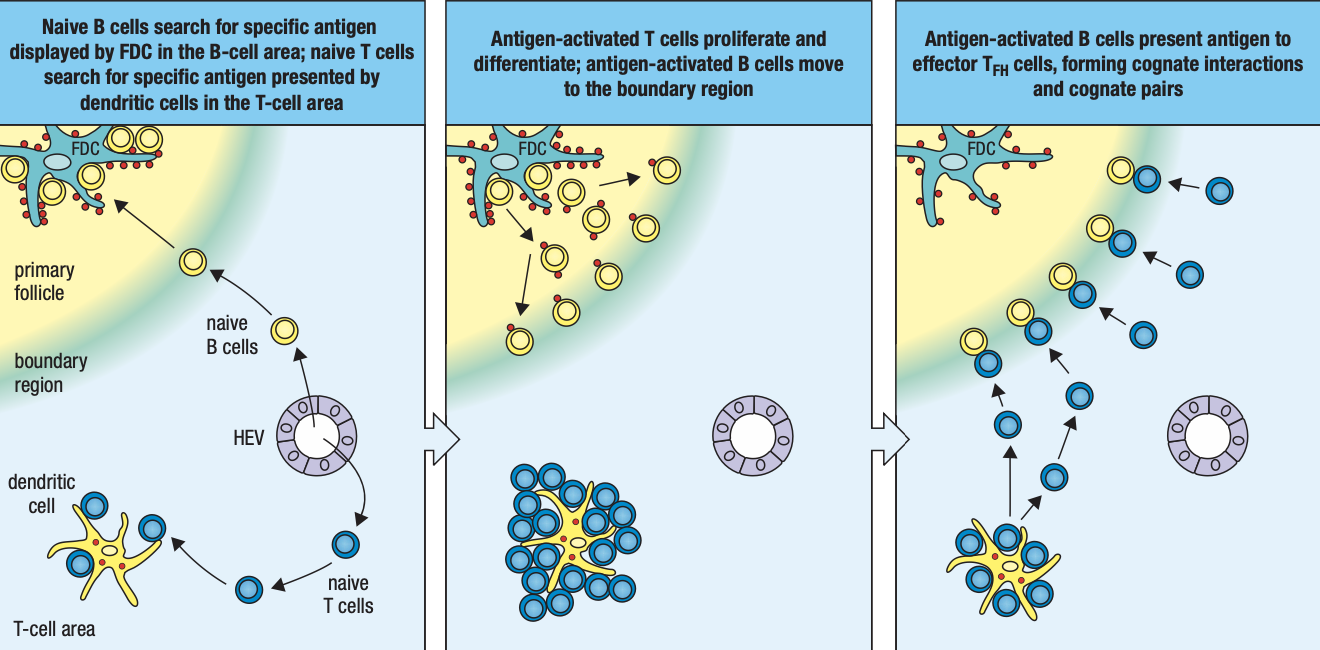
- Naïve B cells from blood will be attracted to T cell area by chemokines CCL21 CCL19 and to B cell follicle by CXCL13
- B cells enter node at subcapsular sinus to screen antigens
- If found, b cell enters B cell area of follicle to interact with TFH cells
- Expresses CD69 which prevents SIP receptor expression
- Stays in the lymphoid tissue to complete differentiation
- Activated B cell begin to endocytose and process the antigen and present peptides on MHC II
- Expression of CCR7 binds to CCL21 and CCL19
- Draws the activated B cell to the boundary between T cell and B cell areas to interact with newly differentiated TFH cells
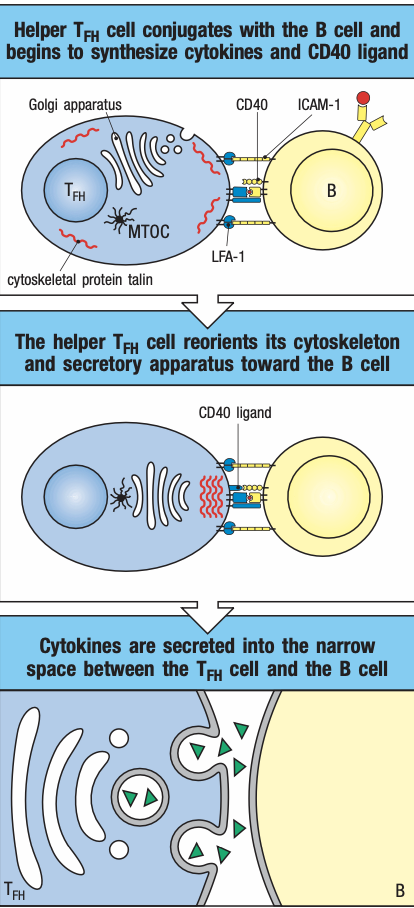
- TFH activated by DCs presenting antigen reduce expression of CCR7
- Moves towards boundary of primary follicle
- Screens antigens presented by activated B cells
- If found, forms conjugate and expresses CD40 ligand
- Activates B cell NFkB and enhances adhesion molecules
- Facilitates delivery of cytokines
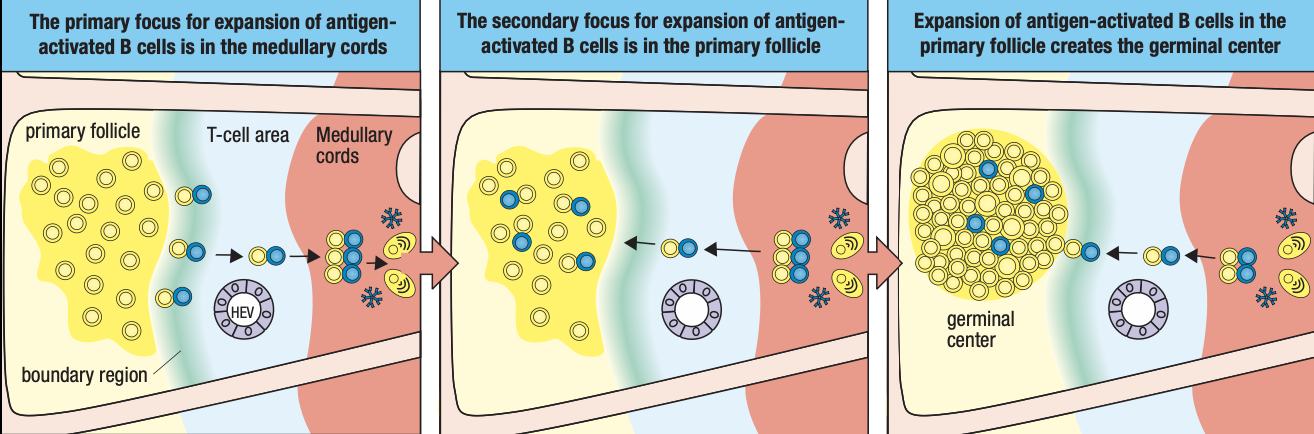
Primary Focus of Clonal Expansion
- Conjugate pair T cells and B cells move together to medullary cords
- They begin to divide to form primary focus
- Gives rise to dividing B lymphoblasts secreting IgM
- Some B cells stay in medullary cords
- Differentiate into palsma cells under the influence of cytokines IL5 and IL6 secreted TFH
- Determined by B-lymphocyte-induced maturation protein 1 (BLIMP-1) which stops transcription for proliferation
- Cells then increase expression of immunoglobulin chains
Somatic Hypermutation and Isotype Switching
- Other B lymphocytes leave the primary focus and move into primary follicles of the B area while still attached to TFH
- Proliferates and forms germinal center
- FDCs secrete IL6 IL15 8D6 and BAFF to start rapid division of B cells to form centroblasts
- TFH also divides and produce more cytokines
- induces B cells to produce activation-induced cytidine deaminase (AID) essential for somatic hypermutation and isotype switching
- during somatic hypermutation and isptype switching, centroblasts no longer express immunoglobulins because focus is on expansion of large population of B cells with switched isotype and V-region mutations not antigen selection
- proliferation of antigen-specific B cells in primary follicles give rise to secondary follicle
- GC no has rapidly dividing B cells and T cells in the center with naïve B cells at the periphery screening for their specific antigens and now forms mantle zone
If B cells produce antibodies, what happens next? What are antibodies exactly for?
Can be simplified into 3 effector functions: neutralization, opsonization, complement actication
Neutralization: antibodies attach to target = pathogen cannot replicate or attach to surface
Opsonization: antibodies has Fc or constant regions that promote recognition to phagocytes
Complement fixation/activation: classical complement pathway; pentameric IgM will bind to C1
Endpoint of classical: MAC to perforate target cell to lead to their death
Humoral is like the highest tech level/latter nag-evolve, para magfunction, need ng innate and t cell immunity
For b cells, soluble antigens are enough while t cells need MHC presentation
Once activated > clonal expansion > differentiation
Memory cells and plasma cells may or may not have the same receptors
B cells need maturation factors from follicular DCs in lymph nodes
Myeloid DCs are APCs
FDCs are fixed in lymph nodes
T cells go to lymph node for activation/ screen antigens while B cells look for maturation factors from FDCs
B cells with maturation factor = Mature B cells
Next step is activation
Soluble antigen attaches to B cell = cross-linking of B cell receptors
Cross-linking: same receptors due to allelic exclusion; more receptors are activated by 1 antigen
ITAMs: needed for signaling by receptors
Pag nag cross-link, mas malakas ang signal
CD3 gen marker of T cells while CD19 for b cells which is required for activation
CR2 complement receptor binds with C3d ligand from alternative pathway cleaved by factor I
B cells cannot function without complement system of innate system: supplementary ang humoral
DAF and MHC stops C3 from proceeding with complement
B1 doesn’t need T cells for activation
Antigens can trigger B1
Kaya mas madali dumami ang B1 kasi cross-linking lang ang need
Drawback: hindi magm-mature ang antibody response
IgD part of development of B cells but effector functions is not known; marker lang for what stage na ang B cell development
IgM is referred as primary antibody kasi siya unang pwedeng isecrete ng lahat ng B cells
Kung mature na B cell, FDCs can hold antigens on their dendrites
Subcapsular sinus macrophage:
Primary follicle: kumpulan ng FDCs waving soluble antigens trying to attract B cells
B cells anergy pag walang infection kahit mag match
B2 cells are not fully activated even after finding target pathogen = they are instructed to go to the boundary of T cell area and present antigens to T cells via MHC II = cognate pairing if match sa T cell receptor
Once nag cognate pair, ITAMs will reorganize intracellular vesicles towards synapse and release of cytokines dictates b cells what to do next
After na-encounter ang antigen and naactivate sila, maturation into plasma cells producing IgM
Kaka-encounter lang ng antigen
Mas malaki ang centroblasts kasi may somatic hypermutation and isotype switching; main enzyme is AID
Somatic recombination and junctional diversity occur during development whereas somatic hypermutation and isotype switching occur during maturation
Activated-induced cytidine deaminase:
Hypervariable regions are on the antigen-binding site
Affinity maturation: sobrang improved ang affinity ng antibody
Antibody-response maturation: improves IgM affinity via somatic hypermutation which is changing hypervariable region which leads to affinity maturation to improve specificity of variable region of antibody
FDCs secreting cytokines has an effect on how the b cell will respond on its antibody maturation
Secondary antibodies: IgG (swiss knife ng antibodies), IgA, IgE
Centrocytes tapos na mag isotype switch and som hypermutation
How can we be sure affinity is improved by som hypermutation? Centrocytes will interact with FDCs
Centrocytes need to returm sa same antigen that activated it sa primary follicle = if som hypermutation is successful, dapat mauna siya bumalik sa antigen sa FDCs kasi may time limit ang centrocytes
Need mag compete kasi nag proliferate nga so only those with the highest affinity will reach the antigen first
This leads to further differentiation
Sa Hyper IgM patients, walang germinal centers sa lymph node bc walang isotype switching na nagaganap
Further maturation of b cells are still guided by FDCs
IL4 and IL10 secreted by FDCs
If IL10 dominant, they become plasma cells but the secreted antibodies are isotype switched
If IL4 dominant, they become memory B cells that retain B cell receptors
Memory b cells: activation process is skipped; theyre targeting the same antigen and has higher affinity; they’ll just secrete secondary antibodies upon infection
Primary immune response: IgM
Secondary immune response: memory B cells
High mutation rates ng pathogens kaya babalik ulit sa primary response
Shingles sa nerve nagtatago at nagfoformat
Narereactivate dahil immunocompromised (i.e. nagka flu prior)
Centrocytes stop dividing but may maititrang memory b cells pero may half life so to make sure na enough pa rin = boosters
Some memory B cells can undergo clonal expansion
IgG can cross the placenta and dominant in circulation
IgA dominant and dimeric on mucosal surfaces but monomeric in circulation
IgE allergic response
Transcytosis: pagtawid sa epithelial tissues
Fc receptors bind to Fc of antibodies = transcytose and reach the tissues where they will let go of the antibodies
Poly-Ig receptors binds to IgA for transcytosis
Passive transfer or passive immunization = binigyan ka ng antibodies na hindi immune system mo ang gumawa
Mothers need to be vaccinated or booster vaccines: IgG will pass thru placenta and received by fetus
IgA can be passed thru breast milk = colostrum highly proteinaceous bc it has IgA
Rabies is 99% mortality
Two types of vaccines are given
Active: promote immune response to make antibodies but this takes time
Passive: contains antibodies and injected near the bite mark
Plasma therapy is also an example of passive immunity
Antibodies are still effective for viruse bc it can neutralize their receptors
Clostridium tetani lives naturally in the soil and anaerobic and will secrete toxin that will cause tetanus
Tetanus can thrive pag lagi binabalot nang masikip gamit gauze
IgM and IgG3 can activate complement
Soluble immune complex pwede tulungan ng complkement receptors
Phagocytes has FcG on their surface
Antibodies can be recognized by FcG receptors to help macrophage to be more specific on their targets
Di lagi for activation ang Fc receptors
Bc of FcG3 which is unique to NK cells, they can have a certain defree of specificity kasi didikit sa kanila ang antibodies which is responsible for specific response
Nabigyan ng specificifty si NK cell kasi may FcG3 receptor to use antibodies
Immunosurveillance:
Lateral immunoflow assay
It might take time to produce antibodies kaya IgG- and IgM- still requires precaution
Maraming blood type classifications pero mas ginagamit lang ang ABO
If we want to test if blood will react, we do cross-matching
The ability of igg to cross placenta now becomes detrimental
Maganda sana kasi it was supposed to give immunity to the fetus
Antibody production
Aseptic technique
- Wash hands
- Proper attire
- Lab gown
- Gloves
- Foot wear
- Sterile materials
- Autoclave sterilization
- 20 mins, 120 C, 15 psi
- Filter sterilization
- For filtering bacteria
Eukaryotic cells are more complex
May TLRs that can recognize PAMPs
Even if the pathogen is dead, their mere presence of their component can trigger a response in the much complex eukaryotic cells
Kaya makakaapekto kung may dead e.coli sa agar plate kahit after ma-autoclave
- Clean bench
- Horizontal flow: laminar flowhood
- Only for protecting workspace
- Vertical flow: biosafety cabinet
- For protecting worker
- Horizontal flow: laminar flowhood
- 70% alcohol
- UV light
- UV will only work with direct contact
- It cannot penetrate glasses
How can you tell if contaminated ang culture
- Color change ng media
- May pH indicator ang media
- Respiration of live organisms = co2 product = carbonic acid = acidic (yellow) color change ang expected if contaminated
- Okay lang pag it took days before nag yellow ang culture kasi it takes days for eukaryotic to multiply
Multiple b cells can recognize a single antigen if it has multiple epitopes
Polyclonal- diff populations of b cells but their targeting the same antigen just different epitopes
It’s the natural way how our immune system produces antibodies
Choice of animal depends kung gaano karami kailangan na antibodies
Why do we need manufactured antibodies?
Fluorescent microscopy staining
Flow cytometry
We use monoclonal as default cuz we are sure bawat antibody target ay same epitope so same results
Hybridoma: joining plasma cell and a multiple myeloma (type of cancer of b cells)
Mas mabilis ang half life ng b cells so they join it with a cancer cell
May hayflick limit ang isang cell that would render is to stop dividing
So need ng cancer cell para tuloy tuloy ang production
They fuse in polyethylene glycol
The cells will open then rejoin
De novo pathway: cells are making nucleic acids out of nothing
Salvage pathway: the cells recycle
In multiple myeloma cells, they cannot acces the salvage pathway
The culture medium used to separate: HAT medium
Adjuvant: signals that tell ur immune system sige mag inflammation ka para masignal mi sa rest na may ongoing infection para mag manifest yung mga APCs ng B7
Can we use manufactured antibodies on ourselves?
Need muna i-modify
ICR mice is outbred: genetically varied
BalbC is inbred: immunodeficient so mas madalas ginagamit kung need patubuan ng cancer