6.1 Protein modifications Note
Posttranslational Protein Modifications
Definition and Biological Role
Post-translational modifications (PTMs)
Covalent modifications of polypeptide chains by enzymes.
Alters protein function, localization, or interactions.
Occurs during and after protein translation.
Translation is the biological process of protein synthesis.
Types of Post-translational Modifications
Glycosylation (and glycation)
Phosphorylation
Proteolysis
Carboxylation
Ubiquitination
Hydroxylation
Lipid modifications
ADP-ribosylation
Acetylation
Other modifications
Glycosylation
Overview
Glycosylation Meaning: Enzymatic addition of sugars to proteins.
What enzymes play a role in glycosylation?
Glycosyltransferase: Enzymes adding sugars.
Glycosidase: Enzymes that remove (cleave off) sugar parts.
Functions of Glycosylation
Plays crucial roles in:
Cell adhesion
Cell-cell and cell-matrix interactions
Activation of receptors
Protein solubility
Protein folding and signaling
Breakdown and secretion of proteins
Amino Acids Involved In Glycosylation
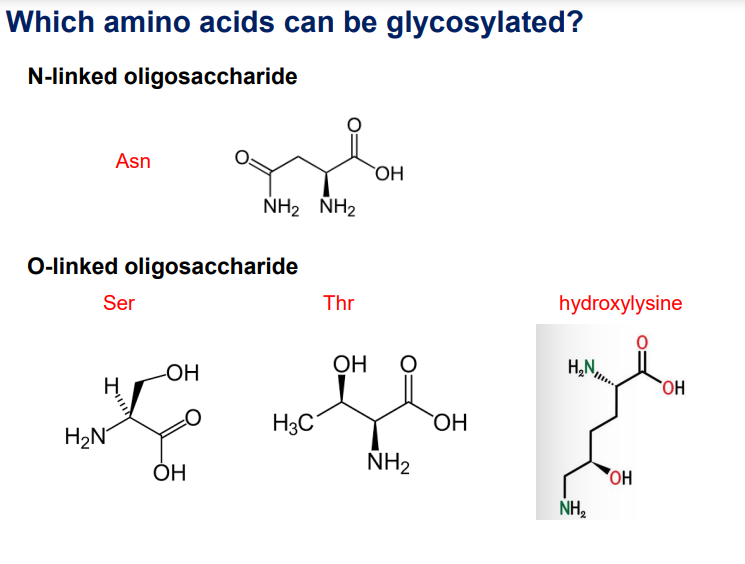
N-linked oligosaccharides: Asparagine (Asn). N-linked oligosaccharides are attached to the nitrogen (N) atom of the amide group in an asparagine (Asn) residue.
O-linked oligosaccharides: Serine (Ser), Threonine (Thr), Hydroxylysine. O-linked oligosaccharides are attached to the oxygen (O) atom of the hydroxyl group in a serine (Ser) or threonine (Thr) residue.
N-Glycosylation
Main Steps
Initiation in the ER lumen using dolichol phosphate.
Addition of N-acetylglucosamine (GlcNAc) and Mannose.
Finalisation of the oligosaccharide chain.
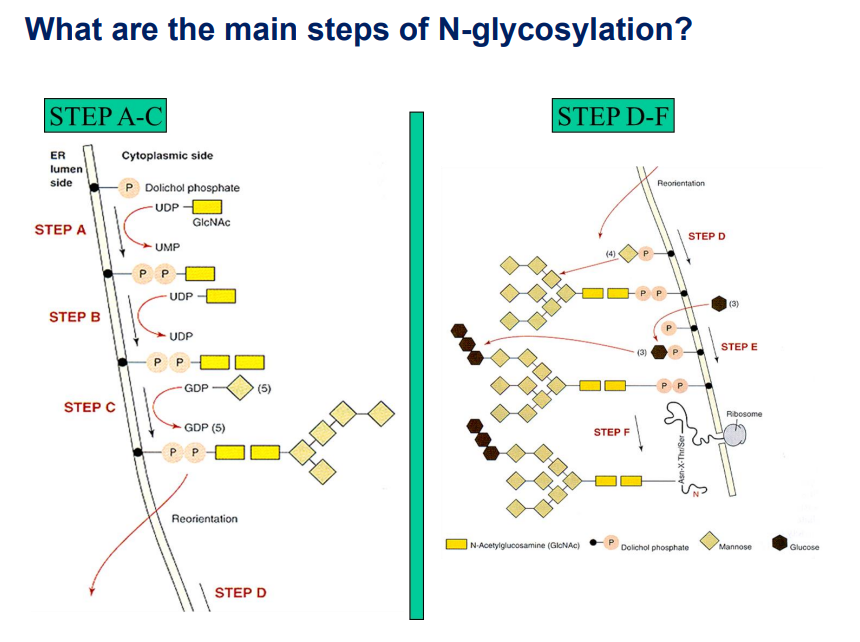
This image explains the main steps of N-glycosylation, which is the process of attaching oligosaccharide chains (glycans) to asparagine (N) residues of proteins. Let's break it down step by step.
STEP A-C: Early Glycan Assembly on the Cytoplasmic Side
These steps occur on the cytoplasmic side of the endoplasmic reticulum (ER) membrane.
Step A: Initiation
The process starts with dolichol phosphate (Dol-P), a lipid carrier embedded in the ER membrane.
UDP-GlcNAc (Uridine Diphosphate-N-acetylglucosamine) donates GlcNAc (yellow rectangle) to dolichol phosphate.
This reaction releases UMP (Uridine Monophosphate).
Step B: Addition of a Second GlcNAc
Another UDP-GlcNAc donates a second GlcNAc residue.
This forms a pyrophosphate (P-P) bond with dolichol, which helps anchor the growing glycan structure.
Step C: Addition of Five Mannose Residues
Mannose (Man) residues (diamond shapes) are added using GDP-Mannose (Guanosine Diphosphate-Mannose) as a donor.
A total of five mannose residues are attached to the growing oligosaccharide structure.
Reorientation (Flipping) to the ER Lumen
The partially assembled oligosaccharide is flipped from the cytoplasmic side into the ER lumen.
This is necessary for further glycan processing.
STEP D-F: Glycan Completion and Transfer
These steps occur inside the ER lumen, where the glycan structure is completed.
Step D: Further Mannose Additions
More mannose residues are added to the growing glycan chain.
The dolichol-linked glycan becomes a high-mannose oligosaccharide.
Step E: Addition of Glucose Residues
Three glucose (Glc) residues (brown hexagons) are added.
These glucose residues play a role in protein folding quality control.
Step F: Transfer to a Nascent Protein
The completed oligosaccharide is transferred from dolichol phosphate to an asparagine (N) residue in the growing polypeptide.
The enzyme oligosaccharyltransferase (OST) catalyzes this transfer.
This occurs in proteins with the sequence Asn-X-Ser/Thr (where X is any amino acid except proline).
Key Takeaways
Step A-C (Cytoplasmic Side) → Dolichol phosphate is loaded with GlcNAc and mannose.
Membrane Flip → The glycan structure moves into the ER lumen.
Step D-F (ER Lumen Side) → The glycan is completed with mannose and glucose, then transferred to a protein.
This process ensures proper protein folding and stability before proteins are sent to the Golgi for further modifications.
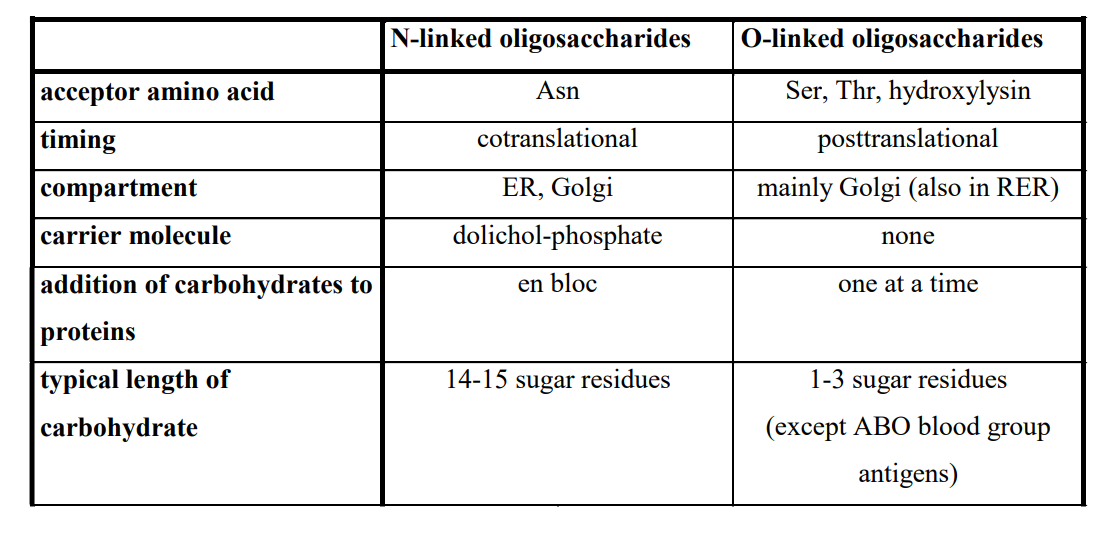
Differences Between N- and O-Glycosylation
N-linked:
Acceptor amino acid: Asn
Timing: Cotranslational
Compartment: ER, Golgi
Carrier: Dolichol phosphate
Sequence: En bloc
O-linked:
Acceptor amino acids: Ser, Thr, Hydroxylysine
Timing: Posttranslational
Compartment: Golgi
Carrier: None
Sequence: One at a time
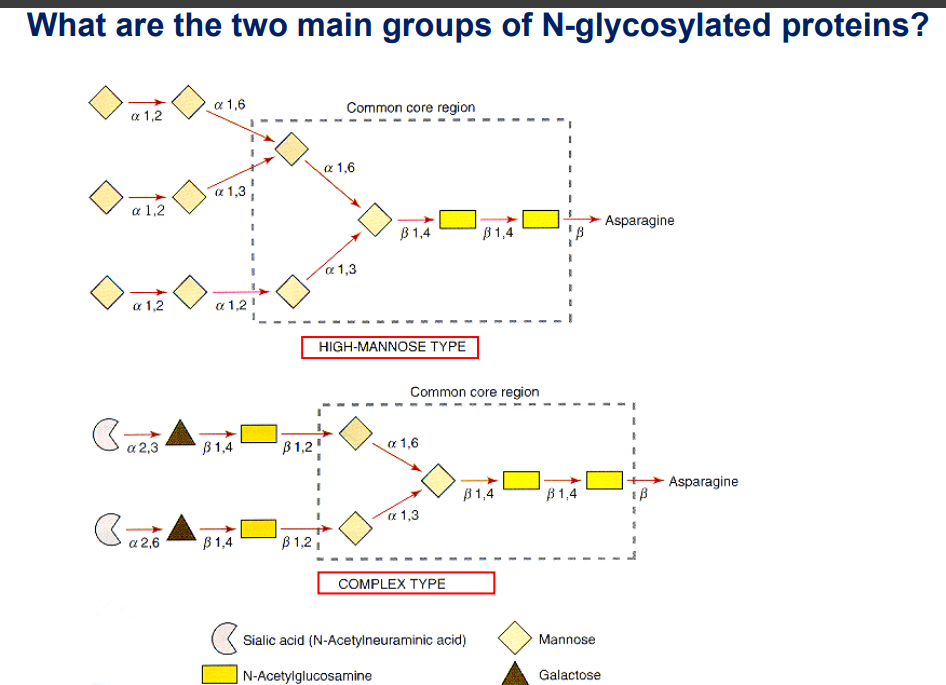
Types of N-Glycosylated Proteins
High-mannose type: Common core region including mannose and N-Acetylglucosamine.
Complex type: Involves sialic acids and various sugars.
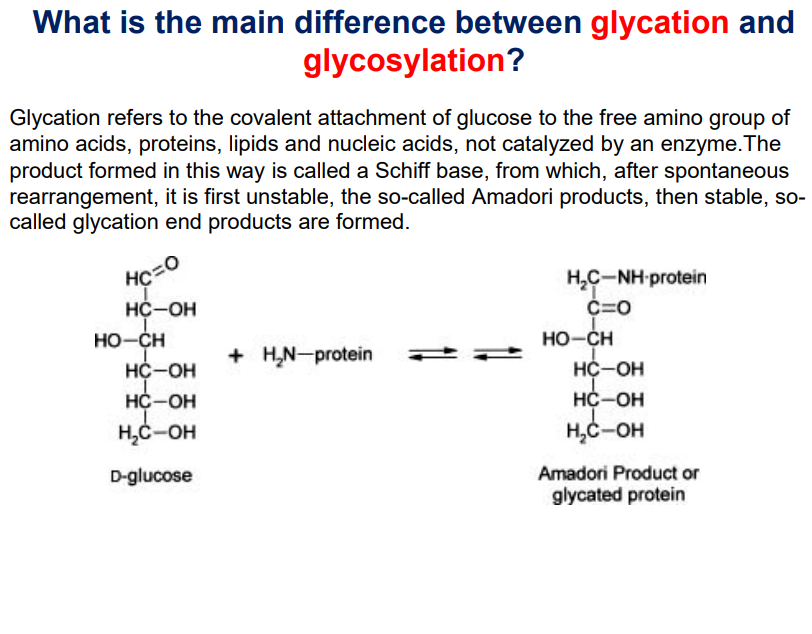
Glycation
Glycation: Non-enzymatic attachment of glucose to the free amino group of amino acids, proteins, lipids and nucleic acids.
During glycation, first a schiff base if formed, then after spontaneous rearrangements, an unstable product called Amadori products are formed and is converted to stable glycation end products
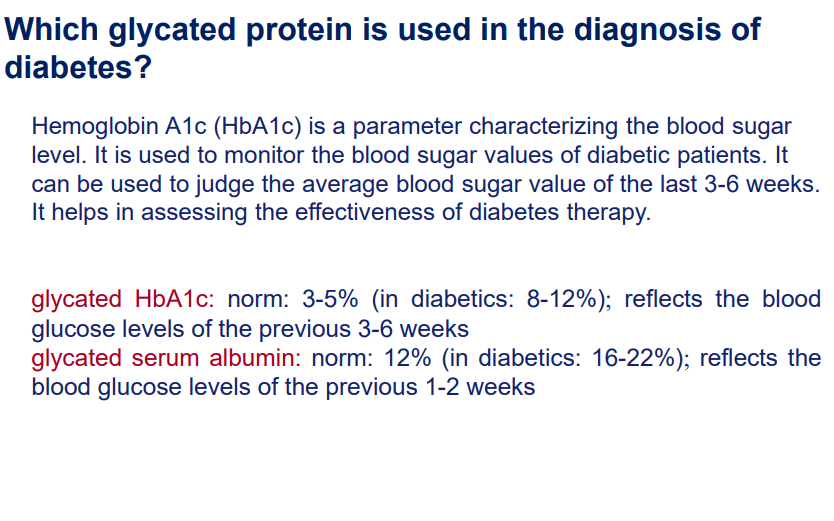
Associated with diabetes, notably Haemoglobin A1c (HbA1c) for monitoring blood glucose.
See diagram below:
Determining the blood glucose levels of a patient using glycated proteins:
Lipid Modifications
General Overview
Occurs at the N-terminal or C-terminal ends of proteins.
Prenylation: Targets/ occurs at the C-terminus (e.g., Farnesylation).
Farnesylation (cysteine)
Geranylgeranylation (Cysteine)
Fatty acylation
Myristoylation: Targets/ occurs at the N-terminus (e.g., Palmitoylation).
Palmitoylation
Aim of lipid modification: Targeting proteins to membranes.
Role of FPP in Farnesylation
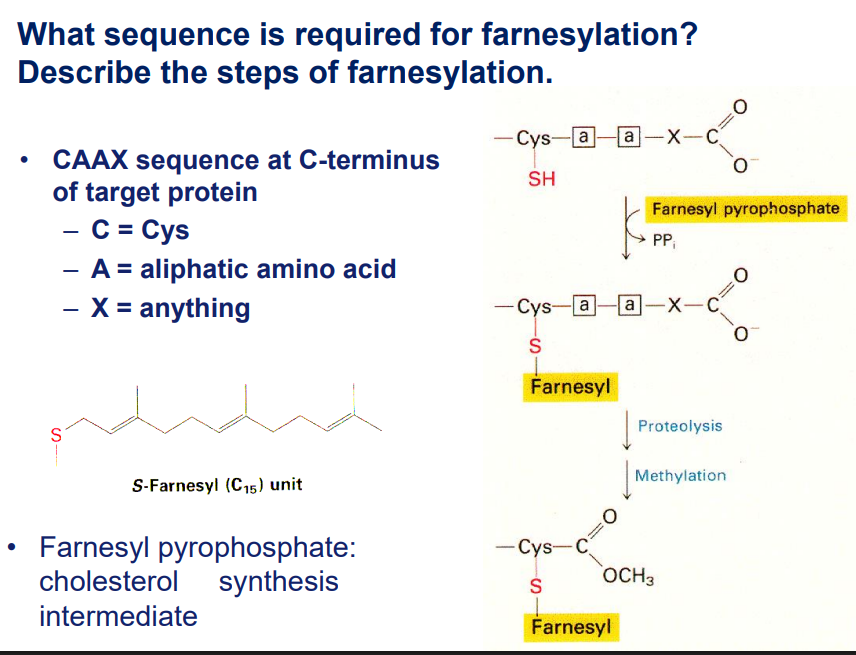
Farnesylation is a lipid modification that helps proteins anchor to membranes. The process occurs in three main steps:
Step 1: Recognition of Target Protein
The enzyme farnesyltransferase (FTase) recognizes specific proteins with a C-terminal CAAX motif (Cysteine-Aliphatic-Aliphatic-X).
The "X" amino acid determines whether a protein gets farnesylated or geranylgeranylated:
If X = methionine, serine, glutamine, or alanine → Farnesylation
If X = leucine → Geranylgeranylation (C20 modification instead of C15)
Step 2: Transfer of the Farnesyl Group
Farnesyltransferase (FTase) catalyzes the transfer of the farnesyl group from FPP (Farnesylation Pyrophosphate) to the cysteine in the CAAX motif.
This forms a thioether bond between the farnesyl group and the cysteine residue.
Step 3: Post-Farnesylation Modifications
The -AAX amino acids are cleaved by the enzyme RCE1 (Ras-converting enzyme 1).
The new C-terminal cysteine is methylated by ICMT (Isoprenylcysteine Carboxyl Methyltransferase).
These modifications increase the hydrophobicity of the protein, helping it associate with membranes.
Ras Protein:
This is a proto-oncogene that is activated during cell division.
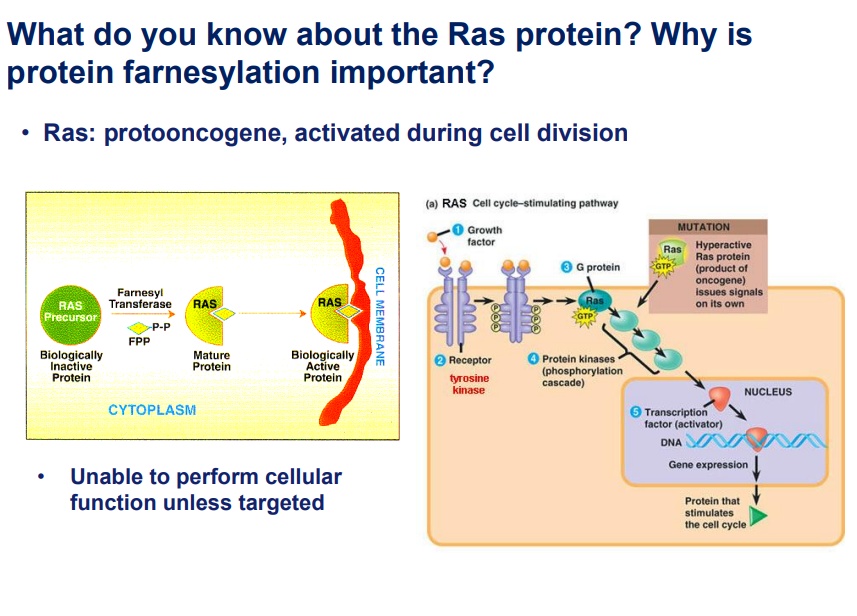
Explanation:
This slide explains the Ras protein, its role in cell division, and the importance of farnesylation in its function. Here’s a breakdown of the key points:
1. What is Ras?
Ras is a proto-oncogene, meaning it is a normal gene that can become an oncogene (cancer-causing) if mutated.
It plays a crucial role in cell signaling during cell division.
Normally, Ras is activated by external signals (e.g., growth factors) and helps regulate the cell cycle.
2. Importance of Protein Farnesylation
Farnesylation is a biochemical modification where a farnesyl group is added to the Ras protein.
This modification is essential for Ras to attach to the cell membrane, where it can properly function.
Without farnesylation, Ras remains in the cytoplasm and cannot perform its role in signaling.
3. The Ras Signaling Pathway
Growth factors bind to their respective receptors (e.g., tyrosine kinase receptors).
This activates G-protein signaling, where Ras plays a central role.
Activated Ras then triggers protein kinase cascades,
leading to the activation of transcription factors in the nucleus.
These transcription factors regulate gene expression and promote cell cycle progression.
4. Mutations in Ras and Cancer
When Ras is mutated, it becomes hyperactive, meaning it continuously signals cell division even in the absence of external growth signals.
This can lead to uncontrolled cell proliferation, which is a hallmark of cancer.
Key Takeaways
Ras is a critical regulator of cell division.
Farnesylation is essential for Ras to be correctly positioned and function properly.
Mutated Ras leads to continuous cell division, which can result in cancer.
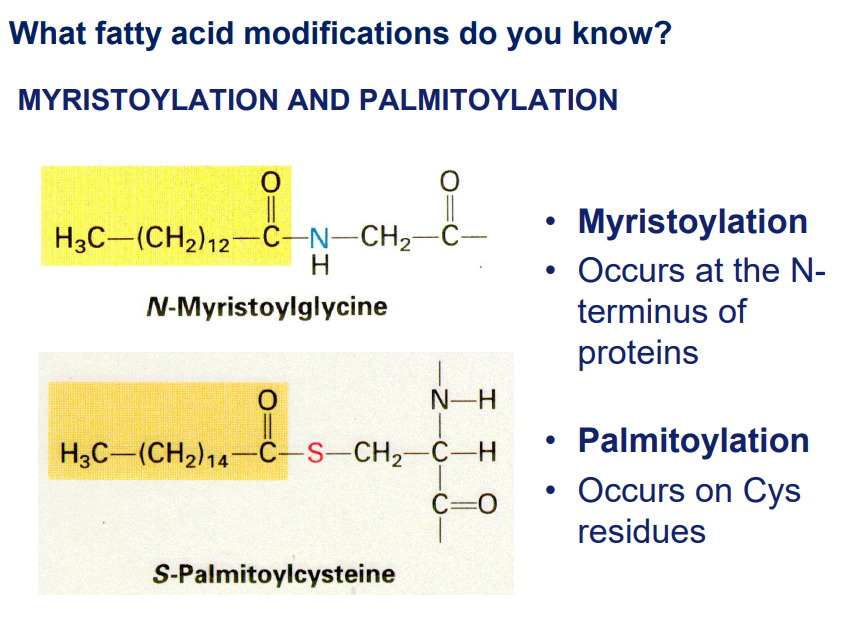
Myristoylation and Palmitoylation
Myristoylation: adding a myristoyl group (C14) to the N-terminal of a protein.
Palmitoylation: adding palmitic acid (16 carbon saturated fatty acid) to specific cysteine residues of a protein via a thioester bond.
Phosphorylation
Process Overview
Addition of a phosphate group, primarily from ATP.
Amino acids affected: Serine, Threonine, Tyrosine.
Enzymes:
Kinases (add phosphates) and Phosphatases (remove phosphates).
Classification of Protein kinase and phosphate
According to the side chain, modified these enzymes are classified:
Protein Kinases:
Serine/Threonine specific kinase
Tyrosine specific kinase
Dual specific kinase: These kinases can phosphorylate both serine/threonine and tyrosine residues in proteins, hence the term "dual specificity."
Phosphatases classification:
Serine/threonine specific phosphatase
Tyrosine specific phosphatase
Dual specific phosphatase: This phosphatase can remove phosphate from both serine/threonine and tyrosine residues in proteins, hence the term "dual specificity."
Purpose of Phosphorylation:
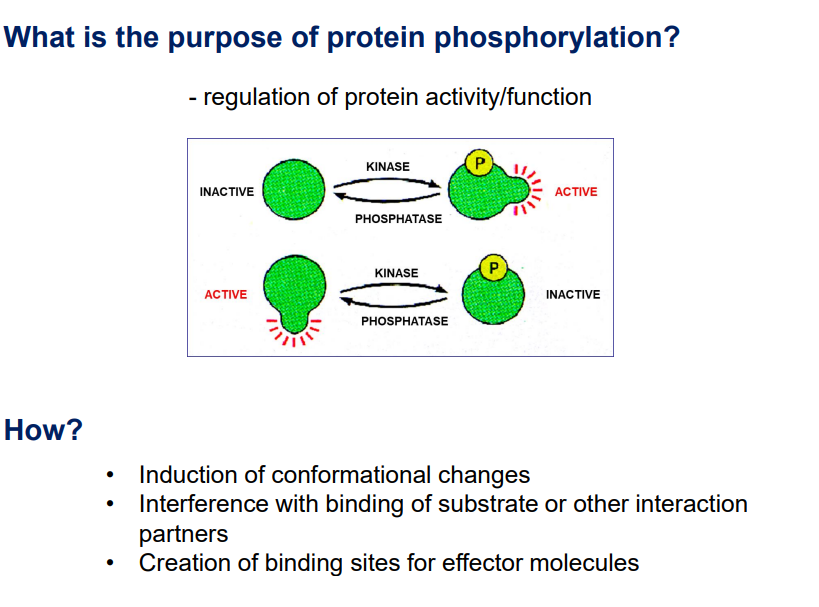
Biological Functions
Regulation of protein activity and function, conformational changes, and creation of binding sites.
Proteolysis
Definition
Cleavage of peptide bonds, resulting in modified protein structures or breakdown into amino acids.
Enzymes involved: Proteases (also called proteinases or peptidases).
Result of Proteolysis:
Cleavage of one or a few peptide bonds————> new protein, new function
Cleavage of several peptide bonds—————> protein breakdown into amino acids/peptides.
Classification of Proteases
Proteases are classified into two:
Exopeptidase: Exopeptidases act on the ends of peptide chains, cleaving terminal peptide bonds. These can be further classified into:
Amino-peptidase
Carboxyl-peptidase
Endopeptidase: Endopeptidases break peptide bonds within the interior of a protein or peptide, rather than at the ends. These can be classified based on the type of active site residues involved in the catalysis:
Serine protease: These proteases have a serine residue at their active site, which plays a key role in the hydrolysis of the peptide bond.
Aspartyl protease: These proteases have aspartic acid residues at their active site, which are involved in the hydrolysis of peptide bonds.
Cystein protease: These proteases use a cysteine residue at their active site to perform the cleavage of peptide bonds.
Metallo-proteases: These proteases require a metal ion, usually zinc, at their active site to perform the cleavage of peptide bonds.
What is the catalytic mechanism of proteases?
Serine and Cysteine proteases use nucleophilic attack for their action.
The nucleophile (serine or cysteine) attacks the carbonyl carbon of the peptide bond, pushing the electron density onto the oxygen of the carbonyl group, which temporarily forms a tetrahedral intermediate. This intermediate is unstable and breaks down.
Aspartyl Proteases and Metalloproteases:
They activate a water molecule to serve as the nucleophile, rather than using a functional group of the enzyme itself.
Biological Roles of Proteolysis
Activation of digestive enzymes
Activation of blood coagulation factors
signaling pathways. (e.g. receptor activation by proteolysis)
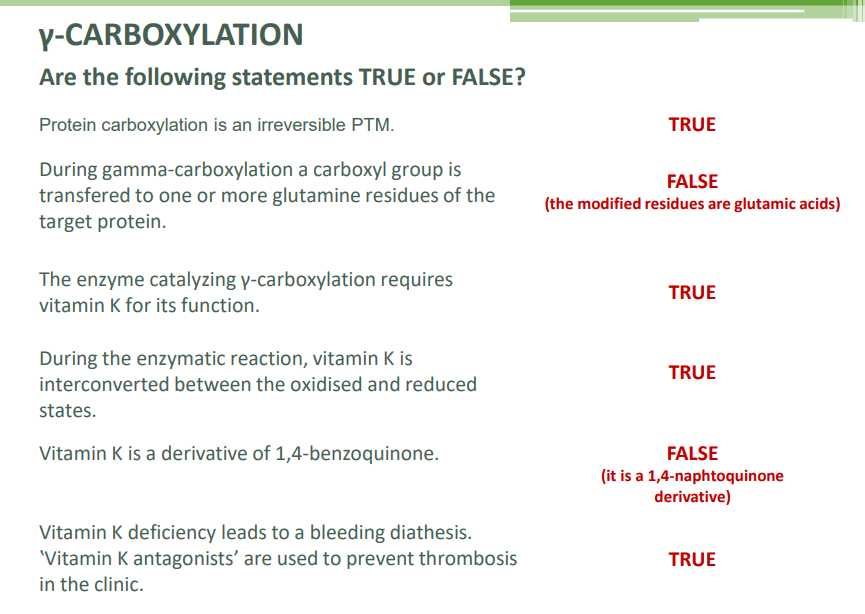
Carboxylation
γ-carboxylation: Involves transferring a carboxyl group to glutamic acids, requiring vitamin K.
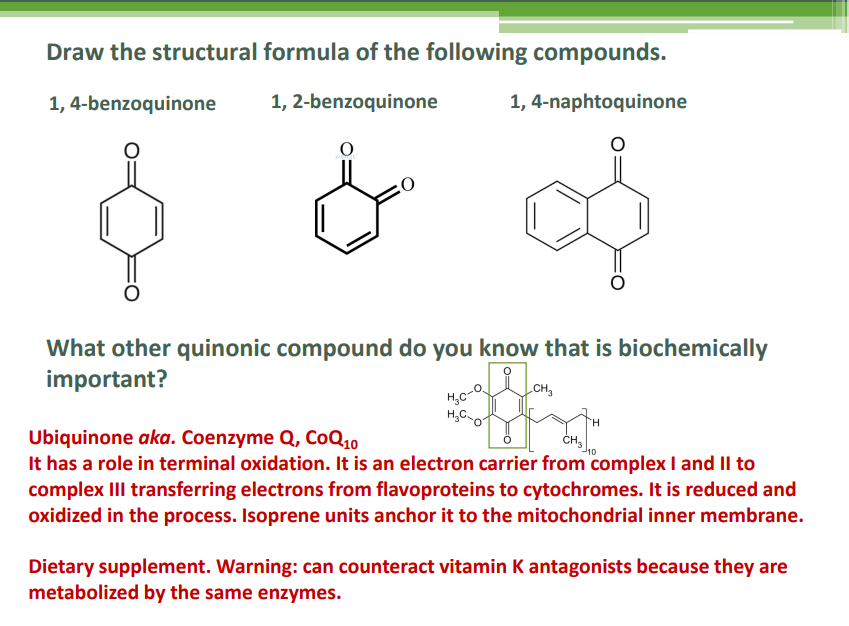
Ubiquitination
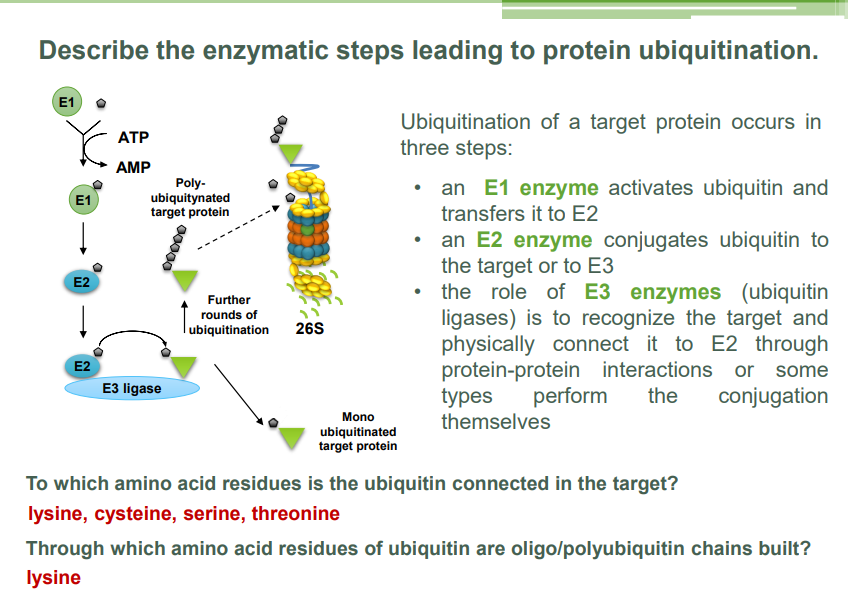
Ubiquitination is a process in which a small protein called ubiquitin is covalently attached to a target protein. This modification plays a crucial role in regulating various cellular processes, especially in marking proteins for degradation by the proteasome, a large protease complex that breaks down proteins.
Steps of Ubiquitination:
Activation of Ubiquitin:
Ubiquitin is first activated by an enzyme called E1 (ubiquitin-activating enzyme). This process requires ATP. The E1 enzyme forms a high-energy thioester bond with ubiquitin.
Conjugation to E2:
The activated ubiquitin is transferred to another enzyme called E2 (ubiquitin-conjugating enzyme), which carries the ubiquitin.
Ubiquitin Transfer to Target Protein:
The E2 enzyme, with the help of a third enzyme called E3 (ubiquitin ligase), catalyzes the transfer of ubiquitin from the E2 enzyme to a lysine residue on the target protein. The E3 enzyme is responsible for recognizing and selecting the appropriate target protein.
Multiple ubiquitin molecules can be added in a chain, usually through a lysine-48 linkage, which signals for proteasomal degradation.
Proteasomal Degradation:
Once a target protein is polyubiquitinated (multiple ubiquitin molecules attached), the proteasome recognizes and binds to the ubiquitin chain. The protein is then unfolded and translocated into the proteasome, where it is degraded into smaller peptides.
Functions of Ubiquitination:
Protein degradation: The most well-known role of ubiquitination is marking proteins for degradation via the proteasome.
Regulation of protein activity: Ubiquitination can regulate a protein's function without breaking it down, such as in the case of kinase activation or DNA repair.
Cellular signaling: It is involved in various signaling pathways, such as cell cycle regulation, immune response, and response to stress.
Summary:
Ubiquitination is the attachment of a small protein (ubiquitin) to a target protein, marking it for degradation or altering its function. It plays a key role in regulating protein levels and functions within cells.
ADP-ribosylation
ADP-ribosylation is a post-translational modification where an ADP-ribose molecule is covalently attached to a target protein. This process involves the transfer of an ADP-ribose group (which is derived from NAD+ — nicotinamide adenine dinucleotide) onto a specific amino acid residue of the protein, typically glutamic acid, asparagine, serine, or tyrosine.
Mechanism
Transfer of ADP-ribose from NAD+ to target proteins, modifying their function.
Enzymes involved: ART family (ADP-ribosyl transferases).
Two major forms of protein ADP-ribosylation
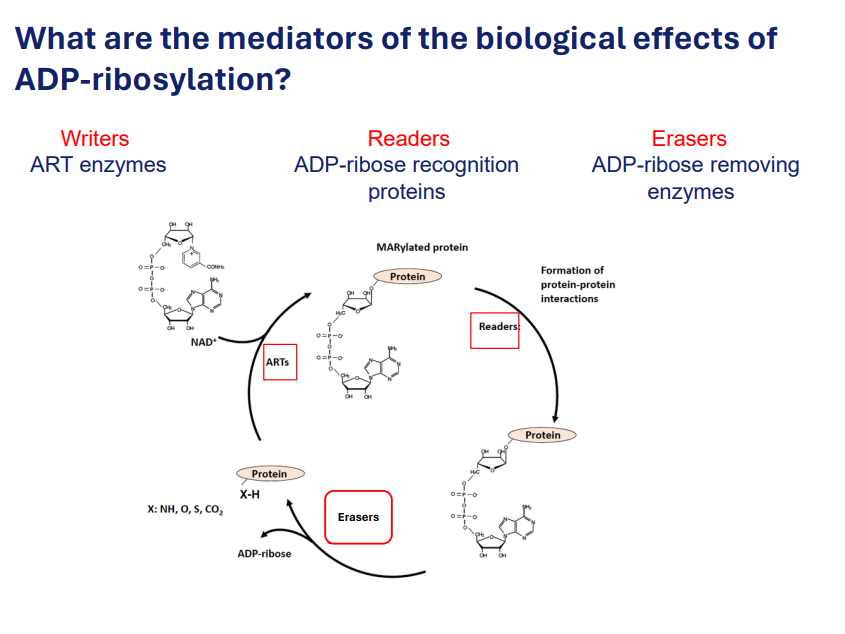
Mediators for the biological effects of ADP-ribosylation:
Writers: ART enzymes
Writers are enzymes responsible for adding ADP-ribose to target proteins. These enzymes are called ADP-ribosyltransferases (ARTs). They catalyse the transfer of the ADP-ribose moiety from NAD+ (nicotinamide adenine dinucleotide) to specific amino acid residues on the target proteins.
Function: ADP-ribosyltransferases can add single ADP-ribose molecules (monoadenylation) or poly(ADP-ribose) chains (polyadenylation) to the target proteins, which can affect the proteins' function, localization, or interactions.
Readers: ADP-ribose recognition proteins
Role: Readers are proteins that recognize and bind to the ADP-ribose modification on target proteins. They contain specific domains or motifs, such as the ARTD (ADP-ribose binding domain), that allow them to interact with ADP-ribose groups and mediate subsequent biological effects.
Function: These reader proteins translate the ADP-ribosylation signal into cellular responses, such as changes in protein activity, protein-protein interactions, or cellular localization. Essentially, they "interpret" the ADP-ribosylation mark and affect cellular processes accordingly.
Erasers: ADP-ribose removing enzymes.
Role: Erasers are enzymes that remove the ADP-ribose modifications from target proteins. These enzymes reverse the effect of ADP-ribosylation, thus regulating the balance of ADP-ribosylation within the cell. Erasers are crucial for controlling the duration and extent of ADP-ribosylation.
Function: By hydrolyzing the ADP-ribose group, erasers deadenylate or remove poly(ADP-ribose) chains from target proteins, reversing the biological effects triggered by the ADP-ribosylation mark. This process allows the cell to reset signaling pathways and maintain homeostasis.
Histone Code
Overview
Histone code is a set of histone protein post-translational modifications that regulate gene transcription by controlling transitions between open and closed states of chromatin.
Modifications include acetylation, methylation, phosphorylation, and ubiquitination.
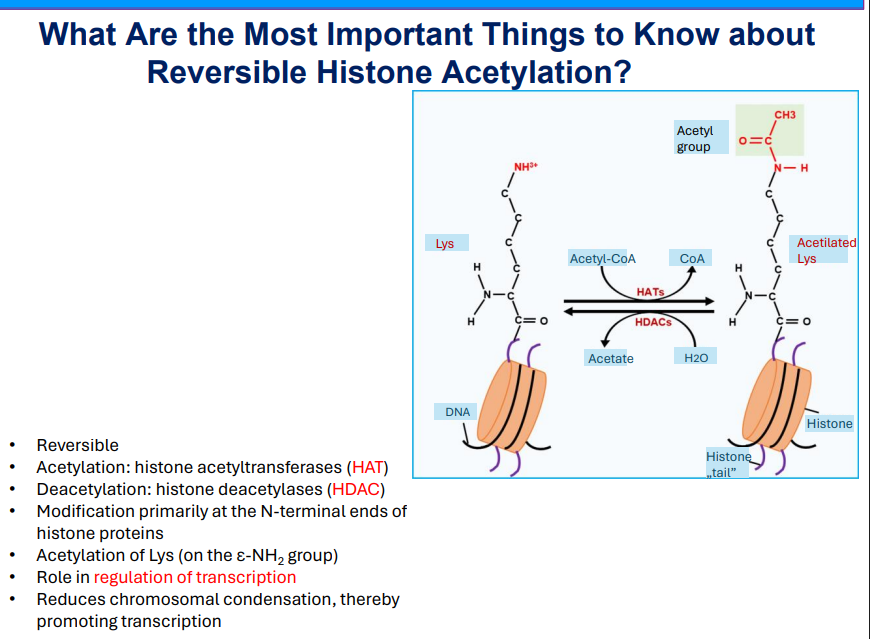
Reversible Histone Acetylation
Enzymatic process involving histone acetyltransferases (HAT) and histone deacetylases (HDAC).