
Biochemical Approaches Underpinning Drug Discovery
8: Assay cascade
IKKs key kinases in NFkB pathway, alpha believed to be involved in cancer, beta involved in normal cell survival so needs to be avoided - values shown are cutoffs
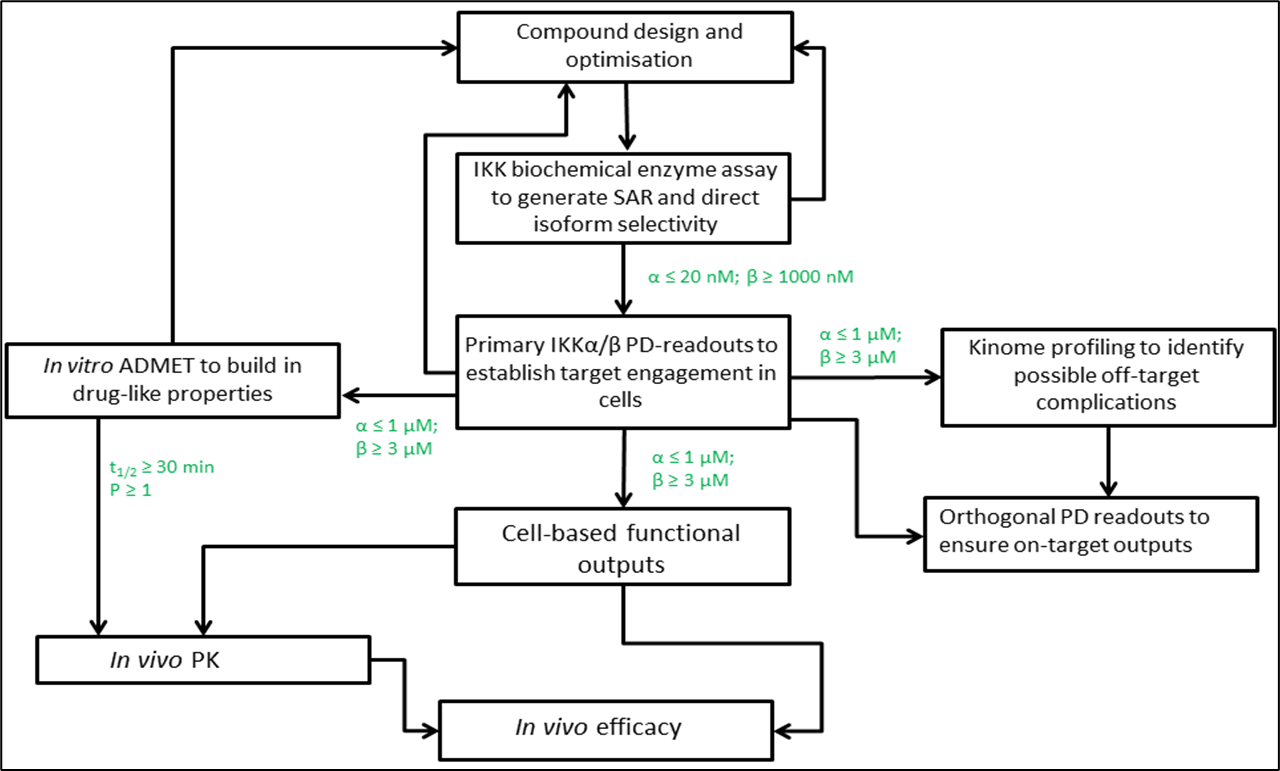
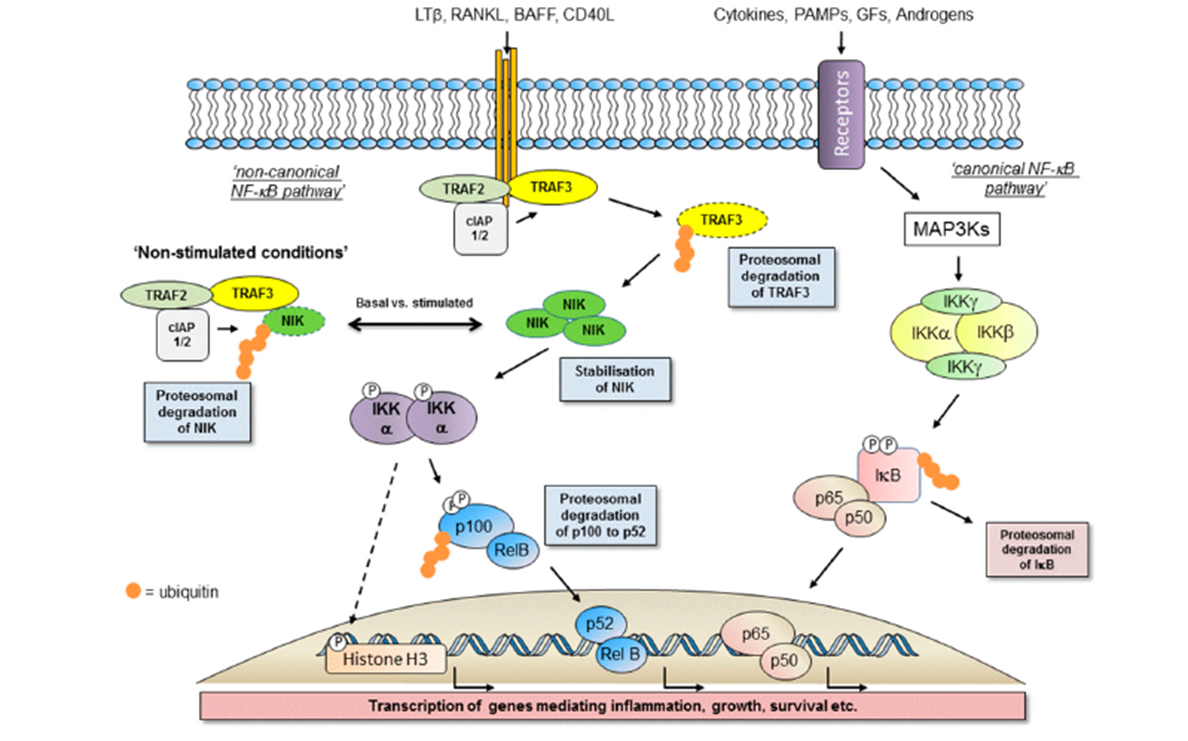
10: IKK alpha target pathway, used to generate SAR and direct isoform selectivity
12: Primary assays are used to test compounds to the identified target, to screen those which could potentially be used as a drug.
Screening assay - isolated enzyme, in test tube. Can be generated for any sort of protein you’re interested eg kinases, phosphatases and x-ases
Primary assays need rapid throughput and measurable output eg through radiolabelled phosphate incorporation, immuno-based antibody detection of modification of substrate or fluorescence (FRET, TR-FRET, FP)
13: Primary kinase screening assays - different formats used to measure direct target in isolated environment
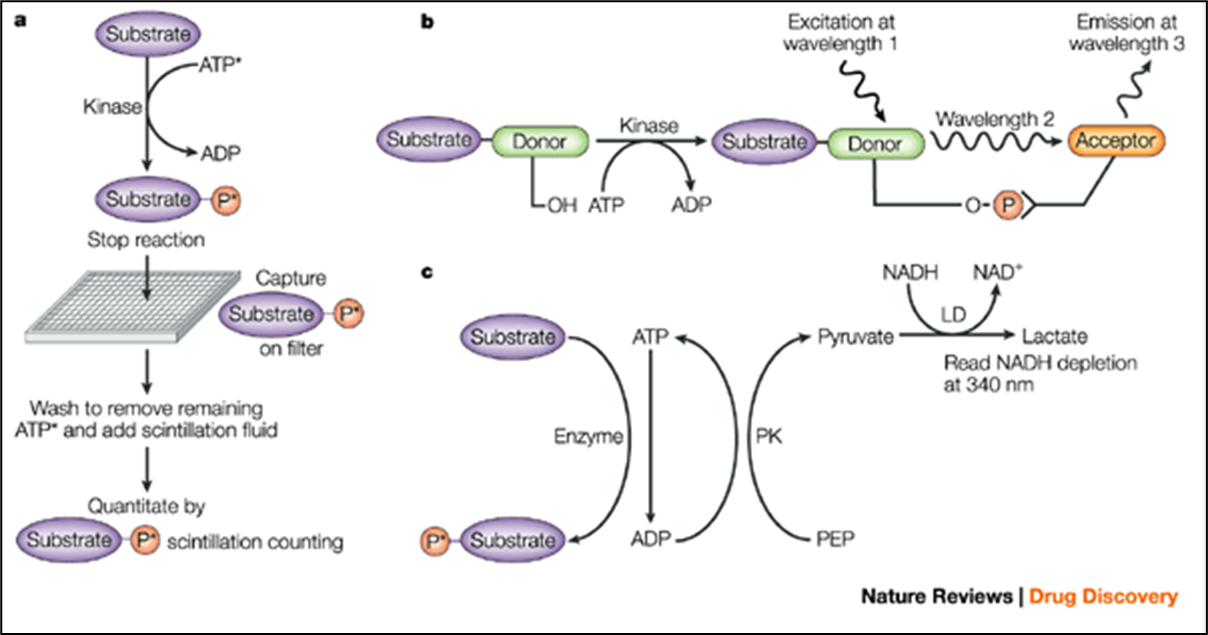
a) radiolabelled ATP transfers a labelled phosphate from ATP to substrate
b) FRET - homogenous assay. Labelled substrate is phosphorylated allowing it to bind a second molecule labelled with an acceptor group
c) conversion of ATP to ADP coupled to conversion of NADH to NAD using two enzymes
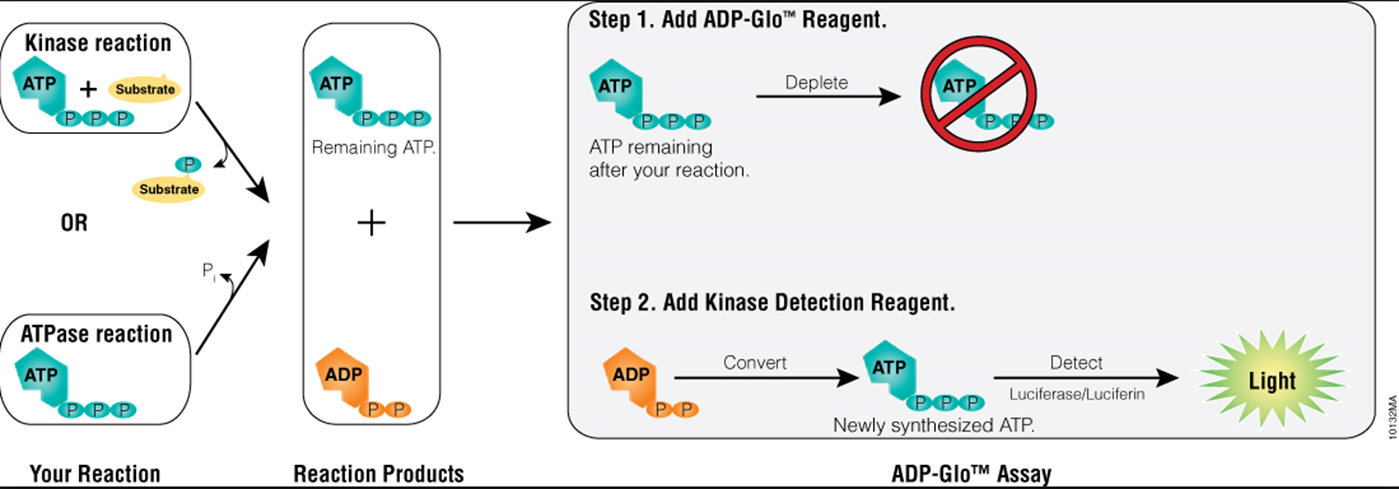
14: Kinase screening assay - following loss of ATP
15: If cutoffs are met, go onto testing in cells rather than isolated enzyme assays. Primary IKKalpha/PD readouts used to establish target engagement in cells
16: Primary assays in cells:
Involves primary PD (?) markers that demonstrate target engagements in a cellular setting. Used to see if compound SAR can be translated from test tube to cellular setting. For a kinase a phosphoantibody based readout may be used - fluorescent in cell western or western blots
17: observed results in primary cell assays
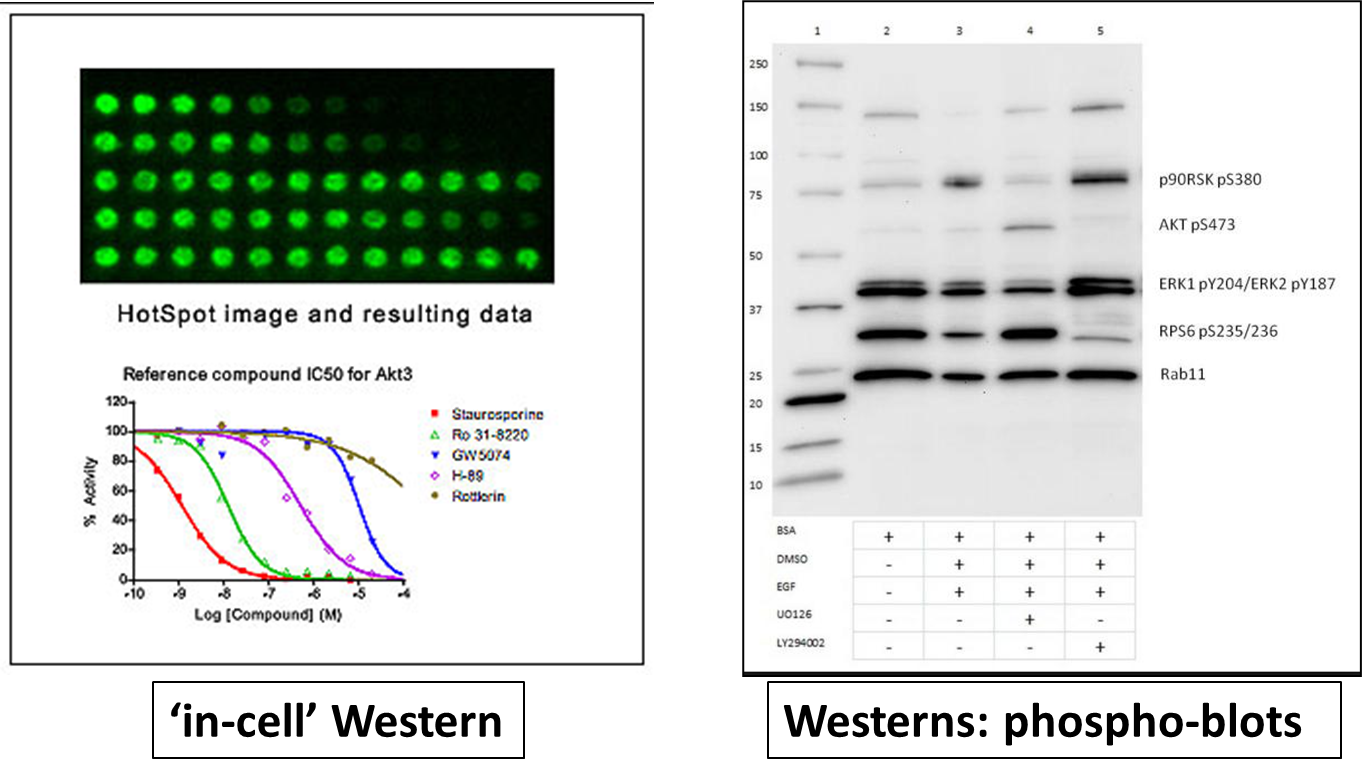
19: Orthogonal PD readouts (see photos for definition) used to ensure on target outputs
20: Molecular biologists often use components/pathways in bacteria and other organisms for their techniques eg:
plasmids to propagate DNA
restriction enzymes for cutting DNA
Cre-LoxP system for genome manipulation
Taq polymerase for DNA amplification
GFP and firefly luciferase to visualise experiments
22: Secondary/orthogonal assays:
High throughput reporter assays of a downstream cellular response are coupled to luciferase, alkaline phosphatase secretion, GFP inductions etc
23:
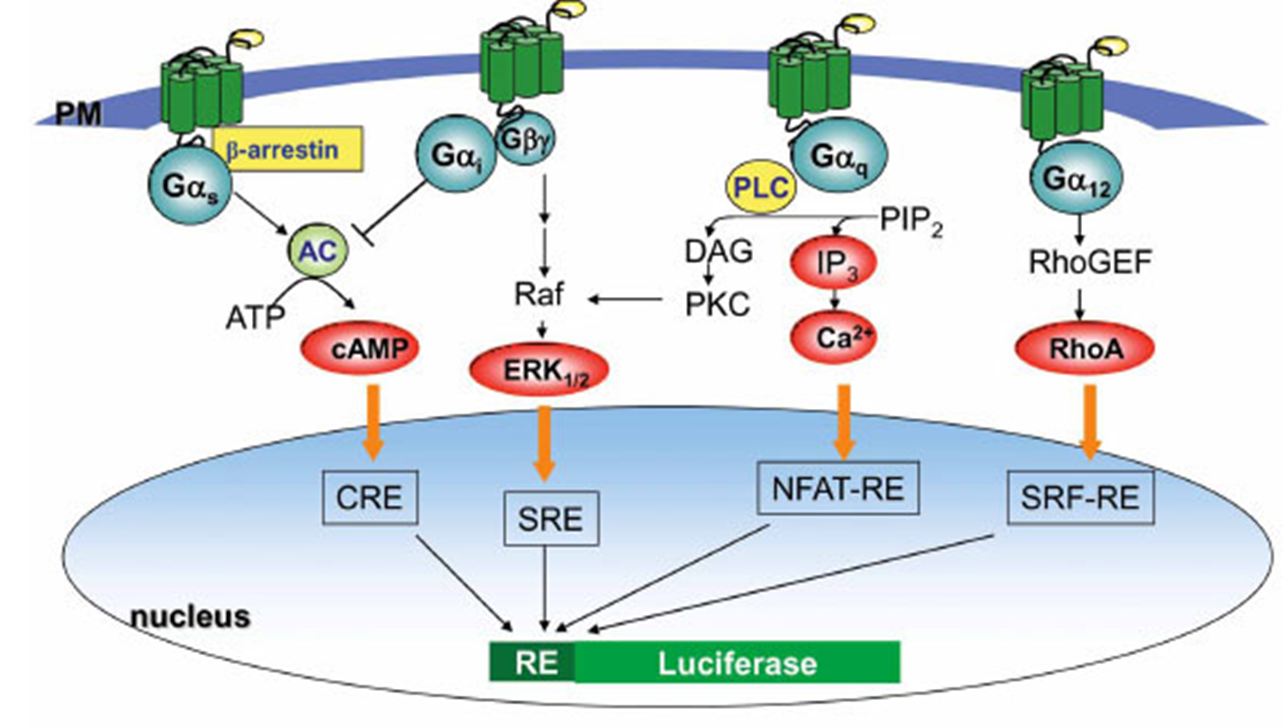
24: Luciferase reporter assays:
regulatory region of target/marker gene cloned upstream of luciferase
vector DNA introduced into cells
transcription
break up cells
add luciferin
light produced in cells containing target gene
25: Orthogonal markers are linked markers regulated specifically by the target protein. Must be able to distinguish inhibition of one isoform over another eg IKK lpha vs IKK beta. An example of an orthogonal marker in this situation is CXCL12 (Plevs lab) which is induced by IKKalpha activity and inhibited by IKKbeta activity therefore the CSCL12 promoter can be fused to a .luc-reporter
26: Phenotypic assays are based on cell based functional outputs
28: Phenotypic outputs are 96 well plate based (or higher eg in automated throughput approach)
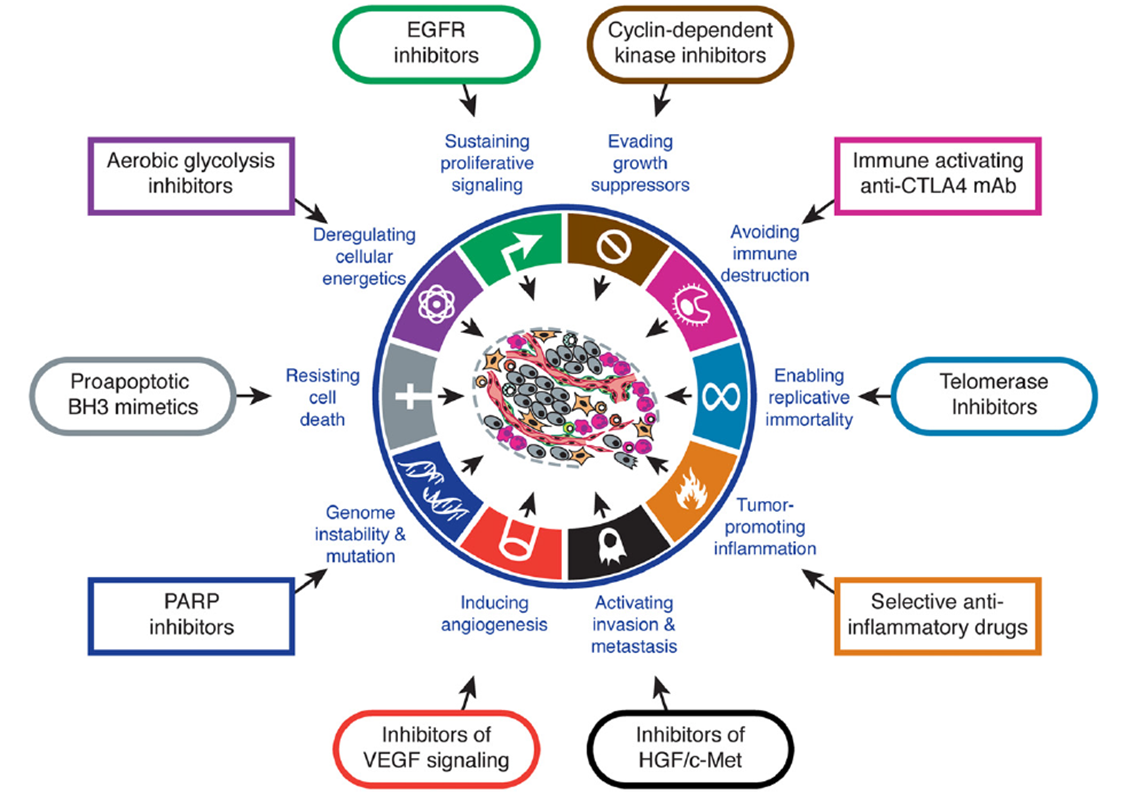
eg for potential cancer drugs different phenotypic assays could be used for each cancer hallmark. Don’t measure protein directly but measure effects tied to target proteins
hallmark | phenotypic assay |
resisting cell death | apoptotic assay |
sustaining proliferative signalling | growth/viability as. say |
activating invasion and metastasis assays | migration/invasion assays |
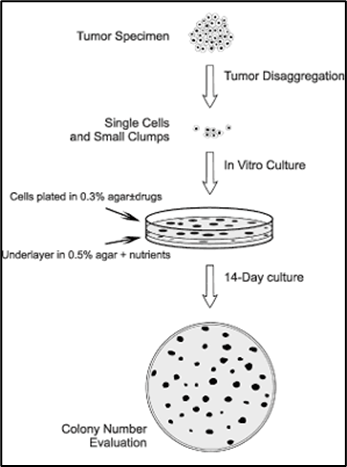
enabling replicative immortality | clonogenic assay |
Other cancer hallmarks are evading growth suppressors and inducing angiogenesis - is there an assay that could be used for these?
29: Apoptotic assay:
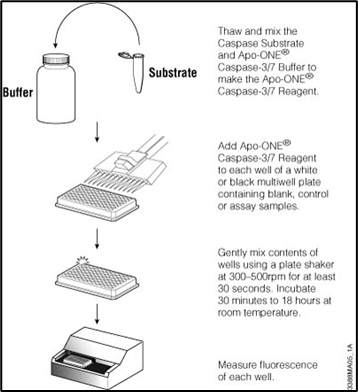
Clonogenic assay:
MTT:
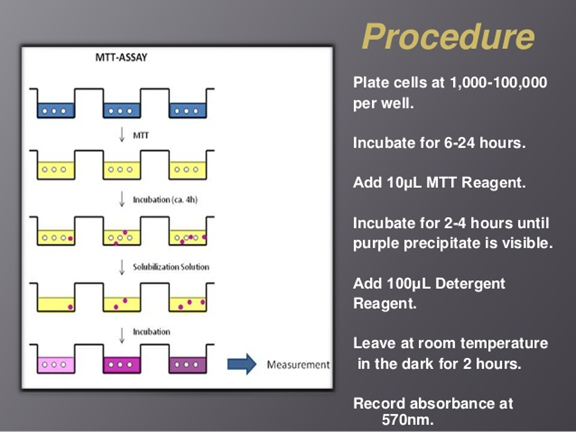
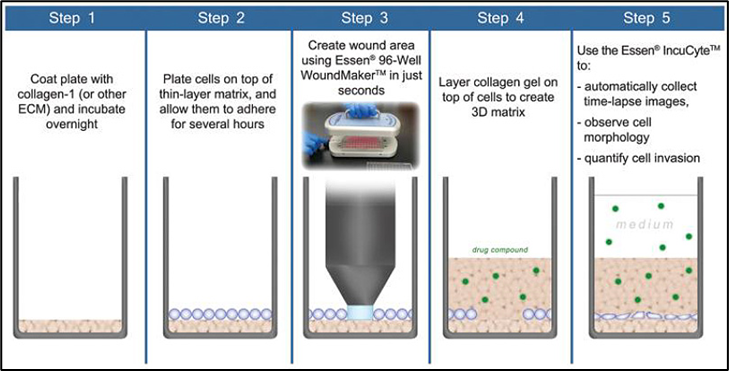
Migration assay:
30: Success in target based approach -therapeutic targeting of hallmarks -
36: Lipinski’s rule of 5 (comes from the fact that each cutoff is a multiple of 5):
Lipinski and coworkers at Pfizer suggested for oral absorption and general cell permeability, a drug needs:
<5 hydrogen bond donors (estimated by counting no. of OH and NH groups in molecule)
fewer than 10 H bond acceptors (estimated by total no. of N and O atoms in molecule)
molecular weight <500
partitioning coefficient (logP) <5
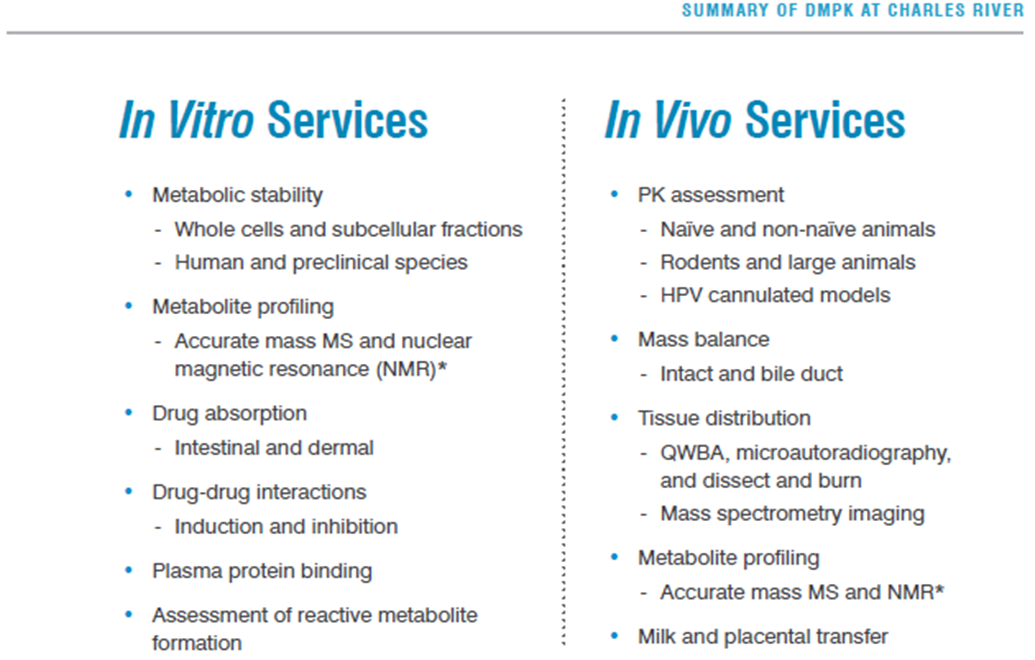
37: ADMET assays can be outsourced to recognised contract research organisations (CROs) eg Charles River Laboratories (Edi based), Cyprotex etc. Carry out a number of services for when academics doesn’t necessarily have the resources
38: not every molecule put through to an in vivo setting, only committed to in vivo testing when drug hitting all characteristics needed so far
42: In vivo models: IVIS. Cells labelled with luciferase enzymes and animals flushed with luciferin output. Greater cancer mass = greater output. Red = most intense output where cancer growing most rapidly
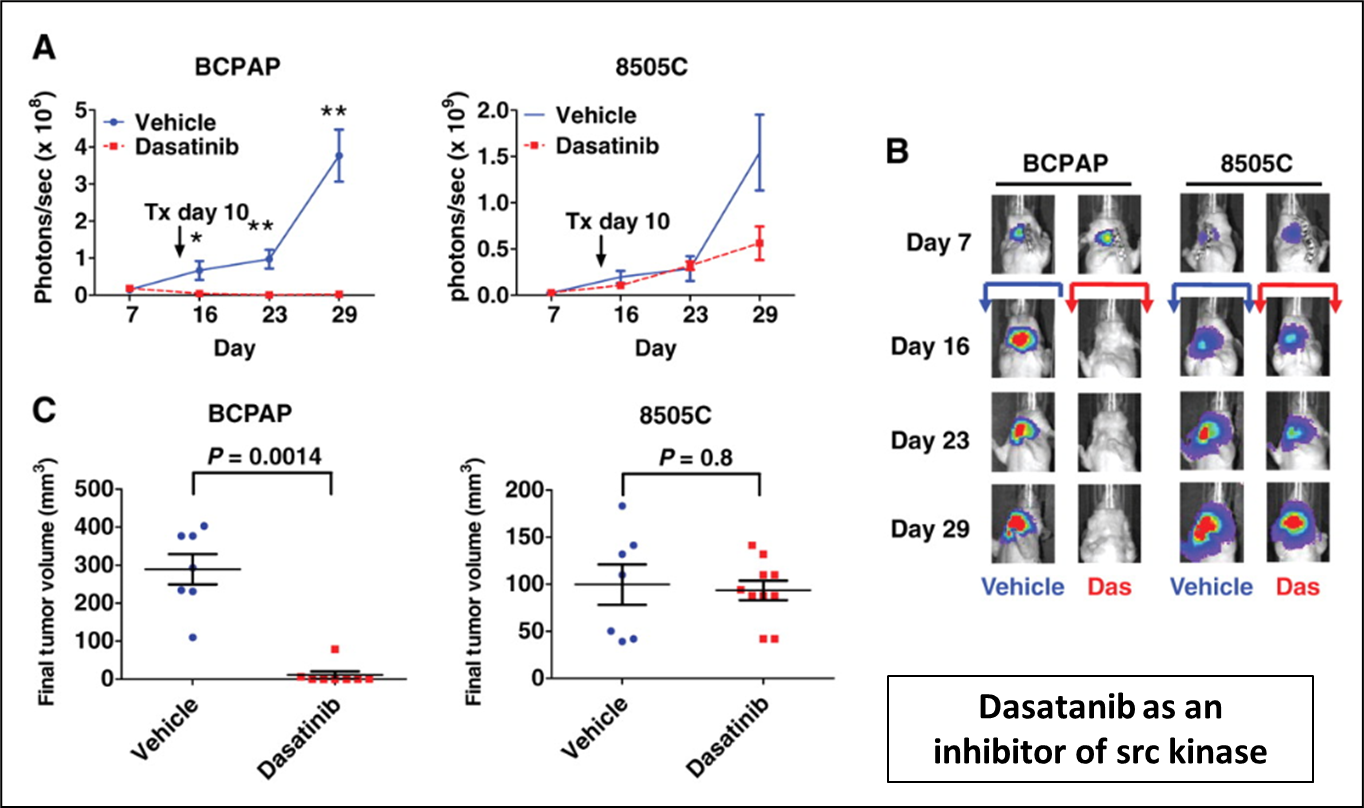
43: In vivo models are used to detect off target activities eg kinase profiling: the kinome consists of 518 kinases vs test tube assay
Kinase profiling can be v expensive depending on how many kinases used
45: test tube assay and in vivo model assays used in parallel to test for potential toxicity. In vivo used to find maximal tolerated dose (ethically should only be doing this type of experiment once).
If no off target activity you have a pre-clinical candidate
Extra reading:
hallmarks of cancer: http://www.cell.com/cell/pdf/S0092-8674(11)00127-9.pdf
Biochemical Approaches Underpinning Drug Discovery
8: Assay cascade
IKKs key kinases in NFkB pathway, alpha believed to be involved in cancer, beta involved in normal cell survival so needs to be avoided - values shown are cutoffs

10: IKK alpha target pathway, used to generate SAR and direct isoform selectivity

12: Primary assays are used to test compounds to the identified target, to screen those which could potentially be used as a drug.
Screening assay - isolated enzyme, in test tube. Can be generated for any sort of protein you’re interested eg kinases, phosphatases and x-ases
Primary assays need rapid throughput and measurable output eg through radiolabelled phosphate incorporation, immuno-based antibody detection of modification of substrate or fluorescence (FRET, TR-FRET, FP)
13: Primary kinase screening assays - different formats used to measure direct target in isolated environment

a) radiolabelled ATP transfers a labelled phosphate from ATP to substrate
b) FRET - homogenous assay. Labelled substrate is phosphorylated allowing it to bind a second molecule labelled with an acceptor group
c) conversion of ATP to ADP coupled to conversion of NADH to NAD using two enzymes
14: Kinase screening assay - following loss of ATP

15: If cutoffs are met, go onto testing in cells rather than isolated enzyme assays. Primary IKKalpha/PD readouts used to establish target engagement in cells
16: Primary assays in cells:
Involves primary PD (?) markers that demonstrate target engagements in a cellular setting. Used to see if compound SAR can be translated from test tube to cellular setting. For a kinase a phosphoantibody based readout may be used - fluorescent in cell western or western blots
17: observed results in primary cell assays

19: Orthogonal PD readouts (see photos for definition) used to ensure on target outputs
20: Molecular biologists often use components/pathways in bacteria and other organisms for their techniques eg:
plasmids to propagate DNA
restriction enzymes for cutting DNA
Cre-LoxP system for genome manipulation
Taq polymerase for DNA amplification
GFP and firefly luciferase to visualise experiments
22: Secondary/orthogonal assays:
High throughput reporter assays of a downstream cellular response are coupled to luciferase, alkaline phosphatase secretion, GFP inductions etc
23:

24: Luciferase reporter assays:
regulatory region of target/marker gene cloned upstream of luciferase
vector DNA introduced into cells
transcription
break up cells
add luciferin
light produced in cells containing target gene
25: Orthogonal markers are linked markers regulated specifically by the target protein. Must be able to distinguish inhibition of one isoform over another eg IKK lpha vs IKK beta. An example of an orthogonal marker in this situation is CXCL12 (Plevs lab) which is induced by IKKalpha activity and inhibited by IKKbeta activity therefore the CSCL12 promoter can be fused to a .luc-reporter
26: Phenotypic assays are based on cell based functional outputs
28: Phenotypic outputs are 96 well plate based (or higher eg in automated throughput approach)
eg for potential cancer drugs different phenotypic assays could be used for each cancer hallmark. Don’t measure protein directly but measure effects tied to target proteins
hallmark | phenotypic assay |
resisting cell death | apoptotic assay |
sustaining proliferative signalling | growth/viability as. say |
activating invasion and metastasis assays | migration/invasion assays |
enabling replicative immortality | clonogenic assay |
Other cancer hallmarks are evading growth suppressors and inducing angiogenesis - is there an assay that could be used for these?
29: Apoptotic assay:

Clonogenic assay:

MTT:

Migration assay:

30: Success in target based approach -therapeutic targeting of hallmarks -

36: Lipinski’s rule of 5 (comes from the fact that each cutoff is a multiple of 5):
Lipinski and coworkers at Pfizer suggested for oral absorption and general cell permeability, a drug needs:
<5 hydrogen bond donors (estimated by counting no. of OH and NH groups in molecule)
fewer than 10 H bond acceptors (estimated by total no. of N and O atoms in molecule)
molecular weight <500
partitioning coefficient (logP) <5
37: ADMET assays can be outsourced to recognised contract research organisations (CROs) eg Charles River Laboratories (Edi based), Cyprotex etc. Carry out a number of services for when academics doesn’t necessarily have the resources

38: not every molecule put through to an in vivo setting, only committed to in vivo testing when drug hitting all characteristics needed so far
42: In vivo models: IVIS. Cells labelled with luciferase enzymes and animals flushed with luciferin output. Greater cancer mass = greater output. Red = most intense output where cancer growing most rapidly

43: In vivo models are used to detect off target activities eg kinase profiling: the kinome consists of 518 kinases vs test tube assay
Kinase profiling can be v expensive depending on how many kinases used
45: test tube assay and in vivo model assays used in parallel to test for potential toxicity. In vivo used to find maximal tolerated dose (ethically should only be doing this type of experiment once).
If no off target activity you have a pre-clinical candidate
Extra reading:
hallmarks of cancer: http://www.cell.com/cell/pdf/S0092-8674(11)00127-9.pdf
 Knowt
Knowt