MICR 3330 Midterm
Introduction
Smallpox
Most dreadful infectious disease in history
May have killed Pharaoh Ramses V
Pivotal role in conquest of North America
Jeffrey Amherst (1763) suggested germ warfare by infected blankets
Captain Simeon Ecuyer distributed blankets to Indians (not confirmed)
Also evidence in French-Indian war
Influenza
1st recorded by Hippocrates 412 BC
Origin
Domesticated animals (likely)
Initially believed as bad air
Seasonal epidemics
Flu shot
Spanish flu
1918-1919
Poliomyelitis & Infantile Paralysis
Transmission
Oral-fecal route
Ebola
Transmission
Direct contact w body fluid, contaminated objects
Origin
Fruit bat or primates (likely)
HIV/AIDS
Attack immune system
Death from secondary infections
Transmission
Body fluid (sex, injection, blood transfusion, etc)
SARS
Outbreak 2002
Common & Manageable Human Viral Diseases
Herpesvirus 1 (cold sore)
Herpes simplex virus 1
Re-activation caused by stress (mental, UV, etc)
Infectious mononucleosis
Epstein-Barr virus
In multiple autoimmune systems
Human papillomaviruses
Cause of most STDs
Age 50, ~80% women (USA) at least 1 type HPV
High-risk 15% in females
High risk types can cause cervical cancer
Other notable viruses
Tobacco mosaic virus
Papaya ringspot virus
Caused papaya devastation
Grapevine leafroll associated viruses
Virus Pathogenicity
Humans are biggest & most effective "vector" for emerging & re-emerging virus
Most viruses not lethal
Co-exist w host through genetic mut'n & co-adaption
Good Things from Viruses
Molecular bio from studying viruses
DNA (in phage) or RNA (TMV) is genetic material
Polyadenylation of mRNA transcripts
Polyomavirus
5' cap of euk mRNAs
Reovirus & alphavirus
Defiance of central dogma, reverse txn, & RTase
Splicing of pre-mRNAs in euk systems
Adenovirus
Nuclear localization signals for prtn targeting
1st complete genome
φX174
1st "mammalian source" genome sequenced
SV40
RNA silencing
RNAi or post-txn'l gene silencing
Viruses as vectors in gene therapy & cancer treatment
Viruses as VIGS vectors for functional genomics
Nature's utility of viruses to maintain the ecosystem
Applications of viruses
Gene therapy
Prtn expression
Oncolytics
Recombinant vaccines
Functional genomics
Revival of phage therapy
Control bact'l infections w multi-drug resistance
Nanoparticles
Questions in Virology
Definition of a virus?
Are viruses living or non-living?
Origin of viruses?
Why can't vaccination programs eradicate all viruses?
How to viruses perpetuate over eternity?
How viruses maximize coding capacity of their genes?
Can viruses be benificial?
How to control viruses (& diseases)?
How to eliminate all viruses?
Is this necessary?
Discovery, Morphology, & Composition
TMV: First Virus Discovered
Adolf Mayer
1879
Extracts from tobacco w mosaic disease were infectious
Fungal pathogen not involved
No culture on Petri dish
1882
Initial conclusion: 'soluble, enzyme-like contagium'
1886
Publication: 'unknown bacterium'
D. Ivanovsky
1892
Passing tobacco extract through bacteria-proof filter
Filtrate still infectious
Reports: Filter must still be infectious
M. Beijerinck
1998
Filter extract -> dilute filtrate -> inoculate healthy tobacco -> replenish
'Contagium vivum fluidum'
Later work on nature of TMV
1935: Stanley
Crystallization of TMV particles
Purified from tobacco
Conclusion: 'virus is proteinaceous in nature'
1936: Bawden & Pirie
TMV particle also contains RNA (5%)
1933: Ernst Ruska
Invented EM
1939: Helmut Ruska
Revealed TMV
FMDV
1st Animal Virus Discovered
Loeffler & Frosch (1898)
Virus from foot-and-mouth disease remained infectious after filtering
Highly contagious in cloven-hoofed animals
Fatal in calves
Repeated vaccination required
Yellow Fever Virus
1st Human Virus Discovered
Natural hosts: monkeys & mosquitos
Symptoms
Damage to liver
Jaundice
Slave brought to America
Philadelphia epidemic (1793)
Killed ~15% of city pop
Discovery: 1901
Human volunteers for vector transmission studies
19 tested
8 infected
3 died
Discovery of Bacteriophages
1915: Frederick Twort
Attempts to grow vaccinia
Petri dish contamination
1915: Felix d'Herelle
Found virus that killed Shigella bacteria (causes dysentery)
Killed bact & plaque formation among soldiers
Wanted phage as therapy to cure bact'l diseases in humans
1939: Delbruck & Ellis
One-step growth experiment
1940: Luria & Delbruck
Make phage workshops
Studies of phages & E. Coli
Foundation for molecular bio & bact'l genetics
DNA as Genetic Material in Phage T2
1952: Hershey & Chase Experiments
Label T2 with 35S or 32P
Mix with bact
Blend
Let culture grow
Centrifuge
Measure radioactivity
Results after centrifugation
35S
(Labelled prtn capsule)
No sulphur detected in cells
32P
(Labelled viral genetic material)
Phosphorus detected
Composition
Nucleic acids
RNA or DNA as genetic material
Not both
Majority RNA
Prtns
Structural & non-structural
Lipids
Enveloped viruses only
Derived from cellular lipid layer
Carbohydrates
In glycoprtns & glycolipids
Recognizing cell receptors & attachment to host cells
---
RNA viruses
dsRNA
Almost always segmented
ssRNA
Segmented or non-segmented
DNA viruses
dsDNA & ssDNA
Linear or circular
---
Naked virus
Nucleic acids inside a capsid
Enveloped virus
Enveloped w glycoprtn spikes
Nucleic acids inside nucleocapsid
--
Hepadnaviridae (dsDNA) & Retroviridae (ssDNA)
Reverse txn to finish replication
Vertebrates viruses
Nanoviridae (ssDNA)
Reverse txn to finish replication
Plant virus
No true dsDNA plant virus discovered
Morphology & Dimensions
Rigid rods / Flexuous filaments
(14 x 71 nm - 80 x 14,000 nm)
TMV: 18 x 300 nm
Filoviridae (Ebola): 80 x 650-1400 nm
Closteroviridae: 12 x 2200 nm
Spherical / Isometric
(17 - 300 nm diameter)
Parvoviridae: 25 nm
Picornaviridae: 30 nm
Adenoviridae: 80 - 110 nm
Herpesviridae: 120 - 300 nm
Irregular / Complex morphology
T-even bacteriophages
Spherical head: contains DNA
Tail: Helical sheath, tail fiber, & tail baseplate
Baculoviruses (insects)
Occluded virion (OV): Survival when released into env'nt
Budded virion (BV): For spread w/in insect
---
Brick Shaped
Vaccinia & pox viruses
Terminology
Virion
Complete viral particle
Capsid (coat)
Prtn shell encasing the viral genome
Nucleocapsid (core)
Nucleic acid + prtn
In virion of enveloped viruses
Subunit (promoter)
Single, folded polypeptide
Individual capsid prtn
Structural unit (capsomer)
Basic unit for building capsid or nucleo-capsid
1+ subunits
Envelope
Lipid mem enclosing nucleo-capsid
(enveloped viruses only)
Structural prtn
Prtns part of virion structure
Non-structural prtn
Encoded by a virus but not part of virion
Enzymes needed for viral replication, movement, & infection
Helical Symmetry
Rod-shaped & filamentous viruses
Capsid prtn subunits have equal binding to another
Except those at both ends
'Open' structure: Unlimited packing capacity (for insertion of foreign DNA/viral vectors)
P (pitch of helix) = µ (# of subunits per helical turn) x p (the axial rise per subunit)
TMV
Length 300 nm |
Diameter 18 nm |
# CP subunits 2130 |
# helical turns 130 |
µ 16.33 |
P (capitol) 2.3 nm |
p 0.14 nm |

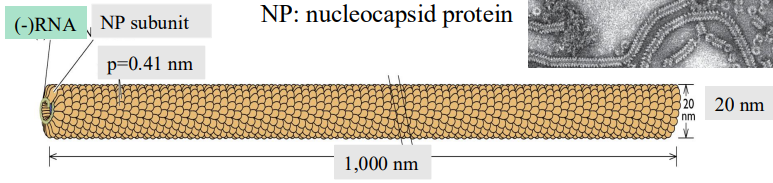
Paramyxovirus (Sendai virus)
Length 1000 nm |
Diameter 20 nm |
# NP subunits 2130 |
µ 13 |
P (capitol) 5.33 nm |
p 0.41 nm |
Icosahedral Symmetry (& linkage to geodesic dome)
More complex structure
'Closed' structure: packaging of only limited genome sizes
Watson & Crick (1956)
Proposed spherical viruses were cubic structures (proven wrong)
Basic design of spherical capsids & nucleocapsids:
Icosahedral symmetry; 20 triangular faces
Pillars:
Triangulation number (T): define possible icosahedral surface lattice
Quasi-equivalence: describe the nearly identical bonding rel'nship among subunits in a spherical capsid
Spontaneous self-assembly of individual CP subunits into virus-like particles (VLPs) to identify the assembly processes
Simplest cases
Capsid built from 60 copies of 1 capsid prtn arranged into 20 triangular faces
Ex. Satellite tobacco necrosis virus, parvoviruses, øX174
All capsid subunits in equivalent bonding rel'nship
3 Types of rotational symmetry: 2-, 3-, and 5-fold
2 axes in 2-fold
20 axes in 3-fold
12 axes in 5-fold

In larger isometric viruses
Triangulation # (T): # of triangles w/in ea of the 20 triangular faces
Only certain multiples of 60 subunits allowed (T= 1, 3, 4, 7)
Quasi-equivalence: in icosahedrons w T>1
Rel'nships b/w capsid prtn subunits similar (NOT identical)
---
β-barrel jelly roll fold
3D structure of capsid prtns
Shared by viruses w icosahedral symmetry (conserved)
Similar fold in 'phaseolin' (storage prtn) of beans
Interactions among CP subunits
CP subunits small in most viruses (20-70 kDa)
# of subunits must incr to let them to exist in a quasi-equivalent position in viruses w large capsids
CP subunits spontaneously assemble into larger structures w or w/out help of viral genome
Ex. Structural units, intact capsid shell
CP subunits stabilized by the max # of non-covalent bonds b/w them
-> lowest free energy state
All sub-sub & sub-RNA bonds are weak
Mainly hydrophobic & van der Waals
Secondary structures in viral RNA = packaging signals
---
T x 60 = # of prtn copies in capsid/nucleocapsid
Exceptions - Do not contain predicted # of subunits
Polyomaviruses
Adenoviruses
Reoviruses
Phage lambda
Icosahedral head
Helical tail made of tail cone (sheath), tail plate, & attachment fibres
Ea part is assembles individually, followed by assembly into an intact virion
Members of the Adenoviridae family
1500 expected copies of subunits (based on T=25)
Actual: 780 copies
Prtn II: 720 copies/virion; forming hexons (240 hexons/virion)
Prtn III: 60 copies; forming pentons (12 pentons/virion)
Simian virus 40
5 VP1 subunits interlock into a pentamer
C-term arms of ea pentamer inserts into neighbouring pentamers
Stabilizes capsid
2 spatial arrangements = deviation from icosahedral symmetry rule
12 pentamers at 5-fold axes, ea surrounded by 5 pentamers
60 pentamers at remainder of capsid, ea surrounded by 6 pentamers
Recap
TMV
1st virus identified (1898) via bacteria-proof Chamberland filter candle
Rod-shaped particle
95% prtn (capsid), 5% RNA (genome)
Major role in understanding genetic info, etc.
FMDV & YFV
Foot-and-mouth disease virus
1st animal virus
YFV
Yellow fever virus
1st human virus
Virus composition
Prtn & nucleic acids (DNA or RNA, not both)
Some also have lipids & carbohydrates in form of glycoprtns & glycolipids
Virus morphology
Sometimes genome split into multiple segments packed into:
Same virion (influenza)
Different virions (some plant viruses)
Categories of viruses
Naked (have capsids)
Enveloped (nucleocapsid w/in a lipid envelope)
Symmetry
Helical
Rigid rods or flexible filaments
Icosahedral
Triangulation # and quasi-equivalence
Describe icosahedral design of viruses w isometric capsids or nucleocapsids
Capsid prtn subunits self-assemble into structural units & intact virions (VLPs)
VLPs= basis for subunit vaccines for human & animal diseases
Classification, Taxonomy, & Nomenclature
Linnaean hierarchical system (1700s): Plants & animals
3 domains system today: Bacteria, Archaea, Eukarya
Homes (1948)
Failed attempt to classify & name viruses using Linnaean syst
Order: Virales
Phaginae (viruses of bact)
Phytophaginae (viruses of plants)
Zoophaginae (viruses of animals & humans)
Classification & Taxonomy
Pre-1930
Based on diseases, signs, & symptoms
No distinction b/w disease & its causative agent
Hep A: Picornaviridae
Hep B: Hepadnaviridae
Hep C: Flaviviridae
Mosaic virus cause mosaic symptoms in plant leaves
TMV: Virgaviridae
Cauliflower MV: Caulimoviridae
Cucumber MV: Bromoviridae
Turnip MV: Tymoviridae
Soybean MV: Potyviridae
---
Virus: agent that causes an infection or disease
Disease: outcome & manifestation of an infection resulting from interactions b/w a virus & its host
Infection: can lead to disease (not necessarily the cause)
---
1930-1966
Emphasis on virus over disease
Based on:
Morphology
Capsid structure
Chemical composition
Type of genome
Viruses classified in 'groups' initially
Herpesvirus group
dsDNA
Icosahedral heads
Large virion
Has envelope
Poxvirus group
dsDNA
Complex & irregular virion
200 nm or larger
Enveloped
Myxovirus group
ssRNA
Spherical virion
Helical nucleocapsids
Enveloped
1966-present
International Committee on the Taxonomy of Viruses (ICTV)
2 approaches considered:
Monothetic: 1 characteristic at a time
Nature of viral genome, symmetry of capsid, presence/absence of envelope, etc…
Problem: assumes all members of a group originate from same ancestor; can't reflect diversity
Polythetic: Considers multiple characteristics
Individuals share most (not all) of a set of common characteristics
Not assume all viruses share same ancestor
Ex. family Closteroviridae
Long & filamentous virion
Large (+)ssRNA genome
HSP70h (viral homolog of cellular HSP70)
Tropism for phloem
Defining a virus species
Pre-2013: "A polythetic class of viruses that constitute a replicating lineage and occupy a particular ecological niche"
Post-2013: "A monophyletic group of viruses whose properties can be distinguished from those of other species by multiple criteria"
Practical:
Defined by relatedness in seq'ces of a specific gene, set of genes, or entire genome
Values of seq'ce identity used for species distinction vary among families
Taxon pre-2019: Order, family, (subfamily), genus, & species
Subspecies designations: Strains, serotypes, genotypes, subtypes, variants, etc
Naming Viruses
Bacterial: Specific code: Qβ, φX, λ, T1, T2…
Plant: Host in which virus identified then descriptor of key symptoms
Eg. Tobacco mosaic virus, beet yellow virus, etc
Mammalian: Based on diseases & symptoms
Eg. Hepatitis virus, measles virus, SARS-CoV, etc
Insect: Latin name of host & effect of infection on virus
Eg. Autographa californica multiple nucleopolyhedrovirus
Characteristics for classifying genus/family
Nature & organization of genomes
DNA or RNA
Strandedness (ss or ds)
Polarity (+/-)
Segmented or non-segmented
Topology (linear/circular; closed/open circle)
Virion morphology (structure of capsids & nucleocapsid)
Helical, icosahedral, complex
Shape, size, surface features
Envelope (yes/no)
Genome structure, strategies for genome replication expression
Enzymes (Pol, RTase, protease, integrase, etc.)
Characteristics for defining species
Natural host range
Cell & tissue tropism
Pathology (host) & cytopathology (cell culture)
Mode of transmission
Physico-chemical properties of virions
Antigenic properties of viral prtns
Seq'ce relatedness of individual genes & whole genomes
Phylogenetic Analysis & Viral Taxonomy
Phylogeny: prediction of evolutionary relatedness among viruses based on comparison of their seq'ces using computer & mathematical algorithms
Analysis based on NT or AA sequences (or both)
Dif methods to generate phylogenetic trees:
Neighbor joining (NJ)
Maximum likelihood (ML)
---
9th Report of ICTV (2011)
Families not yet assigned to an order = 78
Genera not yet assigned to a family = 13
Virus Nomenclature & Taxonomy
1991-2018: 5 rank structure
Since 2019: 15 rank structure
1st attempt to adopt the Linnaean syst to classify all viruses
Intro of 8 more lvls for higher taxa
Changes in 2021
Binomial system
Abolition of type species in all genera
Increased scope: include viruses, viroids, & satellites
Realms ex.
Adnaviria: DNA viruses from Archaea hosts
Duplodnaviria: dsDNA viruses
Varidnaviria: large dsDNA viruses (nucleocytoplasmic)
Riboviria: RNA viruses (including retroviruses)
Types of genome viruses
(+)ssRNA genome viruses
Genome 3-31 kb
Linear genomes (no circular)
Most lack an envelope
Examples
Closteroviridae (Helical)
Coronaviridae (Helical)
(-)ssRNA genome viruses
Helical symmetry
Linear genomes
Some v pathogenic human viruses
May have envelope
ssDNA genome viruses
Small genomes 2-9 kb
Naked (no envelope)
Icosahedral symmetry (except Inoviridae)
Circular genomes (except Parvoviridae)
dsDNA genome viruses
Large variation
Baltimore Classification System: Transcription considered most important aspect for a virus
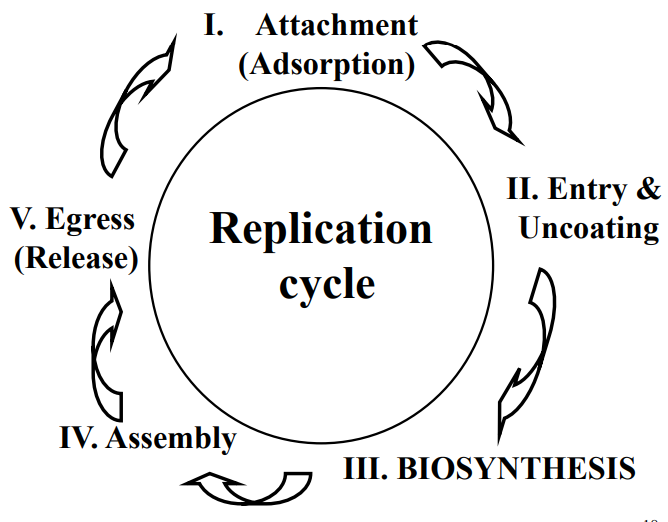
Virus Replication Cycle
Cell culture system
Enders, Weller, Robins (1949)
Primary embryonic cell cultures (from mouse) for measles virus & polio virus
Cultured on plastic surface or as liquid suspensions
Widely used in research & vaccine dev'nt
Types of cell cultures
Primary cell cultures
From live tissues/organs
Multiple cell types
Finite capacity in cell division (5-20x)
Ex. Monkey kidney (for polio vaccine), chicken embryo, mouse embryo
Diploid cell strains
Single cell; epithelial or fibroblast
Normal morphology & # of chromos
Cell division up to 100x
Ex. WI-38 (from female embryonic lung)
Immortal (continuous) cell lines
Homogeneous in cell type
Infinite capacity of cell division
Abnormal chromo morphology & #
Loss of contact inhibition
Detach from surface, piling up, form 'focus'
Tumorigenic
Source: tumors, transformed cell strains, cells mutated by oncoviruses or mutagens
Ex.
HeLa: cervical tumor (Henrietta Lacks, HPV)
Vero: kidney of African green monkey
BHK-21: Baby hamster kidney
BY2: right yellow, tobacco
Cytopathic effects (CPE)
Morphological alterations of a cell due to viral infection
Cell death
Rounding up of cells -> detach from surface
Syncytium = large cell w a lot of nuclei (bc of fusion)
Abnormality in morphology & # of chromosomes
Inclusion bodies:
Polyhedron inclusion bodies (PIBs)
Negri bodies (in rabies-infected cells)
Inclusion bodies in plant viruses
X-bodies, pinwheels, etc
One-step growth cycle
Synchronous infection of all cells w virus
High MOI: 5-10
Time intervals
Eclipse period: from absorption to 1st intra-cellular virion
Latent period: from absorption to 1st extra-cellular virion
Burst size: sum of virions produced in 1 cell
---
MOI: Multiplicity of infection: # of infectious virions added per cell
Not all cels in dish receive same # of viruses
MOI of 5-10 commonly used for synchronous infection
Allocation of virions among cells is calculated by Poisson distribution
B/c random collision b/w viral particles & cells
1. Attachment (adsorption)
Collision b/w viral particles & cells
Via Brownian motion
Weak contact
Via interaction b/w viral particle & -ve charges on cell surface
Specific attachment achieved via interactions b/w:
Attachment prtn (on virus)
Eg. HA in influenza virus; fiber in adenoviruses; surface structure 'canyon' in poliovirus
Some attached to mem by TMD
Some anchored indirectly to PM via fatty acids/alcohol
Types of anchors for these prtns:
Myristic acid
Farnesyl
Glycosyl phosphatidyl inositol-linked prtns
Only external surface of PM
Receptor(s) (on host cell surface)
Eg. Sialic acid for HA; Icam-1 for major group rhinoviruses
~500,000 molecules/cell
Plasma mem has 'lipid rafts' w distinctive structure & fxns
Viruses hijack surface molecules & lipid rafts for entry & replication
Stronger w more interactions b/w attachment prtns & receptors
Naked viruses examples
Attachment via surface features on virion
Polioviruses & rhinoviruses have canyon surrounding ea pentamer - attachment site for cell receptors
Attachment via fibers on virion surface
Adenoviruses & picornaviruses
Fiber is a homo-trimer at ea of the 12 penton bases
Terminal knob on fiber w depression - attachment site for 'Car'
Enveloped viruses examples
Glycoprtns on viral envelope are responsible for attachment
Hemagglutinin of influenza A & B viruses binds sialic acid
Surface glycoprtn (HIV-1) binds CD4 of T-helper cells & macrophages
Co-receptor needed for viral entry
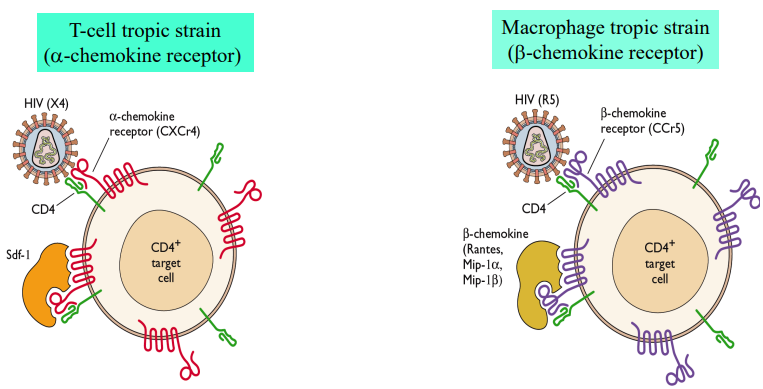
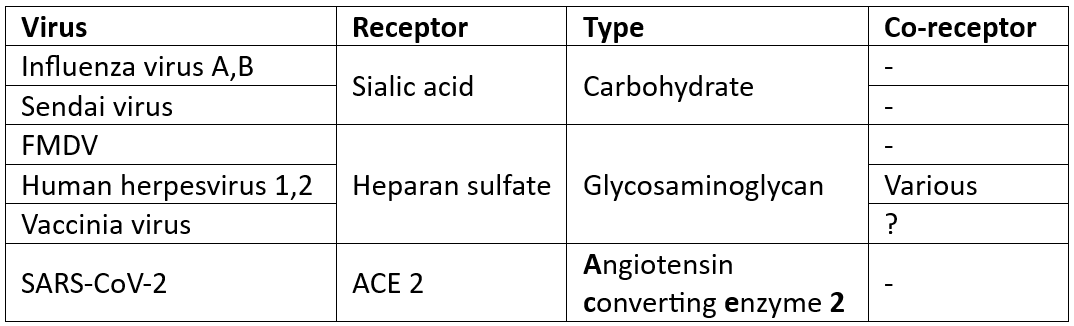
Image: Well-studied receptors & co-receptors
Receptors extra info
For most viruses are unknown
Viruses w carbohydrate receptors tend to have broader host range
Presence of receptor (& co-receptor) determines, in part, the host range & tissue tropism of a virus
2. Entry & Uncoating
Basic tricks
Drilling hole at PM
Eg. Picornaviruses
Fusion b/w viral envelope & PM
At neutral pH
Needs fusion prtns/peptides
Eg. Paramyxoviruses, retroviruses, coronaviruses, baculoviruses
Receptor-mediated endocytosis, followed by uncoating @ an intracellular mem (endosomal or nuclear)
Low pH (endosome, lysosome)
Eg. Orthomyxoviruses, adenoviruses
Models of poliovirus entry
@ plasma or endosomal mem
Steps in entry & uncoating
Attachment to multiple receptor Pvr
N-term of VP1 exposed, inserted into PM (forms channel)
Viral RNA released into ctpsm
HIV-1
Viral entry needs binding to both receptor (CD4) & co-receptor
Triggers exposure of fusion peptide -> fusion b/w viral envelope & cell mem
Receptor-mediated endocytosis
Selective import of extracellular molecules (ligands) into a cell via receptor & mem invagination
Formation of clathrin-coated pits needs ATP hydrolysis
Decrease in pH from early endosome -> late endosome -> lysosome
Ex. Influenza
Receptor mediated endocytosis & uncoating @ endosomal mem
HA attaches to sialic acid
Endocytosis
Fusion pep emerges from HA under low pH
H+ into virion via M2 channel
Matrix layer dismantles, viral envelope fuses w endosomal mem
Release RNPs into cytosol
RNPs enter the nucleus
Ex. Adenoviruses
Receptor-mediated endocytosis & stepwise uncoating
Fiber binds to Car
Penton base interacts w integrin, leading to endocytosis
Losing fibers in endosome
Penton bases dismantle in late endosome
Broken virion release into cytosol
Free ride on microtubule
Dock at nuclear pore
Tug-of-war b/w dynein & kinesin releases viral DNA
Viral DNA enters nucleus
DNA viruses & retroviruses nucleus entry
Adenoviruses:
Partially dissembled virion transported by microtubules
Dock onto nuclear port
Pulling by kinesin break up virion
DNA enters
Herpesviruses:
Release of pressure in inner capsid injects DNA
Polyomaviruses:
Remodeling of nuclear envelope & lamina allows DNA entry
HIV-1:
PIC docks onto nuclear pore due to nuclear localization signals (NLS) on the capsid & integrase proteins
Integration into host chromosome
Phages T4 (dsDNA)
Replication < 30 mins
Tail fibers recognize/bind receptors (LPS & OmpC) on bact'l cell surface
Causes changes in base plate structure
Tail sheath contracts, exposes inside tube
Lysozymes from phage dissolve cell wall
DNA injected into ctpsm
Plant viruses
Most don't need receptors
Many transmitted through insect vectors
Mechanical transmission: via minor wounds/abrasions
Vertical transmission: via reproductive organs (pollen/seeds)
Via ex:
Vegetative propagation materials
Grafting b/w rootstock & scion
3. Biosynthesis
Biosynthesis: Syn of all viral components needed for building next gen of viruses
Transcription: Production of mRNAs from genome (DNA or RNA) (most important aspect of a virus)
Reverse transcription: Generation of cDNA using RNA as template (only in retroviruses & retro-like viruses)
Translation: Production of polypeptides using mRNA; relies on cell tln machinery
Genome replication: Production of nascent viral genomes (DNA or RNA)
Class I: DNA viruses
ssDNA: Parvoviridae (linear), Circoviridae (circular), Geminiviridae
dsDNA: Polyomaviridae (circular), Baculoviridae (circular), Adenoviridae (linear), Herpesviridae (linear)
Class II: RNA viruses
dsRNA: Reoviridae, Birnaviridae, Partitiviridae, Chrysoviridae
(+)ssRNA: Picornaviridae, Togaviridae, Betaflexiviridae, Closteroviridae
(-)ssRNA: Orthomyxoviridae, Paramyxoviridae, Filoviridae, Bornaviridae
Class III: viruses needing reverse txn
Retroviridae (RNA; integrates genome into host chromo)
Caulimoviridae (RNA; doesn't integrate)
Hepadnaviridae (DNA; doesn't integrate)
Sites for biosynthesis
Type Tln Txn Rep'n |
RNA Ctpsm Ctpsm Ctpsm |
DNA Ctpsm Nucleus Nucleus |
Retroviruses & para-retroviruses Ctpsm Nucleus Nucleus |
Exceptions:
Orthomyxoviridae: Need nuc's for txn & rep'n
Poxviridae: Everything in ctpsm of infected cell
NCLDV: need nuc's & ctpsm for rep'n
DNA Viruses
Infect cells that are either dividing or they force dormant cells to enter S phase
Potential oncogenicity
Adenovirus A & C
Herpesviruses: Epstein-Barr virus; Burkitt's lymphoma
Polyomaviruses: SV40
Papillomaviruses: HPV type 16 & 18
Expression
Immediate early (IE) genes
Expressed right after infection
Functions of IE prtns:
Rendering cells toenter S phase
Induce expression of other viral genes
Inhibit host mechanism & biosynthesis
Eg. Adenovirus E1A & E1B bind p23, blocking apoptosis & forcing cell to enter S phase
Early (E) genes
Enzymes & accessary factors required for genome replication
Replication
In nucleus (except Poxviridae - in ctpsm - virions carry all needed prtns/enzymes)
Requires dNTPs, enzymes, host machinery
Strategies vary among families
Late (L) genes
Structural prtns required for assembly
Very late genes
For few viruses (polyhedron, baculoviruses)
RNA Viruses
All of replication cycle in ctpsm
Exceptions: Orthomyxoviridae & Bornaviridae)
Must assoc w intracellular mems
ER - TMV, BYV, PVX, etc.
Endosomal mem - Sindbis virus (Alphavirus, Togaviridae)
Vesicular mem - Poliovirus (Picornaviridae)
Peroxisome mem - Tomato bushy stunt virus (Tombusviridae)
Mitochondria - Grapevine leafroll-associated virus (Ampelovirus)
Chloroplast - Turnip yellow mosaic virus (Tymoviridae)
(+) strand RNA viruses
Txn & rep'n: RNA-dependent RNA Pol (RdRP)
Genomic RNA is 1st (or only) mRNA
Replicase has conserved domains (RdRP, helicase, etc)
5' proximal ORF(s) translated directly on genomic RNA
ORF1 encodes 126 kD prtn w MTR & helicase domains
ORF2 encodes RdRp as part of 183 kD prtn via suppression of leaky stop codon
(-) strand RNA viruses
Genome exists as ribonucleoprotein (RNP) complex and not as naked RNA
Must 1st be transcribed before tln occurs
Virion carries not only RNA but also enzymes
Influenza viruses
RNPs enter the nucleus -> replicate & transcribe
Needs splicing of transcripts made from genome segments
Retroviruses & para-retroviruses
Recall:
Retroviridae (RNA; integrates genome into host chromo)
Caulimoviridae (RNA; doesn't integrate)
Hepadnaviridae (DNA; doesn't integrate)
Discovery of RTase went against central dogma
2 identical (+)RNA molecules, RTase (50-100 copies), integrase
Reverse txn starts in virion on route of entry into host cell
cDNA inserts into host chromo
Becomes part of host genome
Establishes latency
Source of persistent infections
Host cell enzymes transcribes viral mRNAs including gRNA
Assembly & Egress (release)
Formation of individual structural units from 1+ structural prtns
de novo process
Spontaneous; results from interactions among capsid/nucleocapsid subunits
AA sequence decides interlocking among structural prtn subunits
Assembly of capsids occurs in special compartment of infected cell
High [prtns] ensures assembly is correct, efficient, & directional
In ctpsm
RNA viruses (except 1 group), DNA viruses of Poxviridae
In nucleus
DNA viruses, retroviruses, orthomyxoviruses
Examples:
Adenoviruses
Formation of penton requires 2 prtns (fiber, penton base)
Formation of hexon trimer by prtn II needs chaperone
Poliovirus:
Single polyprotein precursor & cleavage
Assembly of capsid shell w structural units
ssRNA viruses
Genome RNA involved in virion assembly (eg. TMV)
Poliovirus RNA may be involved in assembly & final cleavage during virion maturation
Large DNA viruses & DNA phages
Procapsids formed w help of scaffold prtns
Collapse & removal of scaffold structures
Genome DNA inserted into procapsid
Needs ATP hydrolysis for energy
Poliovirus
Proteolytic cleavage of VP0 makes virion infectious
Process may involve genomic RNA
Selective packaging of viral genome & other virion components
Acquisition of a lipid envelope (mostly from PM)
Exit the infected cell
Naked viruses
Lysis of infected cells - cytopathic effects
Shutdown of biosynthesis
Destroy cell structure
Mem alteration
Chromo breakup
Syncytium
Inclusion bodies
Ex. Adenovirus E1B: disrupts nuclear lamina & breakdown of intermediate filaments
Enveloped viruses
Source of envelope determined by glycoprtns of the virus
Budding & pinching off at PM
Maturation of virions
Only some viruses
Needs proteolytic cleavage
---
Host range: Range of hosts that can be infected by a virus
Tissue tropism: A virus' preference for certain types of cell & tissue in hosts
Susceptible cells: Allow attachment & entry of virus b/c of suitable receptor(s)
Permissive cells: Permit replication of a virus, have all factors needed. May/may not be susceptible
Summary
Replication cycle (AEBAE):
Attachment
Entry/uncoating
Biosynthesis
Assembly
Exit
Attachment
Via interactions of specific prtns/cell features on incoming virion & receptors on cell surface
Some viruses need receptor & co-receptor
Factor of host range & tissue tropism
Entry/uncoating
Entry: virion enters cell via PM or internal mem
Direct entry through pore at PM
Direct fusion b/w viral envelope & PM (needs fusion peptide)
Receptor-mediated endocytosis
Uncoating: viral genome released into ctpsm of cell
During or after entry
Biosynthesis
All macromolecules needed for viral rep'n are synthesized
Includes:
Tsl to make structural (& non-structural) prtns
Txn to make mRNAs (& other RNAs w reg'ry fxn)
Genome rep'n
Assembly
Reversal of uncoating
Nascent capsids, nucleocapsids, & intact virions get assembled
de novo process (spontaneous & doesn't need template)
Subunits => structural units => (nucleo)capsids
Viruses w helical symmetry (eg. TMV) - gRNAs involved in virion assembly
Large DNA viruses - empty shells assembled -> packaging of gDNA via insertion of DNA through specialized portal
Egress
Naked viruses
Exit upon lysis of the infected cell
Enveloped viruses
Budding at PM or internal organelle mem
TMV
(+)ssRNA supergroups
Based on phylogenetic rel'nship of RdRP (Pol):
Alphavirus-like (III)
TMV
Picornavirus-like (I)
Polio, FMDV
Flavivirus-like (II)
Yellow fever, Hep C
Class 4
Revised Baltimore system
TMV
Genus: Tobamovirus
Family: Virgaviridae
Easily transmissible; persists for decades
Research Milestones
1898: TMV as first filterable virus conceptualized (contagious living fluid)
1955: Reconstitution of TMV & RNA as genetic material
1971: Role of CP double disks as structural unit in capsid assembly
1978: Discovery of movement prtn in TMV
1986: Infectious RNA transcript from full-length cDNA clones
1986: Coat prtn-mediated resistance against TMV infection
Genome structure & expression strategies
Small, (+)ssRNA, 6.4 kb
gRNA is 1st mRNA to translate replication-related enzymes
5' end: cap structure
3' UTR: secondary & tertiary structures (3 pseudo-knots)
3' end: tRNA-like structure
No Poly-A tail
Aminoacylated w His
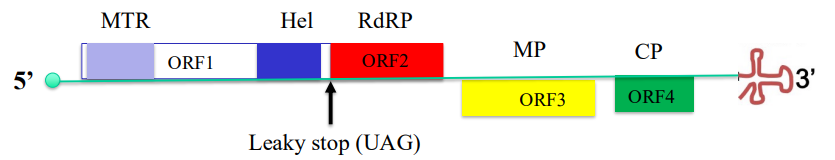
Suppression of leaky stop codon of ORF1 leads to continued tsl of ORF2
126 kDa prtn (has MTR & HEL)
183 kDa prtn (MTR, HEL + RdRP)
*MTR = methyltransferase domain
*HEL = helicase domain
Reconstitution Experiment
Proves RNA is genetic material (not prtns)
Fraenkel-Conrat
CP alone assembles into virion: Not infectious
CP & RNA assembles into virion: Infectious
RNA alone: Infectious
Conclusion
RNA iis genetic material, not capsid prtn
Local lesion assay to quantify TMV
Francis Holmes (1929)
Process:
Spread abrasives on leaf surface
Rub leaf w viral stock dilution
Wait for infection & symptoms
Count # of local lesions
Calculate the titer of OG viral stock
Like plaque assay for bacteriophages & animal viruses
Assembly of capsids & genome packing
Structural unit for TMV: CP double disk
16.3 copies of CP per helical term
2,1300 CP copies per virion
Building-up as double discs
RNA involved in virion assembly
5' genome segment threads through the interior of the elongating helix
One of 1st models on virion assembly
CP-mediated protection in crop plans against viral diseases
1st demonstration of transgenic resistance against viruses by genetic engineering
Transgenic plants expressing TMV CP exhibited delayed onset of disease
Theory: "Coat prtn-mediated protection"
Excess CP from the transgene blocks virus disassembly = virus resistance
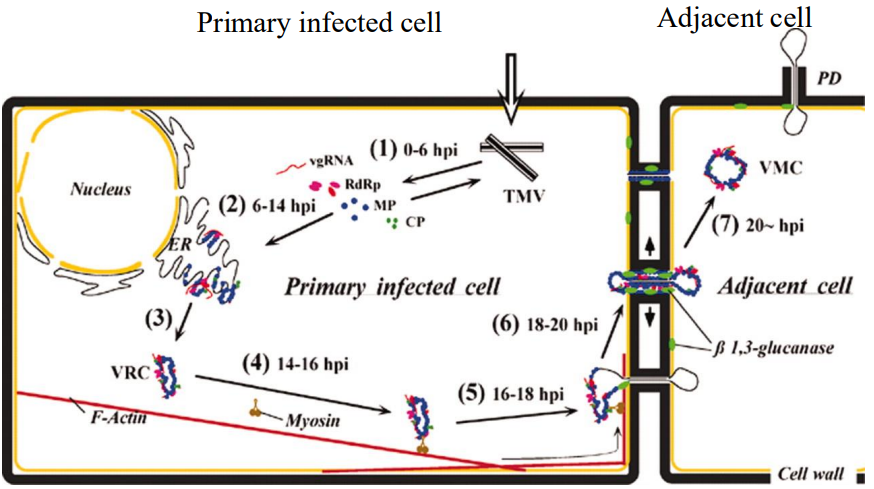
Stages of TMV in plant (virus travels through phloem)
Intracellular movement
Viral rep'n complexes (VRCs) form in assoc'n w ER
Move to other parts of infected cell
Produces multiple VRCs in same cell
Intercellular (cell-to-cell) movement
VRC docks @ plasmodesma traverses it w help of movement prtn (MP)
Completes new rep'n in neighbouring cells
Long distance movement
Virions gain entry into sieve elements of phloem
Move rapidly to distal parts of plants
Causes systemic infection
Viruses move b/w cells via plasmodesmata
Size exclusion limit
Allows passive diffusion (1-7 kDa)
Active process (w ATP) for larger molecule transport through PD
Move w help of MP
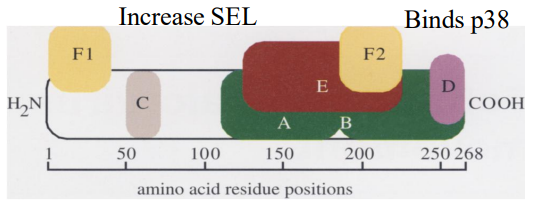
Movement prtns (MP)
1st discovered in TMV, encoded by all plant viruses
Temp sensitive TMV mutant, Ls1
Complementation by transgenic MP
Most plant viruses encode 1 MP (some 2+)
Common properties
Binds RNA
Forms v thin & long RNPs
N-term region increases SEL of PD
Interacts w ER, actin filaments, & microtubes
MPs encoded by non-related viruses may compliment defective MP
Bonds p38 (cell wall assoc'd prtn @ PD, receptor for MP)
Earlier model of TMV cell-cell movement
MP complexes w TMV RNA to form thin thread
Moves on actin filament
Interacts w p38 -> increases SEL
Squeezes through central cavity of PD
Phos'n by host kinase releases MP
Frees viral RNA in 2nd cell for tsln
TMV MP assoc'd w peripheral ER in plants
Immuno-staining w anti-Bip antibody shows ER in red
BiP: luminal prtn of ER, marker for ER network
Composition & structure of VRC
Components:
Replicase prtns
MP
Viral genomic RNA
ER mem
Other viral & cellular prtns
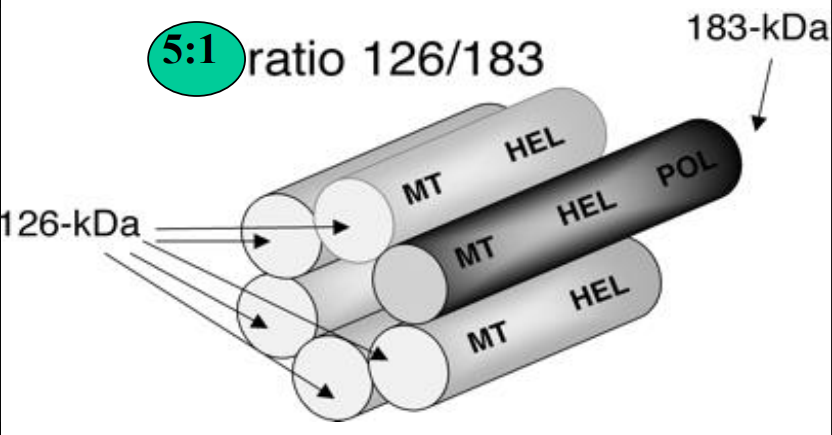
Helicase domains self-interact
Forms hexameric ring-like structures when over-expressed in bact'l cells
Bind ssRNA
Act as ATPase & helicase
Hydrolyzes ATP & unwinds dsRNA
Ea hexamer likely comprised of 5 molecules of 126 kD & 1 molecule 183 kD
VRC aligns w & moves on actin filaments
126:GFP fusion forms punctate structures in cells
DsRed:Talin marks actin filaments (MF) in red
126:GFP co-localizes w DsRed:talin
Suggests localization to microfilaments
VRC (shown by MP:GFP) also aligns w actin filaments
Conclusion: 126 kD aligns on & moves along actin filaments
Recent research in TMV cell-to-cell movement
Aim: test if it takes same amount of time for TMV to replicate in initially infected cells compared to that in secondary infected cells
Tracking of TMV movement w time lapse microscopy in initially infected cells vs. movement in additional cells suggest:
TMV moves as VRC & not as thin MP-RNA threads
Much shorter time to complete same process in neighboring cells
Current model of cell-to-cell movement
TMV as vectors for VIGS & prtn expression in plants
VIGS: virus-induced gene silencing
RNA-based defence syst for gen reg'n & defence against viruses
PSY (phytoene synthase) & PDS (phytoene desaturase) protect chlorophyll
TMV engineered to produce mRNA for PDS or PSY
Triggers RNA silencing in tobacco
Infected leaves exhibit photo bleaching upon light exposure
Summary of TMV
(+)ssRNA
Representative member of the Alphavirus supergroup
Prototype member of genus Tobamovirus in family Virgaviridae
Rod shaped capsid
2130 subunits
Helical symmetry
1 turn = 16.3 subunits
RNA genome
6400 nts
5' cap
No Poly-A tail
V compact
5'UTR
4 ORFs
3' UTR
ORF1&2
Near 5' end
Directly translated from gRNA brought by virion upon entry
ORF1
Translated into 126 kDa prtn
Has conserved methyltransferase (MTR) & helicase (HEL) domains
ORF2
Encodes viral RNA-dependent RNA Pol (RdRp)
Translated as an extension to the 126 kDa via suppression of stop codon (translational read-through)
Ribosome "reads-through" stop codon instead of stopping & makes extension of prtn
Makes 183 kDa prtn
N-term: 126 kDa
C-term : RdRp)
ORF3&4
Expression via translation of 2 sgRNAs
Viral replicase enzymes transcribe sgRNAs -> ea sgRNA has own promoter of the ORF it will be translated into -> sgRNAs used as template for tsln -> production of the prtns
ORF3
Encodes MP (non-structural)
ORF4
Encodes CP (structural)
Used to construct rod-shaped virion
Infection
Via temporary wound on leaf surface
Immediately after entry, ORF 1&2 occur
Makes 2 polypeptides that are needed for rep'n
Viral rep'n in special compartment in ctpsm of infected cell
Viral Replication Complex (VRC)
Made of mem from ER, replicase prtns, gRNA, MP, host factors (unidentified)
Intercellular movement of VRC
Through plasmodesma (PD) (microchannel b/w adjacent cells)
MP interacts w RNA & PD components
Increases size exclusion limit of PD, allowing VRC to go through
Early theory (disproven): RNA-MP thin threads move through
TMV enters vasculature (phloem)
Reaches distal parts of the plants
Results in systemic infection
Recent TMV study
TMV promising in improvement of energy storage & lithium-ion batteries
Picornaviridae
Supergroup I of (+)ssRNA - Picornaviruses
Features
No 5' cap
3' Poly(A) tail
No envelope
V stable
22-30 nm diameter
Icosahedral symmetry
60 copies ea of 4 structural prtns (VP1-4)
VP-3 on surface
VP-4 hidden under
Oral-fecal transmission
Genomic RNA is the only mRNA
Translated into a polyprtn (precursor)
IRES= start site for viral tsln
Triangulation # of pseudo 3
β-barrel jelly roll conserved in icosahedral viruses of plants, insects, animals, humans
Topological family of prtns
Diseases caused by Picornaviridae
Poliomyelitis
Infantile infection w multiple epidemics in North America
Common cold
Upper respiratory tract
~50% of common colds
Hep A
Acute liver infections
Sporadic outbreaks via food & drinks
Heart infections
Myocarditis
Dilated cardiomyopathy (DCM)
Caused by group B Coxsackieviruses
Diabetes & pancreatic disorders
Coxsackieviruses
Encephalomyocarditis virus
Foot-and-mouth disease
Quarantinable
Poliomyelitis
Most polio infections were inapparent & self limiting
Paralytic form most feared
Major infantile & childhood disease in the 1st half of the 20th century
The hygiene hypothesis
Early exposure to microorganisms helps build immune system
United States
1894
1st reported case
1916
27 000 cases
6000 deaths (1/3 from NYC)
1930s
FDR declares national war against polio
National Foundation for Infantile Paralysis
Later, March of Dimes
1952 epidemic
58 000 children infected
21 269 (36%) displayed paralysis of various severity
3145 (5.4%) deaths
Canada
1910
1st reported case (Hamilton, ON)
1937
4000 cases nationally (>50% in ON)
119 deaths (4.7%)
Only single iron lung available - hospital staff rush to make more
1953
~9000 cases
500 deaths (5.5%)
Panic - social distance, isolation, quarantine, etc.
Iron lung
Only treatment & hope of severe polio patients w lung complications
Stay in lung until immune syst heals body
Ex. Barton Hebert
Stayed in iron lung for last 50 yrs of life
Salk vaccine
Jonas Salk (1955)
1st highly effective inactivated polio vaccine
Tested on self & family to convince others
Large clinical trials in USA
Key discoveries made w picornaviruses
FMDV
Loeffler & Frosch (1898)
1st animal virus discovered shortly after TMV
Isolation of PV
Landsteiner (1909)
Via transmission experiment to monkeys
Cell culture
Enders, Robbins, & Weller (1949)
Plaque assay
Dulbecco (1952)
IPV (inactivated vaccine)
Jonas Salk (1955)
OPV (weakened live virus vaccine)
Albert Sabin & Hilary Koprowski (1960)
RdRP
Baltimore (1963)
From polio-infected cell
Polyprtn
Summers & Maizel (1968)
Infectious cDNA clone
Racaniello & Baltimore (1981)
IRES
Pelletier & Sonenberg (1988)
Genome structure & expression strategies
(+)ssRNA
7500 - 8450 nts
5' end
VPg (virion prtn, genome-linked, 22-24 aa residues)
5' UTR: v long secondary structures
Middle
Single large ORF encoding 1 polyprtn as precursor
3' end
47-125 nts UTR
Poly(A) tail essential for infectivity
Polyprtn & proteolytic processing
Viral RNA is only mRNA
Single large ORF encodes a polyprtn
Polyprtn cut by proteases into 11-12 functional prtns needed for rep'n
2Apro cuts once
Separates P1 from rest of polyprtn
3Cpro cuts at 8 places
Produces all final prtns needed for rep'n
Polio infection shuts down tsln of cellular mRNAs
Tsln machinery re-directed for viral prtns only
Prevention of PIC formation at cap of host mRNAs
Cap-dependent tsln
All mRNAs, PIC assembles at 5' cap
Brings 2 ends of mRNA together and scan for AUG
Large ribosome subunit joins & tsln starts
Prtns in tsln:
eIF-4F: tripartite structure (eIF -4A, -4E, and -4G)
eIF-4G: euk initiation factor 4G
Picornaviruses block tsln of host mRNA
Doesn't block own tsln
Inhibit PIC formation at 5' cap via:
2APro (polio) or L protease (FMDV) cleaves eIF-4G
Dephosp'n of 4E-BP1, binds eIF-4E tight, sequestering eIF-4E (used mt FMDV, not PV)
How PV prtns are translated
Secondary structures= stem-loop (5' UTR)
Tertiary structures= pseudoknots (5' UTR)
AUG Is downstream of IRES
Pyrimidine-rich seq'ce upstream of AUG
PIC binds directly at IRES -> lands on AUG -> initiates tsln
No need for cap

Poliovirus replication cycle
Attachment
Canyon: poliovirus, rhinovirus
Surface loop: FMDV
Receptors: PVR, ICAM-1
Uncoating
Sphingosine in hydrophobic pocket of pentamer helps VP1 penetrate mem (forms pore)
Biosynthesis
Genome rep'n & IRES-based prtn syn
Assembly
Maturation
VP0 cleavage into VP2 & Vp4
Control of poliomyelitis
Inactivated polio virus vaccine (Salk, 1955)
Live attenuated vaccines (Sabin, 1960)
Dif Sabin vaccine strains possess mut'ns at multiple sites
The Cutter Incident
April 1955
Patient inoculated in buttock w Cutter vaccine
9 days later, admitted to hospital for flaccid paralysis in both legs
Stats
~400 000 children given same vaccine in 10-day period
Within 2 months:
94 cases of polio among vaccinees
166 cases among family & community contacts
1995, Stalk succeeded with killed polio vaccine
Reconstruction of poliovirus
US department of defence
Syn DNA fragments as oligos
Assembly into genome fragments
Clone vector under T7 promoter
In vitro txn to get viral RNA
Assay for infectivity in mice expressing receptor Pvr
VRC: sites of poliovirus rep'n
Viral rep'n on vesicles from ER
Viral tsln, vesicle formation, & RNA syn are couples
VPg as prtn primes for genome rep'n
Viral 3AB attaches to ER mem
Tyr in 3B undergoes uridylylation
Inserted 'UU' anneals poly(A) tail of viral genome
VPg (3B) cleaved off from 3AB by 3CPro
Syn (-)RNA
Replicative intermediate (RI)
Replicative form (RF)
Summary
Picornaviridae: fam of (+)ssRNA viruses
Order: Picornavirales
Realm: Riboviria
Kingdom: Orthornavirae
Belong to picornavirus-like supergroup of RNA viruses
Members cause various diseases in humans & livestock
Poliomyelitis
Common cold
Heart diseases in humans
FMDV
Polio
Effects
One of most damaging viral diseases of 20th century (after atomic bomb)
Causes epidemics of flaccid paralysis & death in infants due to lack of protecting antibodies in mothers as a result of the practice of better personal hygiene
Development & global use of vaccines (OPV & IPV) resulted in near worldwide eradication
Prototype of Enterovirus genus & family Picornaviridae
RNA genome:
V long 5' UTR
Single ORF
Tsln via cap-independent mechanism by cellular machinery using unique structure designated to the Internal ribosomal entry site (IRES)
Translated into a polyprtn, which is cleaved by proteases (encoded by the virus) into final prtn products
2A: cleaves off P1 (b/w P1 & P2)
3CD & 3C: cleave remaining sites
2A & L cleave the euk tsln initiation factor (eIF-4G) -> shuts down tsln of host RNA upon infection -> infected cell now a factory for progeny viruses
3' UTR
N-term: VPg polypeptide
Prtn primer for syn of gRNA & complimentary RNA (uridylylation)
Research discoveries
Identification of RNA-dependent RNA Pol (RdRP)
Establishment of non-neuronal cell cultures
Plaque assay
Inactivated polio vaccine (IPV) (Salk)
Attenuated oral vaccine (OPV) (Sabin)
Polyprtn & proteolytic processing
Internal ribosome entry site (IRES) for initiation of tsln of viral DNA
1st infectious viral clone for an animal virus
1st synthetic polio viral clone from synthetic biology approach
Flaviviridae
Supergroup of (+)RNA viruses
Phylogeny of Flaviviridae
Based on the helicase domain of NS3
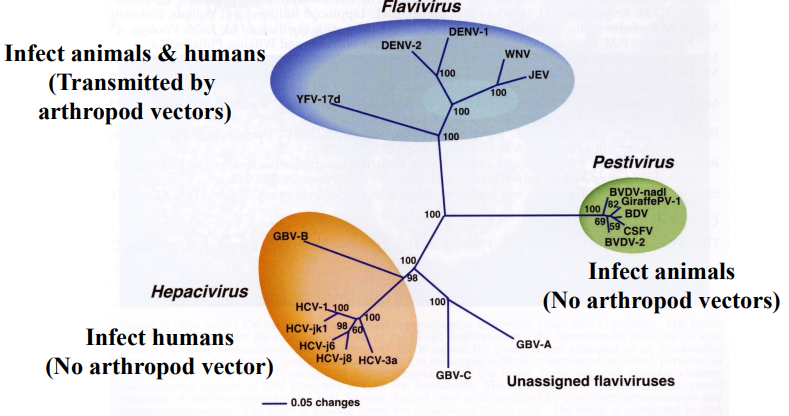
Classification of Flavivirus genus
Viruses transmitted by & replicated in arthropods
Principal hosts & natural reservoirs
Birds, rodents, monkeys, pigs
Defining features
Density map in virion:
Envelope prtn layer: 180 copies of E & M prtns (T=3)
Lipid bilayer (viral envelope)
Nucleocapsid core: icosahedral (T=3), 25-30 nm, CP basic
E prtns lie on top of, & in parallel w, lipid mem
Smooth surface
(+)ssRNA genomes, 10-11 kb
Genome expression: a single ORF, poly-prtn, & proteolytic processing
Many (not all) members are transmitted by arthropod vectors (mosquito or tick) in which they also replicate (hence arboviruses)
Arboviruses
Viruses of humans & animals that are transmitted by, & replicate in, arthropod vectors
Members of the genus Flavivirus transmitted by mosquitos or ticks
Viral families that are also vectored by/replicate in insects:
Flaviviridae
Togaviridae
Bunyaviridae
Arenaviridae
Rhabdoviridae
2 Modes of Transmission
Jungle cycle
Primates
Urban cycle
Humans (epidemic)
Yellow fever
Origin: Africa
America & Europe via slave trades
1648: 1st recorded epidemic
1881: 1st suggested transmission by mosquito (Dr. Finlay)
1901: 1st human virus discovered
Experimentally confirms yellow fever transmission by mosquito
Frequent epidemics in US 1700-1800s
Re-emergence by urbanization & suspension of mosquito control
15% infections develop severe disease
Mortality: 20-50% in severe epidemics
Established by yellow fever commission:
Serum contains the 'virus'
Infectious agent is filterable
Mosquitoes transmit disease
Symptoms
Asymptomatic
Mild flu-like symptoms
Fulminant infections
Fatal
Stages of infections w sever outcomes
Period of infection
3-6 days after catching infection
Fever, chills, myalgia, back pain
Contagious
Period of remission
Period of intoxication
Jaundice, vomiting, viral rep'n in liver, no viremia
Hemorrhagic fever
Renal fever, hemorrhage, shock, multiple organ failure
Control & prevention
Max Theiler (1930s)
Attenuated vaccine strain 17-D
Isolation via passages in monkeys, followed by 176 passages in primary cell cultures
17-D provides immunity in monkeys & humans
Consensus vaccine strains differ from Asibi by 32 aa & 4 nts
Dengue fever
Most prevalent vector-borne viral infection in the world
Spread during WWII
Primary infections for 1-2 weeks
25% of hospitalized patients develop prolonged fatigue & depression
Similar to mononucleosis & long covid
4 genotypes: DENV-1, -2, -3, & -4
Vary by 20-40% in E prtn
Asymptomatic infections
Dengue fever (DF): mild & self-limiting (DENV-4)
Dengue hemorrhagic fever (DHF)
Dengue shock syndrome (DSS)
Secondary Dengue infections & antibody-dependent enhancement
2nd infection in person who previously infected w dif serotype may lead to higher viremia & worse outcomes (DHF & DSS)
Main reason for lack of effective vaccines against DENV
Proposed mechanism:
Cross-reactive antibodies present in the patient from earlier infection bind to virions of new serotype
Assists viral entry into large # of cells expressing Fc receptors for IgG
Instead of conferring partial immunity against secondary infections, pre-existing antibodies help virus enter large # of cells during a new infection
Leads to severe outcomes (DHF & DSS)
Sanofi Pasteur - live attenuated vaccine
West Nile
1937: 1st reported case
Infection of central nervous system (CNS) -> encephalitis (mid-fatal) -> paralysis -> death
1999: Introduction in NYC
2000: Re-appeared in mosquito season
2002: Blood transfusion shown to cause infection in recipients
Humans/mammals = 'dead-end' carriers
Cant transmit back to mosquito
Zika virus
Since 2007: moving from Pacific ocean -> America
Microcephaly in children (small head birth defect)
Feb 2016: WHO declares it as a Public Health Emergency of International Concern
Hepatitis C (HCV)
One of most widespread diseases globally
Transmission: blood, blood products, organ transplants, injection drug use, body piercing
1989: Discovery through molecular cloning and sequencing
Most acute infections become chronic
-> Liver cancer & cirrhosis (liver scarring)
High prevalence Egypt, Asia, & Australia
Reuse of syringes among children when treating schistosomiasis
Infects hepatocytes & lymphocytes
50 virions/day/hepatocyte -> 1012 produced/infected person/day
Infection w one genotype does NOT confer immunity against another
Chromic HCV infections asymptomatic for first decades
Long term outcomes
75-85% infected will develop chronic infection
60-70% develop cirrhosis over 20-30 yrs
1-5% die from cirrhosis or liver cancer
Disease outcomes
Cirrhosis
Increased chance if alcohol or certain prescription drugs
Hepatocellular carcinoma
Liver failure
Death
No vaccines available
Lack of effective experimental systems impeded research & antiviral development until recently
Pre-2005: Slow research bc low viral titer in the liver
Relied on full-length & mini (subgenomic) viral amplicon & humanized mice
Post-2005: JFH-1 isolated
Grows in cell culture without need for adaptive mutants
Treatment w antiviral drugs:
Old: pegylated interferon (INF) α, ribavirin (RBV)
New: direct acting antivirals (DAAs)
Genome structure & expression strategies
No 5' Cap (HEP C ONLY, NOT ALL FLAVIVIRUSES)
Use IRES to initiate txn of viral polyprtn
No 3' Poly-A tail
Has many helices on 3' end
Size: 9.6 kb (HCV)
Members of genus Flavivirus: 11 kb
Shared properties w Flaviviridae family
Single large ORF encoding a protein
Cleavage by viral & host proteases
Virus replication cycle
Attachment
E prtn binds 1+ receptors (possibly glycosaminoglycans)
Entry
Receptor-mediated endocytosis
Genome uncoating
Low pH dependent mem fusion w endosome mem
Prtn synthesis
Polyprtn assoc's w ER
Cleavage into multiple functional prtns
RNA synthesis
In small ER-derived vesicles
Assembly & release
On ctpsmic side of ER
Bud into ER lumen
Exit via exocytosis
Fusion b/w transport vesicle & plasma mem releases virions
Evolution of HCV treatment using antivirals
Traditional treatments relied on combinational therapy w interferon & ribavirin
Problems in low compliance w antiviral treatment
Severe side effects (headache, nausea, fever)
Liver transplants scarce
V costly
Direct acting antiviral (DAA) drugs
Target key viral enzymes (protease & RdRp)
Nucleoside analog RdRP inhibitor drugs offer high % of SVR
High efficiency, short treatment duration
Sofosbuvir
Most effective nucleoside inhibitor analog of RdRP
Cure rate 30-70% (depends on HCV genotypes)
V effective against multiple genotypes
Strong barrier against emergence of resistant mutants
Oral administration
No-mild side effects
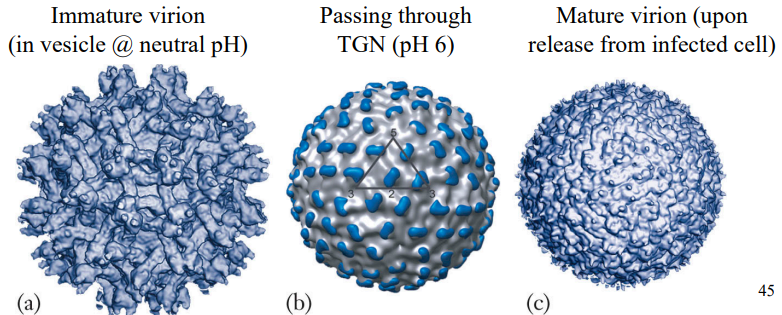
Forms of virion
Virion has lipid mem, studded w envelope prtns (icosahedral symmetry; T=3)
Immature viral particles display spikes on virion surface
Ea spike has 3 pairs of E & M heterodimers
Conformational changes in E & M prtns produce smooth & infectious virions
Homodimer of envelope prtns
Fusion peptide: Hidden by domain III
Domain II: Interaction to form E dimers
Domain III: Ig fold, binding to receptor
W/in endosome:
Low pH changes conformation of E prtn
Exposes fusion peptide
Releases core into ctpsm
Summary
Flaviviridae: Flavivirus-like supergroup of (+)ssRNA viruses
3 genera:
Flavivirus (yellow fever virus)
Hepacivirus (Hep C)
Pestivirus
Flavivirus transmission: arthropod vectors (arboviruses)
Yellow fever
1st human virus identified
In tropics & subtropics
Non-human primates -> humans via mosquito bites
To America via slave trade
1793 epidemic killed 10% of Philadelphia
Caused impediment to Panama Canal Project
Max Theiler - attenuated vaccine strain 17-D (derived from v pathogenic strain)
Hep C (HCV)
Often results in chronic infection
-> cirrhosis, liver cancer, & death
Transmission via blood, organ transplant, contaminated needles, etc
Early 1900s blood screening reduced prevalence of HCV
1989: virus identified through molecular cloning
Cell culture system - HCV isolate (JFH-1)
Was able to replicate in the cell without need for adaptive mutations
No vaccines, treatment uses interferons & antiviral drugs (ribavirin, protease inhibitors)
Recently: direct acting antivirals (DAA)
More efficient
Safer
Shorter treatment times
Less/no side effects
Structure
(+)ssRNA
9.5-11 kb (Flavivirus)
3 layers:
Envelope prtn layer: E & M prtns
Lipid mem layer: icosahedral symmetry
Nucleocapsid core: icosahedral symmetry
1 ORF
Translated into single polyprtn
Proteases cleavage into 10 functional prtns (like Picornaviridae)
Flavivirus only
5' cap
(transmission by arthropod vectors)
Hepacivirus only (Hep C)
No 5' cap (has IRES)