AP BIO Chapter 13 - The Molecular Basis of Inheritance
Introduction -
13.1 DNA is the genetic material
13.2 Many proteins work together in DNA replication and repair
13.3 A chromosome consists of a DNA molecule packed together with proteins
13.4 Understanding DNA structure and replication makes genetic engineering possible
BIG IDEAS: The evidence that DNA and RNA carry genetic information in all lifeforms (Big Idea 1) began with a series of historical experiments (Big Idea 3) that led to an understanding of how the structure and interaction of molecules (Big Idea 4) facilitate the replication and transfer of DNA and RNA across membranes (Big Idea 3)
Hershey and Chase concluded that DNA is the source of heritable information. Select the data from their experiments that support that claim (Big Idea 3)
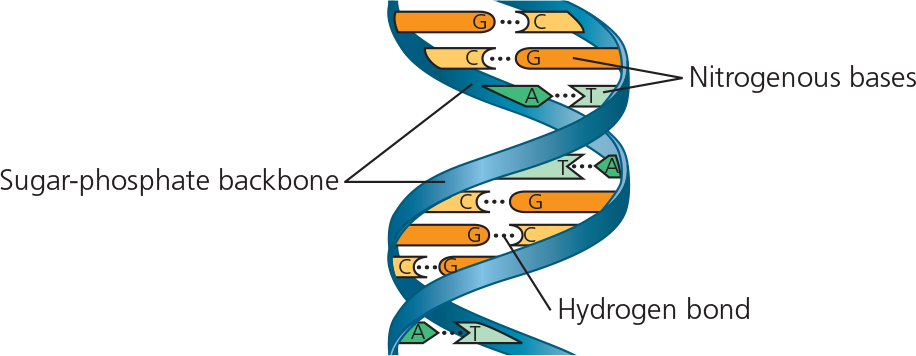
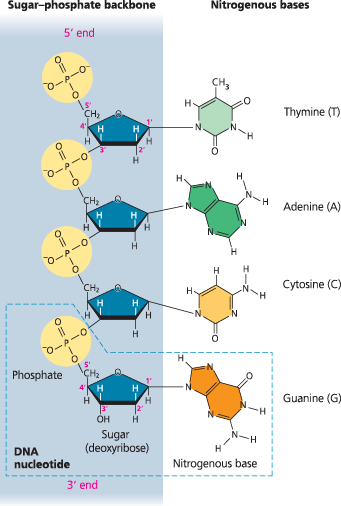
Structure of DNA - Sugar phosphate backbone and base pairs

James Watson and Francis Crick with their DNA model

}}DNA Replication:}}
The process by which a DNA molecule is copied; also called DNA synthesis
What are nucleotides made of?
P group, 5 carbon sugar (deoxyribose or ribose), a nitrogen base
What are the nitrogen-containing bases of DNA?
Adenine, thymine, cytosine, and guanine (A+T and C+G pair)
What basic elements makeup DNA?
CHONP
What basic elements makeup proteins?
CHONS
^^13.1 - DNA is the genetic material^^
Experiments with bacteria and phages provided the first strong evidence that the genetic material is DNA
Watson and Crick deduced that DNA is a double helix and built a structural model. Two antiparallel sugar-phosphate chains wind around the outside of the molecule; the nitrogenous bases project into the interior, where they hydrogen-bond in specific pairs: A with T, G with C
}}Streptococcus Pneumoniae:}}
A bacterium that causes pneumonia in mammals
}}Pathogenic:}}
Disease-causing
}}Nonpathogenic:}}
Harmless
When Griffith killed the pathogenic bacteria with heat and then mixed the cell remains with living bacteria of the nonpathogenic strain, some of the living cells became ___
- pathogenic
What apparently caused the heritable change?
A chemical component of the dead pathogenic cells caused it (the substance was not known)
}}Transformation:}}
The process by which a cell in culture acquires the ability to divide indefinitely, similar to the division of cancer cells; a change in genotype and phenotype due to the assimilation of external DNA by a cell. When the external DNA is from a member of a different species, transformation results in horizontal gene transfer
Experiment - Frederick Griffith studied two strains of the bacterium Streptococcus pneumoniae. The S (smooth) strain can cause pneumonia in mice; it is pathogenic because the cells have an outer capsule that protects them from an animal’s immune system. Cells of the R (rough) strain lack a capsule and are nonpathogenic. To test for the trait of pathogenicity, Griffith injected mice with the two strains
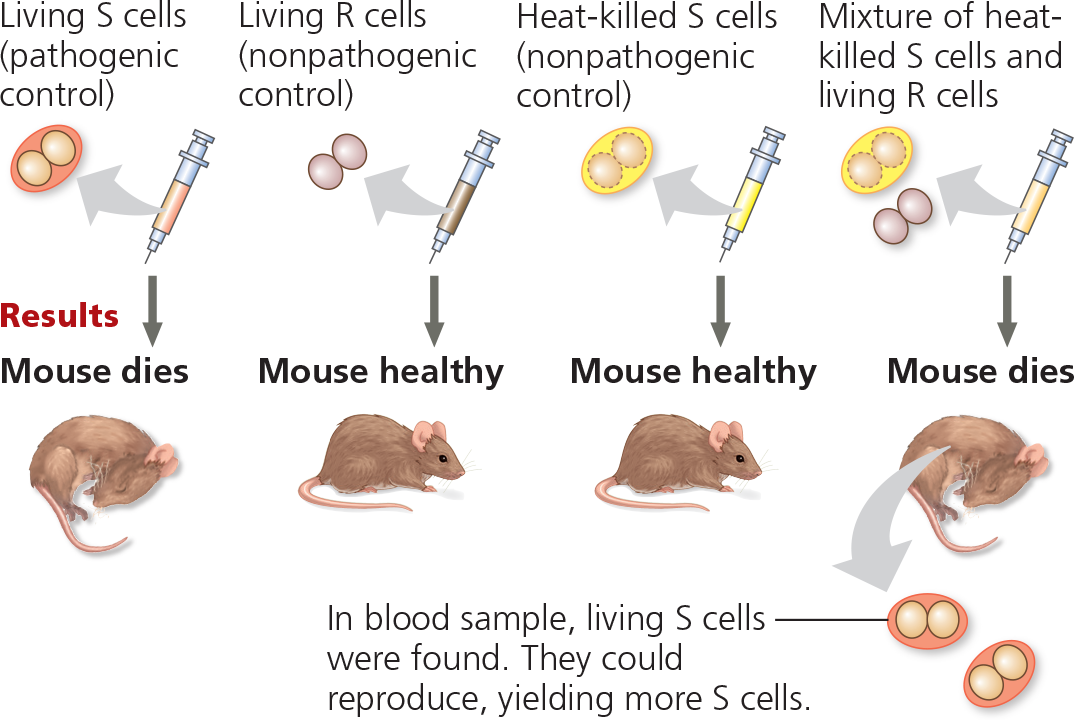
What did Griffith conclude?
The living R bacteria had been transformed into pathogenic S bacteria by an unknown, heritable substance from the dead S cells that enabled the R cells to make capsules
How did this experiment rule out the possibility that the R cells simply used the dead S cells’ capsules to become pathogenic?
The living S cells found in the blood sample were able to reproduce to yield more S cells, indicating that the S trait is a permanent, heritable change, rather than just a one-time use of the dead S cells’ capsules
}}Bacteriophage:}}
A virus that infects bacteria; “bacteria-eaters“
}}Phage:}}
A virus that infects bacteria; short for bacteriophage
}}Virus:}}
An infectious particle incapable of replicating outside of a cell, consisting of an RNA or DNA genome surrounded by a protein coat (capsid) and, for some viruses, a membranous envelope
A virus infecting a bacterial cell - A phage called T2 attaches to a host cell and injects its genetic material through the plasma membrane while the head and tail parts remain on the outer bacterial surface (colorized TEM)
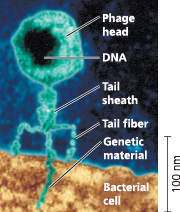
}}Escherichia Coli (E. Coli):}}
A bacterium that normally lives int eh intestine of mammals and is a model organism for molecular biologists.
T2, like many other phages, was composed almost entirely of ___
- DNA and protein
What did Hershey and Chase want to find out?
They wanted to show that only one of the two components of T2 actually enters the E. coli cell during infection
Experiment - Alfred Hershey and Martha Chase used radioactive sulfur and phosphorus to trace the fates of protein and DNA, respectively, of T2 phages that infected bacterial cells. They wanted to see which of these molecules entered the cells and could reprogram them to make more phages
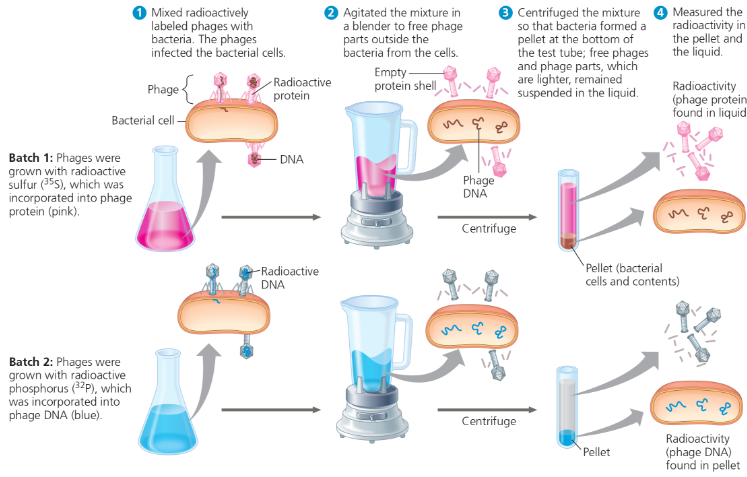
What were the results of the experiment?
When proteins were labeled (batch 1), radioactivity remained outside the cells, but when DNA was labeled (batch 2), radioactivity was found inside the cells. Cells containing radioactive phage DNA released new phages with some radioactive phosphorus
What did Hershey and Chase conclude?
Phage DNA entered bacterial cells, but phage proteins did not. DNA, not protein, functions as the genetic material of phage T2. DNA injected by the phage must be the molecule carrying the genetic information that makes the cells produce new viral DNA and proteins
How would the results have differed if proteins carried the genetic information?
The radioactivity would have been found in the pellet when proteins were labeled (batch 1) because proteins would have had to enter the bacterial cells to program them with genetic instructions. It’s hard for us to imagine now, but the DNA might have played a structural role that allowed some of the proteins to be injected while it remained outside the bacterial cell (thus no radioactivity in the pellet in batch 2)
The structure of a DNA strand - Each DNA nucleotide monomer consists of a nitrogenous base (T, A, C, or G), the sugar deoxyribose (blue), and a phosphate group (yellow). The phosphate group of one nucleotide is attached to the sugar of the next by a covalent bond, forming a “backbone” of alternating phosphates and sugars from which the bases project. The polynucleotide strand has directionality, from the 5′ end (with the phosphate group) to the 3′ end (with the —OH group of the sugar). 5′ and 3′ refer to the numbers assigned to the carbons in the sugar ring
DNA base composition ___ between species
- varies
The percentages of A and T bases are ___, as are those of G and C bases
- roughly equal
Data from base pair experiments
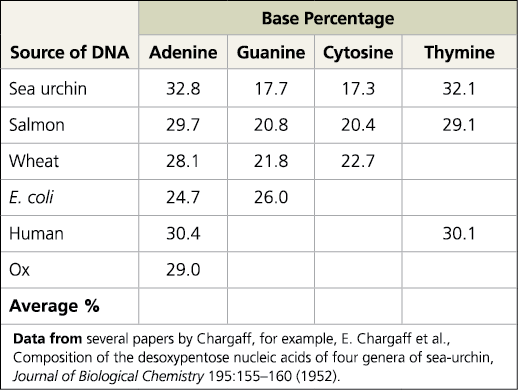
Explain how the sea urchin and salmon data demonstrate both of Chargaff’s rules
They demonstrate the first rule (DNA base composition varies between species) because the percentages of A, G, C, and T are different in sea urchins and salmon. They demonstrate the second rule (The percentages of A and T bases are roughly equal, as are those of G and C bases) because the percent of A and T in sea urchins and salmon was around 32.45 and 29.4 respectively. For G and C in sea urchins and salmon, it was around 17.5 and 20.6 percent respectively
By the early 1950s, the arrangement of ___ bonds in a nucleic acid polymer was well established
- covalent
}}Double Helix:}}
The form of native DNA, referring to its two adjacent antiparallel polynucleotide strands wound around an imaginary axis into a spiral shape
Rosalind Franklin and her X-ray diffraction photo of DNA - Franklin, a very accomplished X-ray crystallographer, conducted critical experiments resulting in the photo that allowed Watson and Crick to deduce the double-helical structure of DNA

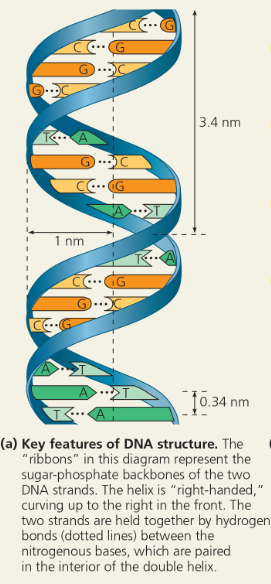
The structure of the double helix - Key features of DNA structure
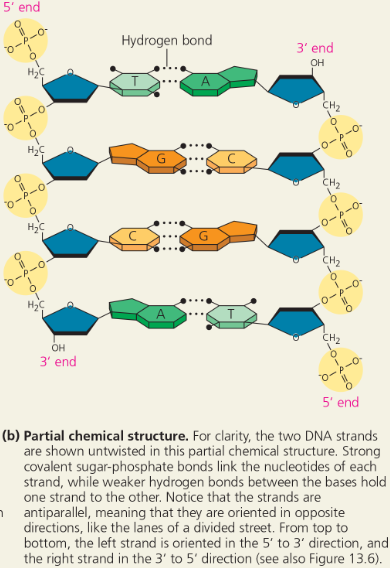
The structure of the double helix - Partial chemical structure
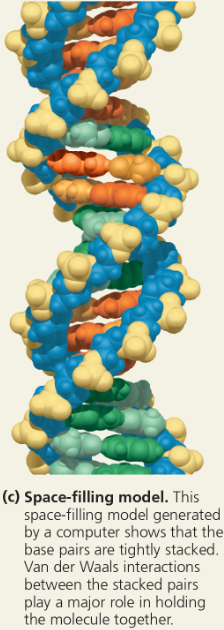
The structure of the double helix - Space-filling model
Why are the sugar-phosphate backbones were on the outside of the DNA molecule?
It puts the negatively charged phosphate groups facing the aqueous surroundings, while the relatively hydrophobic nitrogenous bases were hidden in the interior
}}Antiparallel:}}
Referring to the arrangement of the sugar-phosphate backbones in a DNA double helix (they run in opposite 5’ → 3’ directions)
With the bases stacked just ___ apart, there are _ “rungs” of base pairs in each full turn of the helix
- .34 nm; ten
}}Purines:}}
Nitrogenous bases with two organic rings (A and G)
}}Pyrimidines:}}
Nitrogenous bases with one organic ring (C and T)
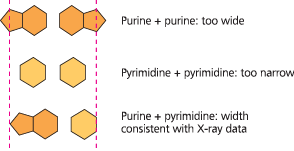
Possible base pairings in the DNA double helix - Purines (A and G) are about twice as wide as pyrimidines (C and T). A purine-purine pair is too wide and a pyrimidine-pyrimidine pair is too narrow to account for the 2-nm diameter of the double helix, while a purine-pyrimidine pair fits the data well
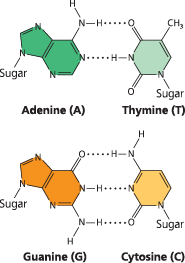
Base pairing in DNA - The pairs of nitrogenous bases in a DNA double helix are held together by hydrogen bonds, shown here as black dotted lines
Given a polynucleotide sequence such as GAATTC, explain what further information you would need in order to identify which end is the 5’ end
In order to tell which end is the 5′ end, you need to know which end has a phosphate group on the 5′ carbon (the 5′ end) or which end has an —OH group on the 3′ carbon (the 3′ end)
Griffith did not expect transformation to occur in his experiment. What results was he expecting?
Griffith expected that the mouse injected with the mixture of heat-killed S cells and living R cells would survive since neither type of cell alone could kill the mouse
What does it mean when we say that the two DNA strands in the double helix are antiparallel? What would the end of the double helix look like if the strands were parallel?
Each strand in the double helix has polarity, the end with a phosphate group on the 5′ carbon of the sugar being called the 5′ end, and the end with an —OH group on the 3′ carbon of the sugar being called the 3′ end. The two strands run in opposite directions, one running 5′ → 3′ and the other running 3′ → 5′. Thus, each end of the molecule has both a 5′ and a 3′ end. This arrangement is called “antiparallel.” If the strands were parallel, they would both run 5′ → 3′ in the same direction, so an end of the molecule would have either two 5′ ends or two 3′ ends
^^13.2 - Many proteins work together in DNA replication and repair^^
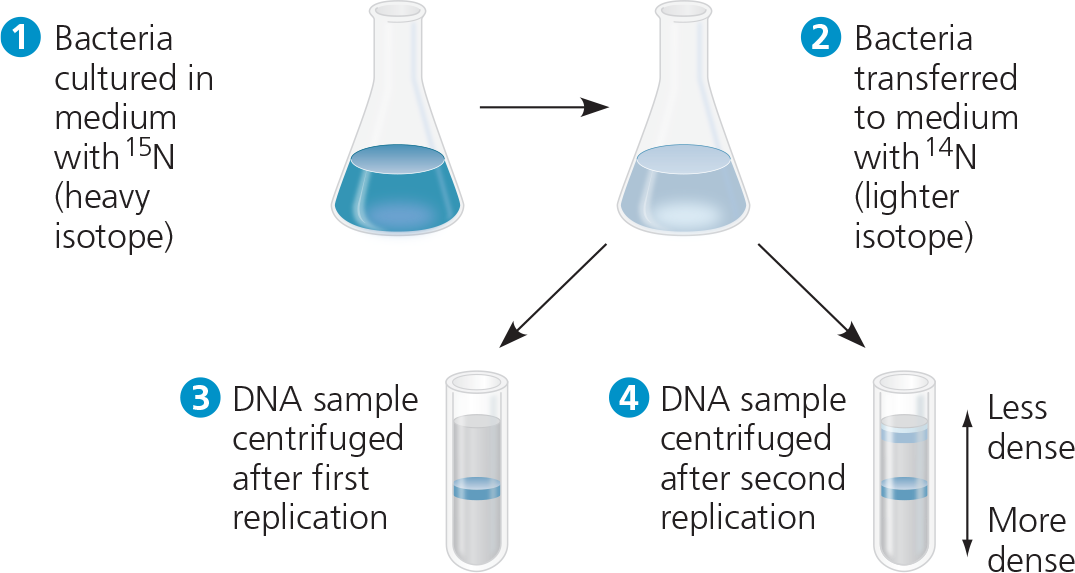
The Meselson-Stahl experiment showed that DNA replication is semiconservative: The parental molecule unwinds, and each strand then serves as a template for the synthesis of a new strand according to base-pairing rules
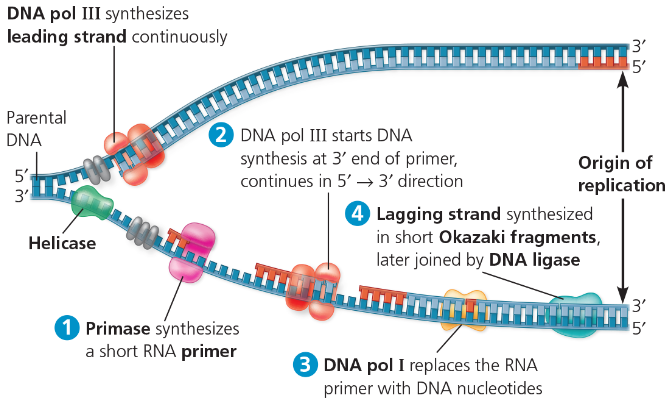
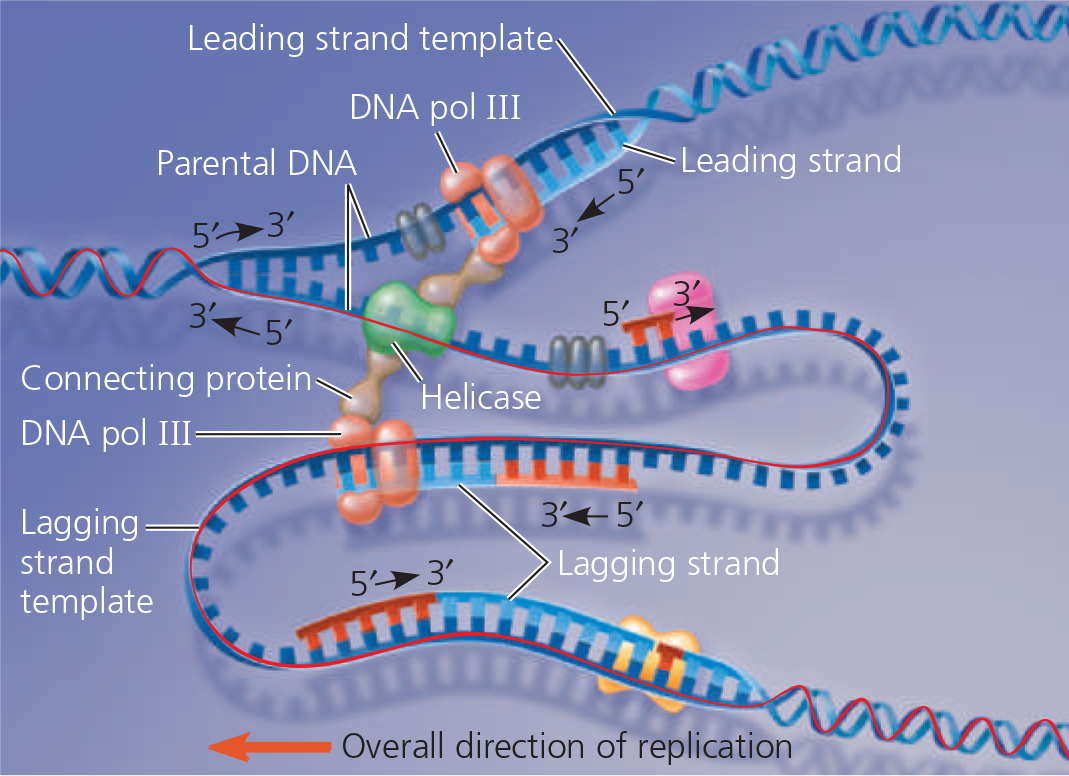
DNA replication at one replication fork is summarized here:
DNA polymerases proofread new DNA, replacing incorrect nucleotides. In mismatch repair, enzymes correct errors that persist. Nucleotide excision repair is a general process by which nucleases cut out and replace damaged stretches of DNA
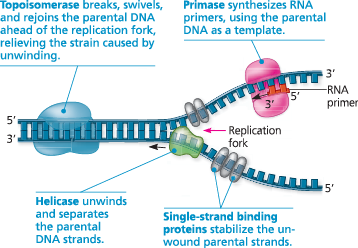
^^Helicase^^ - breaks hydrogen bonds of DNA to separate it
^^DNA polymerase^^ - catalyzes addition of nucleotides to RNA primer (5’ to 3’), replaces RNA primers with DNA
- limitations: only 5’ to 3’, cannot initiate synthesis on its own
^^Single-Strand Binding Protein^^ - saves space for Jesus cause the strands are still attracted to each other after being broken apart
^^Topoisomerase^^ - relieves strain from helicase and prevents too much unwinding
^^Primase^^ - synthesizes RNA primers (to start new DNA strand)
^^DNA Ligase^^ - the glue (between lagging strands/okasaki fragments)
^^Nuclease^^ - cuts DNA or RNA
^^Telomerase^^ - lengthens telomeres in germ cells (to prevent shortening of DNA and loss of genes)
- can make cells malignant (cancerous)
^^Error Rate and Consequences^^ - very little errors but can lead to permanent mutations in future generations
Prokaryotic vs Eukaryotic - In prokaryotic cells, there is only one point of origin, replication occurs in two opposing directions at the same time, and takes place in the cell cytoplasm. Eukaryotic cells on the other hand, have multiple points of origin, and use unidirectional replication within the nucleus of the cell
A model for DNA replication: the basic concept - In this simplified illustration, a short segment of DNA has been untwisted. Simple shapes symbolize the four kinds of bases. Dark blue represents DNA strands present in the parental molecule; light blue represents newly synthesized DNA
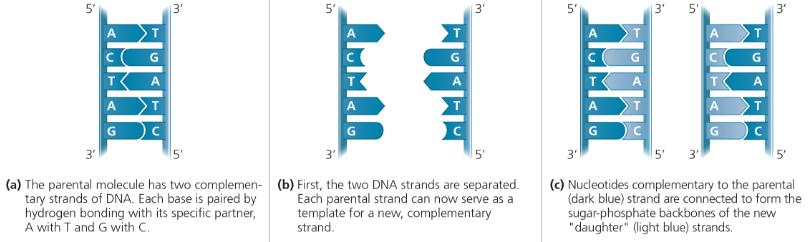
Watson and Crick’s replication hypothesis
Now our model for deoxyribonucleic acid is, in effect, a pair of templates, each of which is complementary to the other. We imagine that prior to duplication the hydrogen bonds are broken, and the two chains unwind and separate. Each chain then acts as a template for the formation on to itself of a new companion chain, so that eventually we shall have two pairs of chains, where we only had one before. Moreover, the sequence of the pairs of bases will have been duplicated exactly
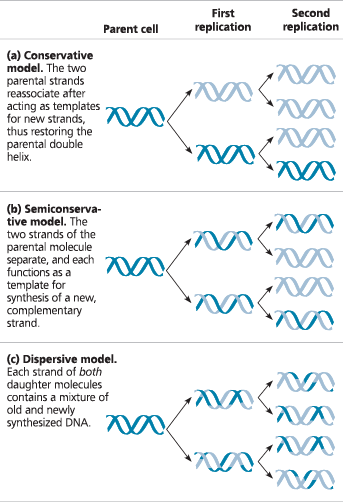
}}Semiconservative Model:}}
Type of DNA replication in which the replicated double helix consists of one old strand, derived from the parental molecule, and one newly made strand
}}Conservative Model:}}
Two parental strands come back together after the process (that is, the parental molecule is conserved)
}}Dispersive Model:}}
All four strands of DNA following replication have a mixture of old and new DNA
Three alternative models of DNA replication - Each segment of double helix symbolizes the DNA within a cell. Beginning with a parent cell, we follow the DNA for two more generations of cells—two rounds of DNA replication. Parental DNA is dark blue; newly made DNA is light blue
Experiment - Matthew Meselson and Franklin Stahl cultured E. coli for several generations in a medium containing nucleotide precursors labeled with a heavy isotope of nitrogen, 15N. They then transferred the bacteria to a medium with only 14N, a lighter isotope. They took one sample after the first DNA replication and another after the second replication. They extracted DNA from the bacteria in the samples and then centrifuged each DNA sample to separate DNA of different densities
Conclusion - Meselson and Stahl compared their results to those predicted by each of the three models. The first replication in the 14N medium produced a band of hybrid (15N -14N) DNA. This result eliminated the conservative model. The second replication produced both light and hybrid DNA, a result that refuted the dispersive model and supported the semiconservative model. They therefore concluded that DNA replication is semiconservative
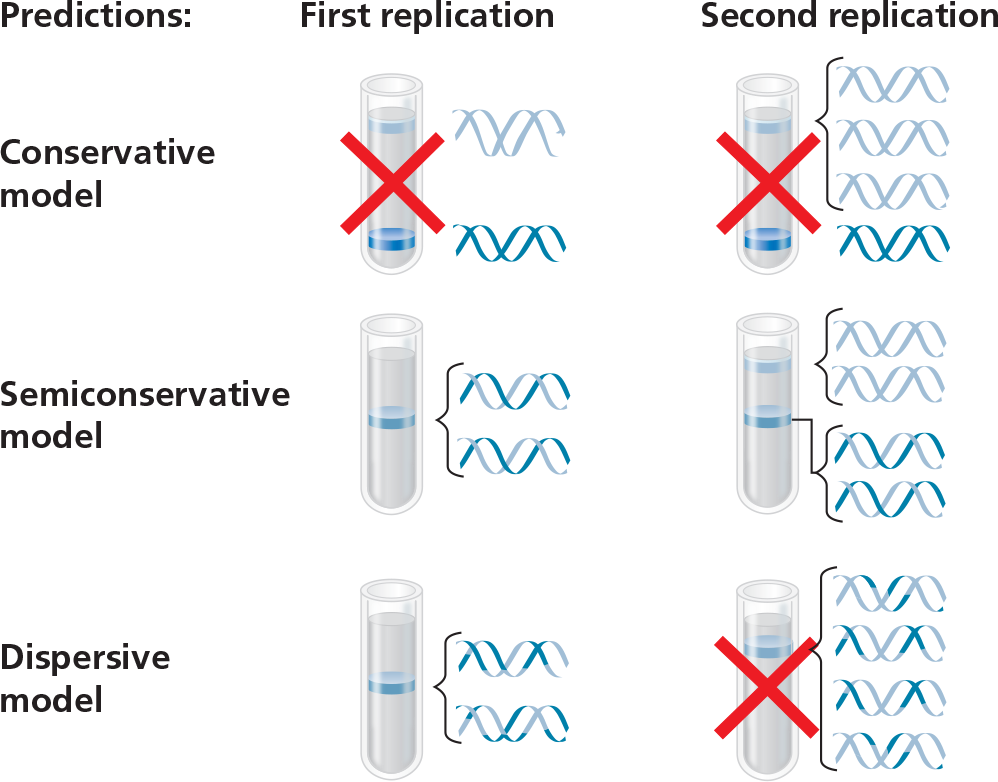
If Meselson and Stahl had first grown the cells in 14N-containing medium and then moved them into 15N-containing medium before taking samples, what would have been the result?
The tube from the first replication would look the same, with a middle band of hybrid 15N - 14N DNA, but the second tube would not have the upper band of two light blue strands. Instead, it would have a bottom band of two dark blue strands, like the bottom band in the result predicted after one replication in the conservative model
Each of your cells has ___ DNA molecules in its nucleus, _ long double-helical molecule per chromosome
- 46; one
Describe the speed and accuracy of DNA copying
It takes one of your cells just a few hours to copy all of this DNA, and it is achieved with very few errors—only about one per 10 billion nucleotides
}}Origin of Replication:}}
Site where the replication of a DNA molecule begins, consisting of a specific sequence of nucleotides
}}Replication Fork:}}
A Y-shaped region on a replicating DNA molecules where the parental strands are being unwound and new strands are being synthesized
}}Helicases:}}
An enzyme that untwists the double helix of DNA at replication for, separating the to strands and making them available as template strands (think zipper)
}}Single-Strand Binding Protein:}}
A protein that binds to the unpaired DNA strands during DNA replication, stabilizing them and holding them apart while they serve as templates for he synthesis of complementary strands of DNA
}}Topoisomerase:}}
A protein that breaks, swivels, and rejoins DNA strands. During DNA replication, topoisomerase helps to relieve strain in the double help ahead of the replication fork
Some of the proteins involved in the initiation of DNA replication - The same proteins function at both replication forks in a replication bubble. For simplicity, only the left-hand fork is shown, and the DNA bases are drawn much larger in relation to the proteins than they are in reality
Origins of replication in E. coli and eukaryotes - The red arrows indicate the movement of the replication forks and thus the overall directions of DNA replication within each bubble
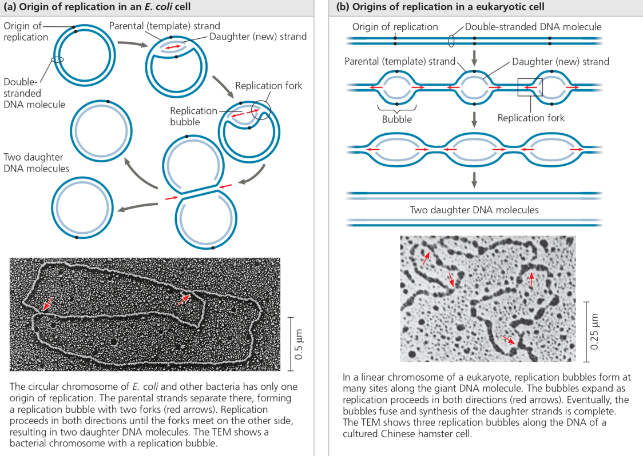
In the TEM above, add arrows for the third bubble
In the bubble at the top of the micrograph in (b), arrows should be drawn pointing left and right to indicate the two replication forks
The enzymes that synthesize DNA cannot ___ the synthesis of a polynucleotide; they can only _
- initiate; add nucleotides to the end of an already existing chain that is base-paired with the template strand
}}Primer:}}
A short stretch of RNA with a free 3’ end, bound by complementary base pairing to the template strand and elongated with DNA nucleotides during DNA replication
}}Primase:}}
An enzyme that joins RNA nucleotides to make a primer during DNA replication, using the parental DNA strand as a template
}}DNA polymerases:}}
An enzyme that catalyzes the elongation of new DNA (for example, at a replication fork) by the addition of nucleotides to the 3’ end of an existing chain.
DNA polymerase III and DNA polymerase I play major roles ___
- in DNA replication in E. coli
}}dATP:}}
The adenine nucleotide used to make DNA
How is dehydration and hydrolysis used in growing DNA?
As each monomer joins the growing end of a DNA strand in a dehydration reaction catalyzed by DNA polymerase, two phosphate groups are lost as a molecule of pyrophosphate (℗—℗i). Subsequent hydrolysis of the pyrophosphate to two molecules of inorganic phosphate (℗i) is a coupled exergonic reaction that helps drive the polymerization reaction
Addition of a nucleotide to a DNA strand - DNA polymerase catalyzes the addition of a nucleotide to the 3′ end of a growing DNA strand, with the release of two phosphates
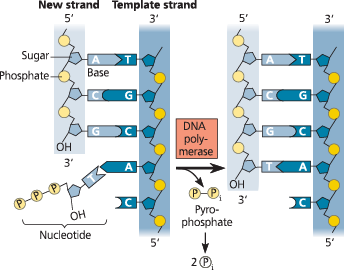
Use this diagram to explain what we mean when we say that each DNA strand has directionality
Looking at any of the DNA strands, we see that one end is called the 5′ end and the other the 3′ end. If we proceed from the 5′ end to the 3′ end on the left-most strand, for example, we list the components in this order: phosphate group → 5′ C of the sugar → 3′ C → phosphate → 5′ C → 3′ C. Going in the opposite direction on the same strand, the components proceed in the reverse order: 3′ C → 5′ C → phosphate. Thus, the two directions are distinguishable, which is what we mean when we say that the strands have directionality. (Review Figure 13.6 if necessary.)
The two new strands formed during DNA replication must also be ___ to their template strands
- antiparallel
DNA polymerases can add nucleotides only to the free ___ end of a primer or growing DNA strand, never to the _ end
- 3’; 5’
}}Leading Strand:}}
The new complementary DNA strand synthesized continuously along the template strand toward the replication fork in the mandatory 5’ → 3’ direction
Synthesis of the leading strand during DNA replication - This diagram focuses on the left replication fork shown in the overview box. DNA polymerase III (DNA pol III), shaped like a cupped hand, is shown closely associated with a protein called the “sliding clamp” that encircles the newly synthesized double helix like a doughnut. The sliding clamp moves DNA pol III along the DNA template strand
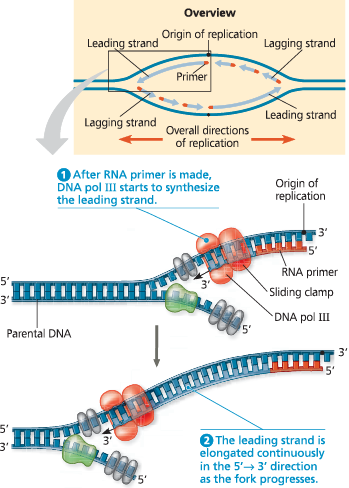
}}Lagging Strand:}}
A discontinuously synthesized DNA strand that elongates by means of Okazaki fragments, each synthesized in a 5’ → 3’ direction away from the replication fork
}}Okasaki Fragments:}}
A short segment of DNA synthesized away from the replication fork on a template strand during DNA replication. Many such segments are joined together to make up the lagging strand of newly synthesized DNA
Whereas only one primer is required on the leading strand, each Okazaki fragment on the lagging strand must be primed ___
- separately
}}DNA Ligase:}}
A linking enzyme essential for DNA replication; catalyzes the covalent bodnin of the 3’ end of one DNA fragment (such as an Okazaki fragment) to the 5’ end of another DNA fragment (such as a growing DNA chain)
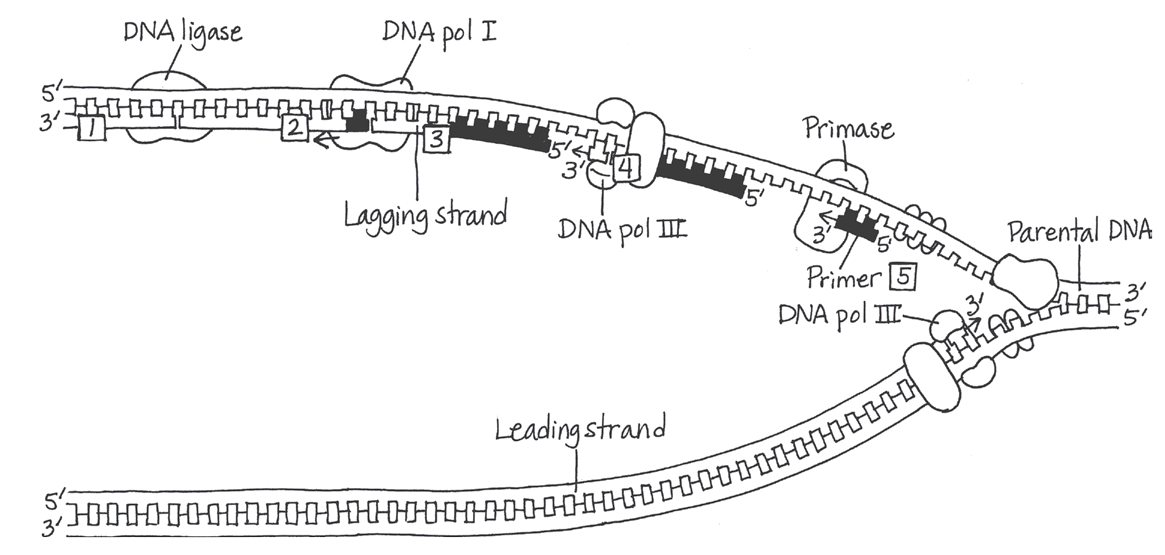
Synthesis of the lagging strand
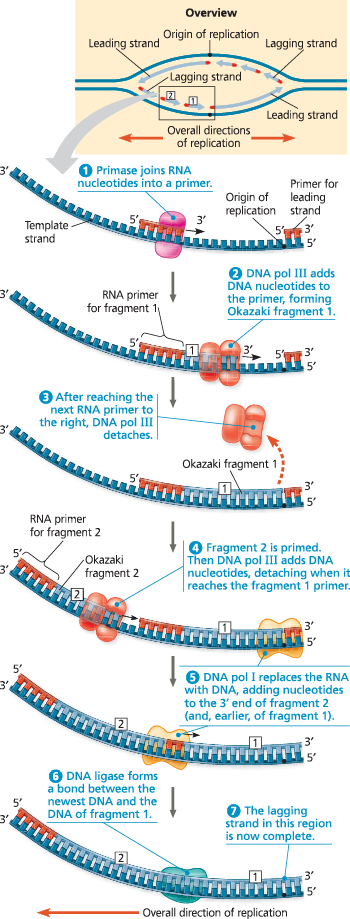
A summary of bacterial DNA replication - The detailed diagram shows the left-hand replication fork of the replication bubble in the overview (upper right). Viewing each daughter strand in its entirety in the overview, you can see that half of it is made continuously as the leading strand, while the other half (on the other side of the origin) is synthesized in fragments as the lagging strand
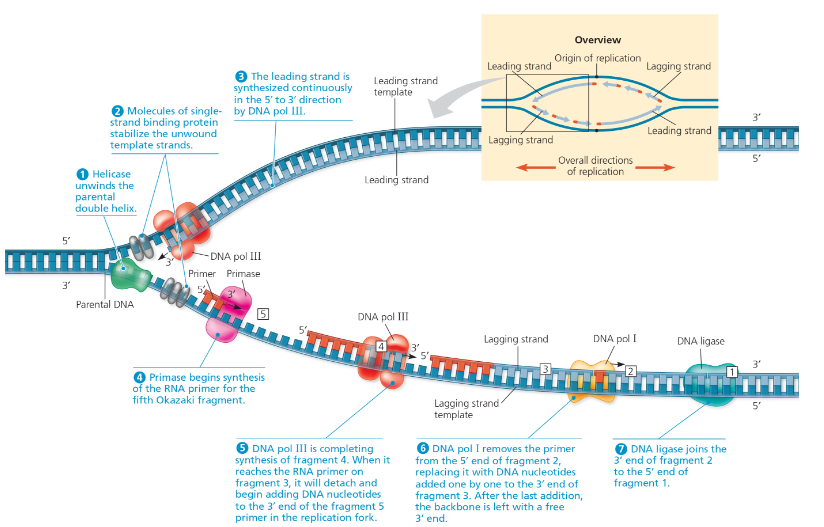
Draw a diagram showing the right-hand fork of the bubble in Figure 13.19. Number the Okazaki fragments and label all 5′ and 3′ ends
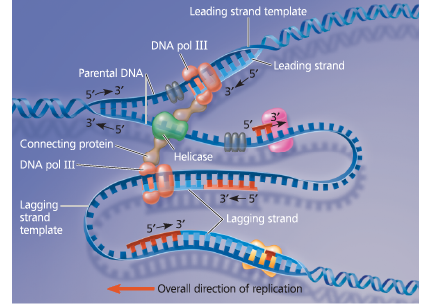
The “trombone” model of the DNA replication complex - Two DNA polymerase III molecules work together in a complex, one on each template strand. The lagging strand template DNA loops through the complex, resembling the slide of a trombone
Draw a line tracing the lagging strand template along the entire stretch of DNA shown here
BioFlix - DNA Replication
https://mediaplayer.pearsoncmg.com/assets/_SBpDyyv0FfgdmC2W6IoTLxWQp3MzETp
DNA polymerases ___ each nucleotide against its template as soon as it is added to the growing strand. Upon finding an incorrectly paired nucleotide, the polymerase _
- proofread; removes the nucleotide and then resumes synthesis
}}Mismatch Repair:}}
The cellular process that uses specific enzymes to remove and replace incorrectly paired nucleotides
What’s the importance of repair enzymes?
Researchers found that a hereditary defect in one of them is associated with a form of colon cancer allowing cancer-causing errors to accumulate in the DNA faster than normal
What are some potentially harmful chemical and physical agents?
Cigarette smoke and X-rays can cause damage to existing DNA after replication; DNA bases often undergo spontaneous chemical changes under normal cellular conditions
}}Mutations:}}
Permanent changes in DNA
}}Nuclease:}}
An enzyme that cuts DNA or RNA, either removing one or a few bases or hydrolyzing the DNA or RNA completely into its component nucleotides
}}Nucleotide Excision Repair:}}
A repair system that removes and then correctly replaces a damaged segment of DNA using the undamaged strand as a guide
Nucleotide excision repair of DNA damage
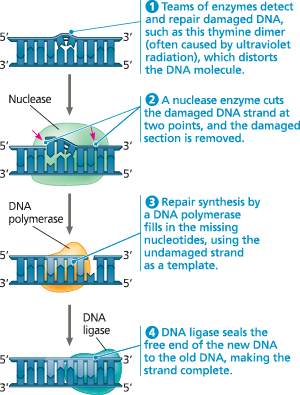
}}Thymine Dimers:}}
Adjacent thymine bases on a DNA strand become covalently linked causing DNA to buckle
___ are the source of the variation on which natural selection operates during evolution and are ultimately responsible for the appearance of new species
- Mutations
Telomeres - Eukaryotes have repetitive, noncoding sequences called telomeres at the ends of their DNA. Telomeres are stained orange in these mouse chromosomes (LM)
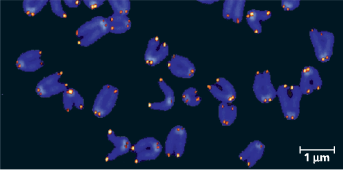
Describe the properties of telomere
Telomeric DNA acts as a buffer zone that protects the organism’s genes. Telomeres do not prevent the erosion of genes near ends of chromosomes; they merely postpone it.
}}Telomere:}}
The tandemly repetitive DNA at the end of a eukaryotic chromosome’s DNA molecule. Telomeres protect the organism’s genes from being eroded during successive rounds of replication (see also repetitive DNA)
An enzyme called ___ catalyzes the lengthening of telomeres in eukaryotic germ cells, thus restoring their original length and compensating for the shortening that occurs during DNA replication
- telomerase
Where is telomerase, and what does it do?
Telomerase is not active in most human somatic cells, but shows inappropriate activity in some cancer cells that may remove limits to a cell’s normal life span. Thus, telomerase is under study as a target for cancer therapies
What role does base pairing play in the replication of DNA?
Complementary base pairing ensures that the two daughter molecules are exact copies of the parental molecule. When the two strands of the parental molecule separate, each serves as a template on which nucleotides are arranged, by the base-pairing rules, into new complementary strands
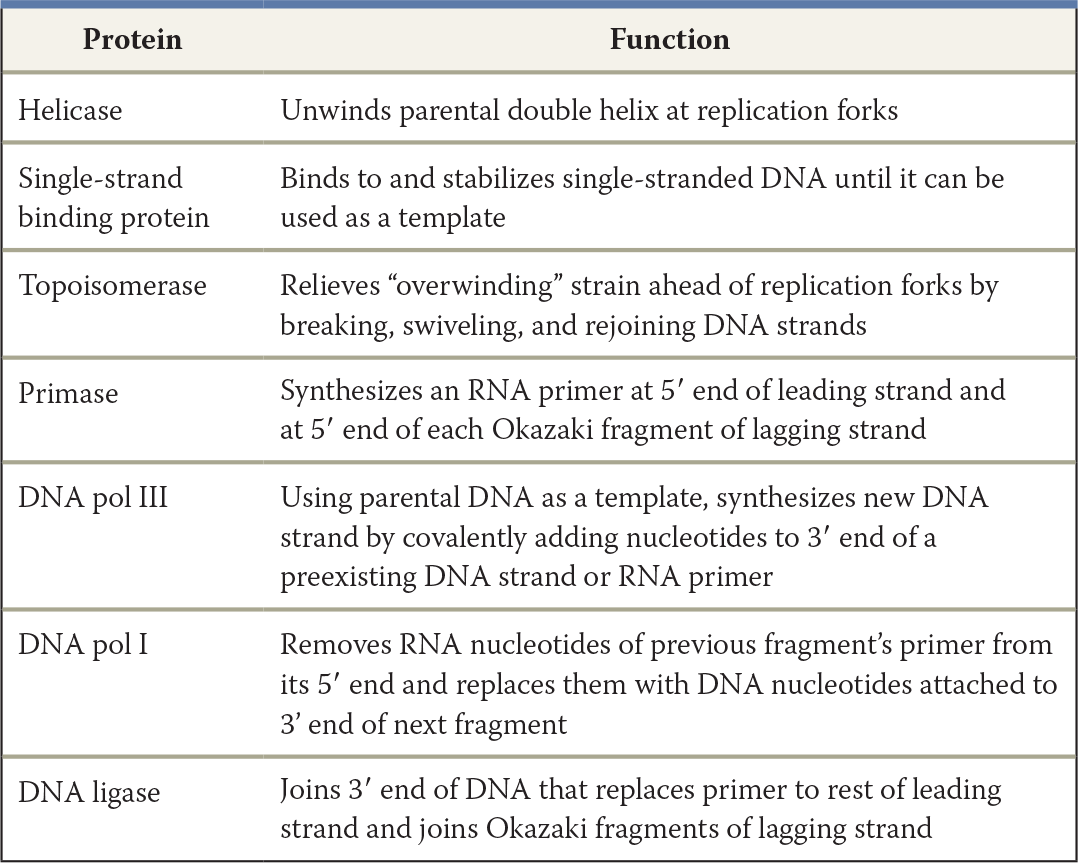
Make a table listing the functions of seven proteins involved in DNA replication in E. coli
What is the relationship between DNA replication and the S phase of the cell cycle?
In the cell cycle, DNA synthesis occurs during the S phase, between the G1 and G2 phases of interphase. DNA replication is therefore complete before the mitotic phase begins
Compare DNA replication on the leading and lagging strands
On both the leading and lagging strands, DNA polymerase adds onto the 3′ end of an RNA primer synthesized by primase, synthesizing DNA in the 5′ → 3′ direction. Because the parental strands are antiparallel, however, only on the leading strand does synthesis proceed continuously into the replication fork. The lagging strand is synthesized bit by bit in the direction away from the fork as a series of shorter Okazaki fragments, which are later joined together by DNA ligase. Each fragment is initiated by synthesis of an RNA primer by primase as soon as a given stretch of single-stranded template strand is opened up. Although both strands are synthesized at the same rate, synthesis of the lagging strand is delayed because initiation of each fragment begins only when sufficient template strand is available
^^13.3 - A chromosome consists of a DNA molecule packed together with proteins^^
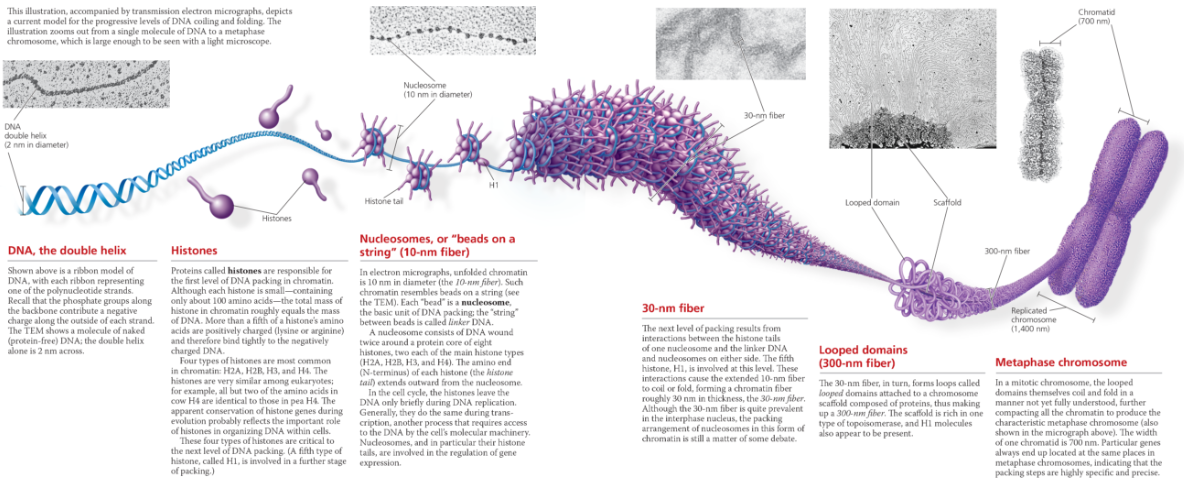
- The chromosome of most bacterial species is a circular DNA molecule with some associated proteins, making up the nucleoid. The chromatin making up a eukaryotic chromosome is composed of DNA, histones, and other proteins. The histones bind to each other and to the DNA to form nucleosomes, the most basic units of DNA packing. Additional coiling and folding lead ultimately to the highly condensed chromatin of the metaphase chromosome. In interphase cells, most chromatin is less compacted (euchromatin), but some remains highly condensed (heterochromatin). Euchromatin, but not heterochromatin, is generally accessible for transcription of genes
}}Nucleoid:}}
A non-membrane-enclosed region in a prokaryotic cell where its chromosome is located
}}Chromatin:}}
The complex of DNA and proteins that makes up eukaryotic chromosomes. When the cell is not dividing, chromatin exists in its dispersed form, as a mass of very long, thin fibers that are not visible with a light microscope
In interphase cells stained for light microscopy, the chromatin usually appears as a ___, suggesting that the chromatin is highly extended
- diffuse mass within the nucleus
As a cell prepares for mitosis, its chromatin ___, eventually forming a characteristic number of short, thick metaphase chromosomes that are distinguishable from each other with the light microscope
- coils and folds up (condenses)
Exploring Chromatin Packing in a Eukaryotic Chromosome
}}Heterochromatin:}}
Eukaryotic chromatin that remains highly compacted during interphase and is generally not transcribed
}}Euchromatin:}}
The less condensed form of eukaryotic chromatin that is available for transcription; “true chromatin“
The looser packing of euchromatin makes its DNA accessible to this machinery, so the genes present in euchromatin can be ___
- transcribed
Describe the structure of a nucleosome, the basic unit of DNA packing in eukaryotic cells
A nucleosome is made up of eight histone proteins, two each of four different types, around which DNA is wound. Linker DNA runs from one nucleosome to the next.
What two properties, one structural and one functional, distinguish heterochromatin from euchromatin?
Euchromatin is chromatin that becomes less compacted during interphase and is accessible to the cellular machinery responsible for gene activity. Heterochromatin, on the other hand, remains quite condensed during interphase and contains genes that are largely inaccessible to this machinery
^^13.4 - Understanding DNA structure and replication makes genetic engineering possible^^
Gene cloning (or DNA cloning) produces multiple copies of a gene (or DNA segment) that can be used to manipulate and analyze DNA and to produce useful new products or organisms with beneficial traits
In genetic engineering, bacterial restriction enzymes are used to cut DNA molecules within short, specific nucleotide sequences (restriction sites), yielding a set of double-stranded restriction fragments with single-stranded sticky ends
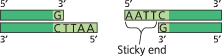
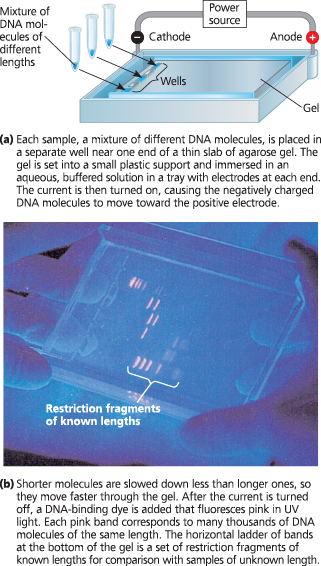
DNA fragments of different lengths can be separated and their lengths assessed by gel electrophoresis
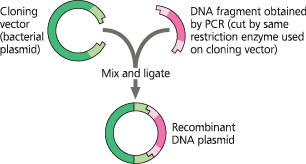
The sticky ends on restriction fragments from one DNA source—such as a bacterial plasmid or other cloning vector can base-pair with complementary sticky ends on fragments from other DNA molecules; sealing the base-paired fragments with DNA ligase produces recombinant DNA molecules
The polymerase chain reaction (PCR) can amplify (produce many copies of) a specific target segment of DNA for use as a DNA fragment for cloning. PCR uses primers that bracket the desired segment and requires a heat-resistant DNA polymerase
The rapid development of fast, inexpensive techniques for DNA sequencing is based on sequencing by synthesis: DNA polymerase is used to replicate a stretch of DNA from a single-stranded template, and the order in which nucleotides are added reveals the sequence
The CRISPR-Cas9 system allows researchers to edit genes in a specific, desired way. This may ultimately be used for treatment of genetic diseases
}}Nucleic Acid:}}
A polymer consisting of many nucleotide monomers; serves as a blueprint for proteins and, through the actions of proteins, for all cellular activities
What are the types of nucleic acid?
DNA and RNA
}}Nucleic Acid Hybridization:}}
The base pairing of one strand of a nucleic acid to a complementary sequence on another strand
}}Genetic Engineering:}}
The direct manipulation of genes for practical purposes
E. coli is a ___, and like many other bacteria, it has _
- large, circular molecule of DNA; plasmids
}}Plasmid:}}
A small, circular, double-stranded DNA molecule that carries accessory genes separate from those of a bacterial chromosome; in DNA cloning, can be used as a vector carrying up to about 10,000 base pairs (10kb) of DNA
}}Recombinant DNA Molecule:}}
A DNA molecule made in vitro with segments from different sources
To clone pieces of DNA using bacteria, researchers first obtain a ___ and inset _
- plasmid; DNA from another source
}}Gene Cloning:}}
The production of multiple copies of a gene
}}Cloning Vector:}}
In genetic engineering, a DNA molecule that can carry foreign DNA into a host cell and replicate there. Cloning vectors include plasmids
An overview of gene cloning and some uses of cloned genes - In this simplified diagram of gene cloning, we start with a plasmid (originally isolated from a bacterial cell) and a gene of interest from another organism. Only one plasmid and one copy of the gene of interest are shown at the top of the figure, but the starting materials would include many of each
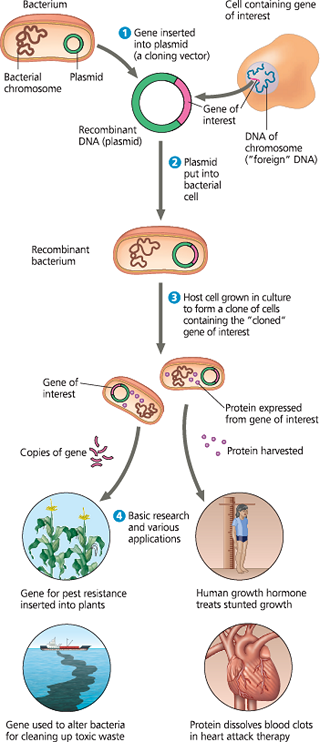
What are the two basic purposes of gene cloning?
To make many copies of, or amplify a particular gene and to produce a protein product from it
}}Genetically Modified Organisms:}}
Those in which DNA from the desired organism is inserted in the laboratory
}}Restriction Enzyme:}}
An endonuclease (type of enzyme) that recognized and cuts DNA molecules foreign to a bacterium (such as phage genomes). The enzyme cuts at specific nucleotide sequences (restriction sites)
Restriction enzymes ___ the bacterial cell by cutting up foreign DNA from other organisms or phages
- protect
}}Restriction Site:}}
A specific sequence on a DNA strand that is recognized and cut by a restriction enzyme
}}Restriction Fragment:}}
A DNA segment that results form the cutting of DNA by a restriction enzyme
Using a restriction enzyme and DNA ligase to make a recombinant DNA plasmid - The restriction enzyme in this example (called EcoRI) recognizes a single six-base-pair restriction site present in the plasmid. It makes staggered cuts in the sugar-phosphate backbones, producing fragments with “sticky ends.” Foreign DNA fragments with complementary sticky ends can base-pair with the plasmid ends; the ligated product is a recombinant plasmid. (If the two plasmid sticky ends base-pair, the original non-recombinant plasmid would re-form.)
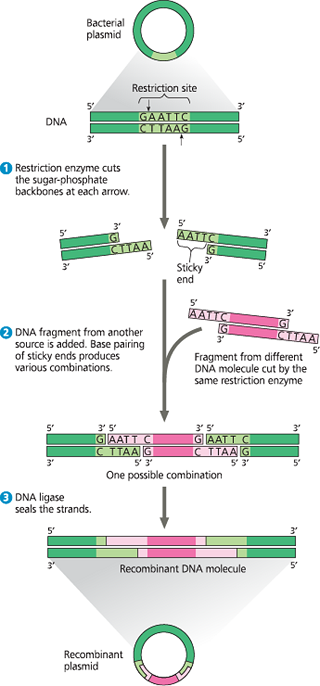
The restriction enzyme Hind III recognizes the sequence 5*′-AAGCTT-3′*, cutting between the two A’s. Draw the double-stranded sequence before and after the enzyme cuts
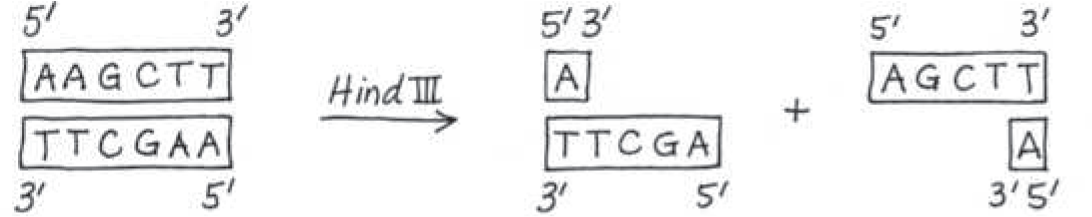
}}Sticky End:}}
A single-stranded end of a double-stranded restriction fragment
These short extensions can form ___ with _ on any other DNA molecules cut with the same enzyme
- hydrogen-bonded base pairs (hybridize); complementary sticky ends
The associations formed in this way are only temporary but can be made permanent by ___, which catalyzes the formation of covalent bonds that close up the sugar-phosphate backbones of DNA strands
- DNA ligase
}}Gel Electrophoresis:}}
A technique for separating nucleic acids or proteins on the basis of their size and electrical charge, both of which affect their rate of movement through an electric field in a gel made of agarose or another polymer
Gel electrophoresis - A gel made of a polymer acts as a molecular sieve to separate nucleic acids or proteins differing in size, electrical charge, or other physical properties as they move in an electric field. In the example shown here, DNA molecules are separated by length in a gel made of a polysaccharide called agarose
}}Polymerase Chain Reaction (PCR):}}
A technique for amplifying DNA in vitro by incubating it with specific primers, a heat-resistant DNA polymerase, and nucleotides
The PCR - Application
With PCR, any specific segment—the so-called target sequence—in a DNA sample can be copied many times (amplified) within a test tube
Technique - PCR requires double-stranded DNA containing the target sequence, a heat-resistant DNA polymerase, all four nucleotides, and two 15- to 20-nucleotide DNA strands that serve as primers. One primer is complementary to one end of the target sequence on one strand; the second primer is complementary to the other end of the sequence on the other strand
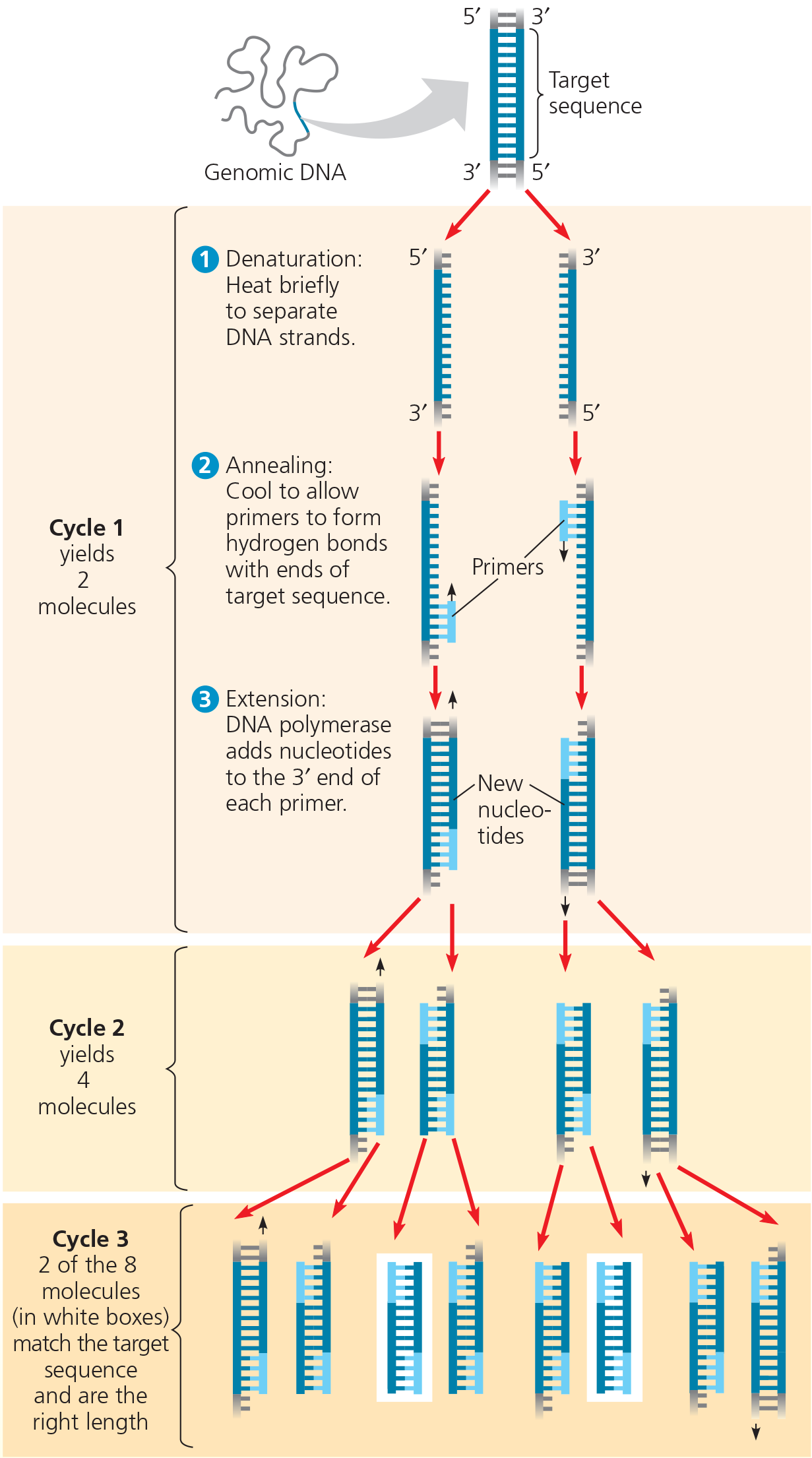
The PCR - Results
After three cycles, two molecules match the target sequence exactly. After 30 more cycles, over 1 billion (109) molecules match the target sequence
Describe PCR
It is speed and very specfic
Describe the primers
They have high specificity; the sequences are chosen so they hybridize only with complementary sequences at opposite ends of the target segment
Why can’t PCR substitute for gene cloning, and what’s its role instead?
Occasional errors during PCR replication limit the number of good copies and the length of DNA fragments that can be copied. Instead, PCR is used to provide the specific DNA fragment for cloning. PCR primers are synthesized to include a restriction site at each end of the DNA fragment that matches the site in the cloning vector, and the fragment and vector are cut and ligated together. The resulting plasmids are sequenced so that those with error-free inserts can be selected
Use of a restriction enzyme and PCR in gene cloning - PCR is used to produce the DNA fragment or gene of interest that will be ligated into a cloning vector, in this case a bacterial plasmid. Both the plasmid and the DNA fragments are cut with the same restriction enzyme, combined so the sticky ends can hybridize, and ligated together. The resulting plasmids will then be introduced into bacterial cells
}}DNA Sequencing:}}
Determining the order of nucleotide bases in a gene or DNA fragment
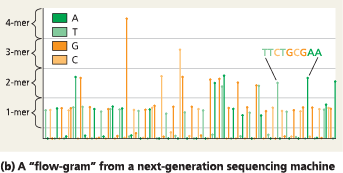
Next-generation sequencing - Next-generation sequencing machines use “sequencing by synthesis” to sequence many 300-nucleotide fragments in parallel. In this way, one machine can sequence about 2 billion nucleotides in 24 hours. (b) The results for one fragment are displayed as a “flow-gram,” where the nucleotides are identified by color, one by one. The sequences of the entire set of fragments are analyzed using computer software, which “stitches” them together into a whole sequence—often, an entire genome

An example of a third-generation sequencing technique - In this approach, a single strand of an uncut DNA molecule would be passed, nucleotide by nucleotide, through a nanopore in a membrane. Here, the pore is a protein channel embedded in a lipid membrane. The nucleotides are identified by slight differences in the amount of time they interrupt an electrical current across the opening
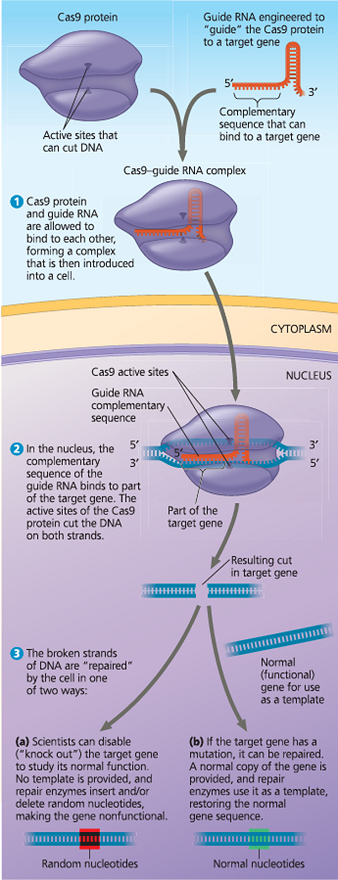
}}CRISPR-Cas9:}}
A technique for gene editing
}}Cas9:}}
A bacterial protein that helps defend bacteria against bacteriophage infection; a nuclease that cuts double-stranded DNA molecules
Gene editing using the CRISPR-Cas9 system
The restriction site for an enzyme called PvuI is the following sequence:
5′-C G A T C G-3′
3′-G C T A G C-5′
Staggered cuts are made between the T and C on each strand. What type of bonds are being cleaved?
The covalent sugar-phosphate bonds of the DNA strands
One strand of a DNA molecule has the following sequence: 5′-CCTTGACGATCGTTACCG-3′. Draw the other strand. Will PvuI (see question 1) cut this molecule? If so, draw the products
Yes, PvuI will cut the molecule (at the position indicated by the dashed red line) 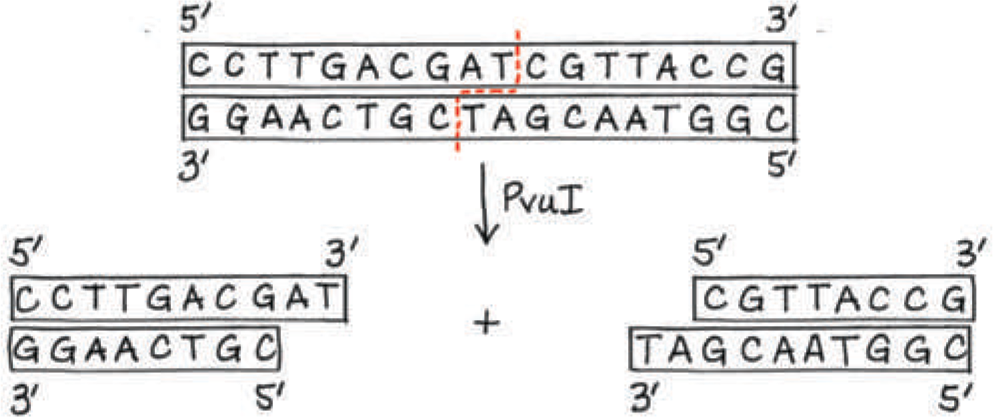
Describe the role of complementary base pairing during cloning, PCR, DNA sequencing, and gene editing using the CRISPR-Cas9 system
Cloning requires joining two pieces of DNA—a cloning vector, such as a bacterial plasmid, and a gene or DNA fragment from another source. Both pieces must be cut with the same restriction enzyme, creating sticky ends that will base-pair with complementary ends on other fragments. (The sugar-phosphate backbones will then be ligated together by ligase.) In PCR, the primers must base-pair with their target sequences in the DNA mixture, bracketing one specific region among many, and complementary base pairing is the basis for the building of the new strand during the extension step. In DNA sequencing, primers base-pair to the template, allowing DNA synthesis to start, and then nucleotides are added to the growing strand based on complementarity of base pairing. Using the CRISPR-Cas9 system to edit genes involves an RNA used as a guide that is complementary to the sequence to be edited. Complementary base pairing anchors the Cas9 protein to the target DNA sequence it will then cut
Describe how the process of gene cloning results in a cell clone containing a recombinant plasmid
A plasmid vector and a source of foreign DNA to be cloned are both cut with the same restriction enzyme, generating restriction fragments with sticky ends. These fragments are mixed together, ligated, and reintroduced into bacterial cells, which can then make many copies of the foreign DNA or its product