AP Bio: Cell Energetics (unit 3)
ENZYMES
ENZYME PROPERTIES
proteins; biological catalysts; reusable
essential (without them, biological reactions would progress too slowly to sustain life)
enzymes help overcome low concentration of substrates by holding onto them and giving them more time to react (more of a chance than spontaneous collisions)
lower activation energy of reactions
highly specific (each one acts on a specific narrow range of chemical reactions)
induced fit hypothesis:
explains how enzymes catalyze similar chemical reactions. the active site changes shape to conform to each substrate, forming an enzyme-substrate complex.
is like a handshake
disproves the previous “lock and key” hypothesis that enzymes and substrates are in a 1-1 specific relationshi
appearance:
3º or 4º structures
active site
substrate binds here
cofactor “site”
cofactors: molecules that enhance enzyme activity; like vitamins
allosteric sites
binding sites for regulatory molecules; switches enzymes on/off to regulate its activity
“ase”
kinases → enzymes that phosphorylate substrates (and more)
factors affecting enzyme activity:
general protein-related factors:
pH
temperature
freezing: enzyme activity slows down a lot, but low temp. does not denature the protein. enzyme activity resumes when temperatures heat up again
heat: excessive heat denatures the enzyme, as the bonds break and the amino acids unravel, changing the active site
salinity
the presence of extra ions change the attraction of ionic bonds holding tertiary structures together
inhibitors
competitive
compete with the substrate for the active site
block the substrate from accessing the active site
reaction rate slows
reduces maximum reaction rate
non-competitive
binds to an allosteric site away from the active site
causes the active site to change shape, so that the substrate no longer fits and can’t react
permanent/temporary binding
shuts off the enzyme
maximum reaction rate can be reduced to zero
concentration of enzyme/substrate
^ enzyme: faster reactions (more processors/locations that substrates can react)
^ substrate: longer overall reaction → more things that are needed to be processes
enzyme kinetics:
rate = substrate consumed/time OR products/time
an enzyme has a maximum rate and it can’t go any faster than that
ENZYME REGULATION
feedback inhibition
the end product plays a role in regulating the first enzyme
in some pathways, the end product may turn off the first enzyme by acting as a competitive inhibitor
ATP → ADP + Pi → AMP + Pi
ATP is an inhibitor
too much ATP? don’t make more! ATP bind to the enzyme and blocks the process
AMP is an activator
AMP rises after ATP is burned—it activates enzymes to make more ATP after the ATP is used up
allosteric regulation
inhibit and activate enzyme activity
occurs in enzymes with 4º structures in the metabolic pathway
enables finer control of the on/off mechanism
milliseconds!! of on/off to conserve energy
cooperativity
makes it easier for a substrate to bind to an active site
once the first substrate binds, the following substrates have an easier time binding to their active sites
spontaneous binding to the active site
only some enzymes do this
most common in 4º structures with more than 1 active site
but…can happen in tertiary structures (but it must have more than one active site)
EX: hemoglobin
4 subunits (2 alpha chains and 2 ß chains)
binding of one O2 happens spontaneously, which causes favorable changes in the other subunits
now, the other O2 molecules have an easier time binding to their active sites
INTRO TO METABOLISM
metabolism: a metabolic pathway involving enzyme-catalyzed steps
catabolism: breaking down substances
anabolism: synthesizing substances
OIL RIG: oxidation is loss, reduction is gain
redox reactions:
reduction-oxidation reactions transfer energy by adding/removing electrons.
C6H12O6 + O2 → CO2 + H2O
glucose: oxidation (glucose loses electrons and H)
O2: reduction (CO2 gains electrons and H)
biological redox reactions: electrons “fall” from molecules containing lots of hydrogen
oxygen is reduced—gain e- and H+—becoming H2O
glucose carbons are oxidized—they lose H and e-
cellular respiration
a process that generates ATP by metabolizing food using O2
turns one glucose molecule → ~36 ATP
equation: C6H12O6 + 6O2 + 6H2O → 6CO2 + 12H2O + energy (ATP)
glucose vs ATP: ATP is important because glucose releases too much energy per molecule. Too much energy all at once causes cellular overheating and results in the loss of excess heat and energy
C-H bonds in glucose hold most of the chemical energy
cellular respiration: glucose oxidizes into CO2
H+ and electrons are carried away by NAD+ and FAD
energy is transferred and held onto by ATP
ATP (adenosine triphosphate)
transfers energy released from exergonic reactions to endergonic reactions
holds onto potential energy
renewable and recyclable
hydrolysis of ATP produces energy
7.3 kcal per mole of ATP
NAD+/NADH
metabolic energy carrier
accepts one H+ and 2e- from C-H bond → transfer that energy during ETC
then transfers it to ADP, then ATP
common oxidizing agent; oxidizing agents get reduced
NAD+ is an “electron bus”
NAD+ + 2e- + H+ → NADH
FAD/FADH2
metabolic energy carrier
accepts 2H+ and 2e- from C-H bond → transfer that energy during ETC
then transfers it to ADP, then ATP
common oxidizing agent; oxidizing agents get reduced
FAD + 2e- + 2H+ → FADH2
GLYCOLYSIS
glucose → 2 pyruvic acid
in: 6-carbon glucose, 2 ADP+2P, 2NAD+
out: 2 3-carbon pyruvic acids, 2ATP, 2NADH & 2H+
location: cytoplasm
no organelles are needed
math:
“energy investment”: 2 ATPs used in 1st steps
“energy payout”: 2 ATPs generated in last steps
net production of 2 ATP
ADPs get re-phosphorylated
glucose → 2 pyruvate + 2 ATP + 2 NADH
history:
no O2 for billions of years…so glycolysis evolved
inputs (glucose/simple sugars) matched to early earth’s conditions
most widespread metabolic pathway today (almost everything undergoes this)
FERMENTATION
the step that follows glycolysis—used to clear up NADH that builds up from glycolysis
cells need a way to regenerate NAD+ (after it’s used up by glycolysis)
otherwise, glycolysis cannot continue and no more ATP can be made
fermentation: a way to regenerate NAD+ from NADH; it strips the electrons from NADH
DOES NOT PRODUCED ATP
alcohol (ethanol) fermentation:
common in fungi, yeast, and some bacteria
used to make bread, wine, beer, hard cider, old soft drinks
equation:
2 pyruvate (3C) → 2 acetaldehyde (2C) + 2 CO2 (1C)
2 acetaldehyde (2C) + 2 NADH → 2 ethanol (2C) + 2 NAD+
input: 2 pyruvate
output: 2 CO2, 2 ethanol, 2 NAD+
NAD+ is the end product goal; the thing that we mainly want from this reaction
diagram:

lactic acid fermentation:
common in bacteria, animals
used to make cheese, vinegar, sauerkraut, kimchi, pickles, sour cream, yogurt, etc.
acidity curdles milk and crates a sour taste. the high acid content limits enzymes in the pathway.
strenuous exercise → anaerobic muscle cell response
lactic acid builds up in muscle cells, causing fatigue (and then pain)
lactic acid is eventually moved from muscle cells → bloodstream → liver
liver cells connect lactic acid back to pyruvate, and then breaks it down aerobically
equation:
2 pyruvate (3C) + 2 NADH → 2 lactate (3C) + 2 NAD+
no intermediate reaction; pyruvate accepts electrons from NADH
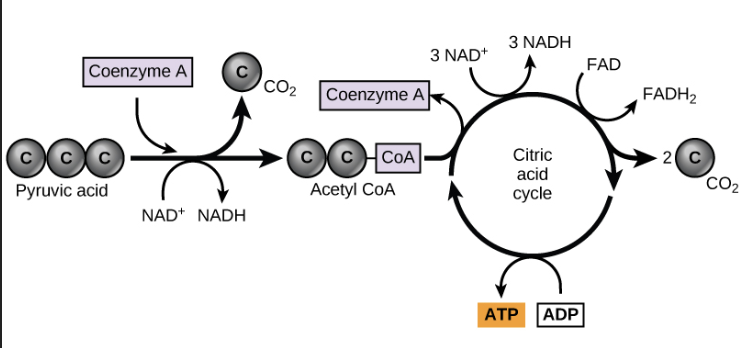
KREBS TRANSITION AND KREBS CYCLE
krebs transition:
moves 2 pyruvate made from glycolysis in the cytoplasm into the matrix
enters 2 acetyl-coA into the Krebs cycle
equation: 2 pyruvate → 2 Acetyl-coA + 2CO2 + 2NADH
krebs cycle:
extracts energy from Acetyl-coA
carbons are given off as CO2
makes ATP
energy stored in NAD+ and FA
it takes 2 krebs cycles to metabolize one glucose molecule (2 pyruvate)
yield per cycle: 3 NADH, 1 FADH, 1 ATP
total yield: 6 NADH, 2 FADH, 2 ATP
steps:
acetyl-coA binds to oxaloacetate → citrate
7 more steps to remove 2 carbons from acetyl-coA and generate oxaloacetate
byproducts: 6NADH, 2FADH2, 4CO2, 2ATP
diagram:
ELECTRON TRANSPORT CHAIN (ETC)
electron transport chain: a series of protein complexes and other molecules that transfer electrons and create an electrochemical gradient
after krebs cycle:
__NADH (2e, one H+ each) and __FADH2 (2e, 2H+ each) carry over their cargo to the ETC
this remaining energy in NADH and FADH2 is used to convert ADP → ATP
yield: 3 ATP per NADH, and 2 ATP per FADH2
FADH yields less because it is more electronegative and holds onto its electrons longer during the ETC
process:
ETC uses proteins in the inner mitochondrial membrane
NADH and FADH are oxidized (lose electrons) and give electrons to these proteins
passing electrons down the ETC pumps H+ ions into intermembrane space
the proton gradient provides the energy to convert ADP → ATP (chemiosmosis)
final electron acceptor: oxygen
this is the main reason for why we breathe!!
oxygen accepts the electrons from the integral proteins, freeing them up to keep cycling more electrons through
oxygen accepts the electrons and protons and use them to form water
without oxygen, the ETC can’t continue because their hands are full with electrons that they can’t let go of
H+ ions in intermembrane space flow back into the matrix through ATP synthase, which has a rotor mechanism that connects ADP back to P, creating ATP
ATP Synthase:
a channel protein for H+ ions to pass through.
the movement of H+ through the protein makes the rotor spin and phosphorylate ADP (creating ATP)
this is an ancient molecule that’s also used in photosynthesis
chemiosmosis: movement of H+ ions through ATP synthase
H+ gradient is established by the ETC
H+ easily flows back into the matrix through ATP Synthase (facilitated diffusion)
yield:
input:
10 NADH (2 from glycolysis, 2 from 2 krebs transitions, 6 from 2 krebs cycles)
2 FADH2 (2 from 2 krebs cycles)
output:
max 38 ATP
3 × (10 NADH)
yield 3 ATP per NADH
2 × (2 FADH2)
yield 2 ATP per FADH2
2 ATP from glycolysis
2 ATP from krebs cycle
 Knowt
Knowt