Study Guide For MicroBio
Study Guide: Microscopes
Parts of the Microscope and Their Functions
Eyepiece (Ocular Lens): The lens you look through, typically has a magnification of 10x.
Objective Lenses: Found on a rotating nosepiece, these lenses can have various magnifications (e.g., 4x, 10x, 40x, 100x) and are used to observe the specimen.
Stage: The flat platform where the slide is placed for observation.
Stage Clips: Hold the slide in place on the stage.
Condenser: Focuses light onto the specimen, improving clarity.
Diaphragm: Adjusts the amount of light that reaches the specimen.
Light Source: Provides illumination, can be a mirror or an electric bulb.
Arm: Part of the microscope that connects the base and the head; used for carrying the microscope.
Base: The bottom support of the microscope.
Coarse Focus Knob: Used for initial focusing of the specimen.
Fine Focus Knob: Allows for precise focusing once the specimen is in general focus.
Magnification
Total Magnification Calculation:
Formula: Ocular Lens Magnification x Objective Lens Magnification = Total Magnification.
Example: If the ocular lens is 10x and the objective lens is 40x, then Total Magnification = 10x * 40x = 400x.
Resolution
Resolution: The ability to distinguish two close points as separate. Higher resolution means clearer and more detailed images.
How to Use a Microscope
Setup: Place the microscope on a stable surface and plug it in if it has an electric light source.
Preparation: Place the slide on the stage and secure it with stage clips.
Magnification: Start with the lowest power objective lens (4x) for initial focusing. Use the coarse focus knob to bring the stage up to the lowest objective lens.
Focusing: Once the specimen is in view, switch to a higher power objective lens (10x, 40x) and use the fine focus knob to sharpen the image.
Adjust Light: Use the diaphragm to adjust the light intensity for better visibility of the specimen.
Returning the Microscope
When returning the microscope, make sure to:
Lower the stage completely.
Rotate the nosepiece to the lowest power lens.
Turn off the light source.
Clean any oil off the objective lenses (for oil immersion lenses).
Replace the dust cover on the microscope and return it to its designated storage area.
Study Guide: Staining
Gram Staining
Purpose: To classify bacteria as gram-positive (+) or gram-negative (-) based on differences in their cell wall structure, which influences their susceptibility to certain antibiotics and their pathogenicity.
Principle: Involves a series of staining steps:
Crystal Violet Stain: All bacteria initially take up this dye, which stains them purple.
Iodine Treatment: The iodine acts as a mordant, forming a complex with crystal violet that gets trapped in the thick peptidoglycan layer of gram-positive bacteria.
Decolorization: Typically done with ethanol or acetone. This step is critical; gram-positive bacteria retain the dye and remain purple because of their thick peptidoglycan layer, while gram-negative bacteria, which have a thinner peptidoglycan layer and an outer membrane, lose the dye and become colorless.
Counterstaining: Usually with Safranin. After decolorization, gram-negative bacteria will take up the red dye and appear pink, whereas gram-positive bacteria remain purple.
Interpretation:
Gram-positive (+): Retain crystal violet stain, appear purple under the microscope. Typically have thicker peptidoglycan walls and include genera such as Staphylococcus and Streptococcus.
Gram-negative (-): Do not retain the crystal violet stain, and appear pink/red. They often have additional layers, including an outer membrane, and include bacteria like Escherichia coli and Salmonella.
Identification of Shape and Arrangement of Bacteria
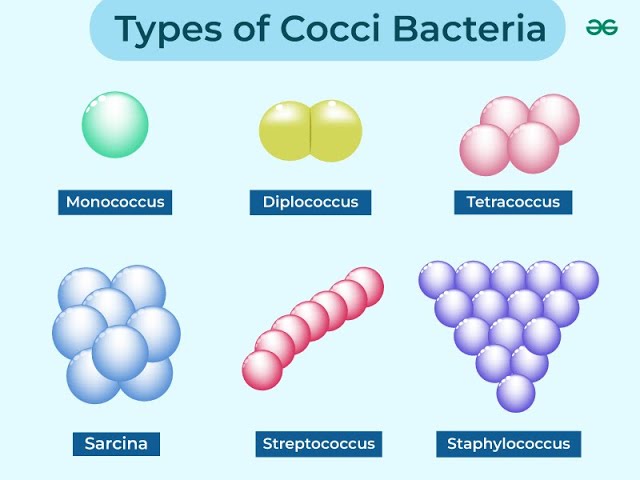
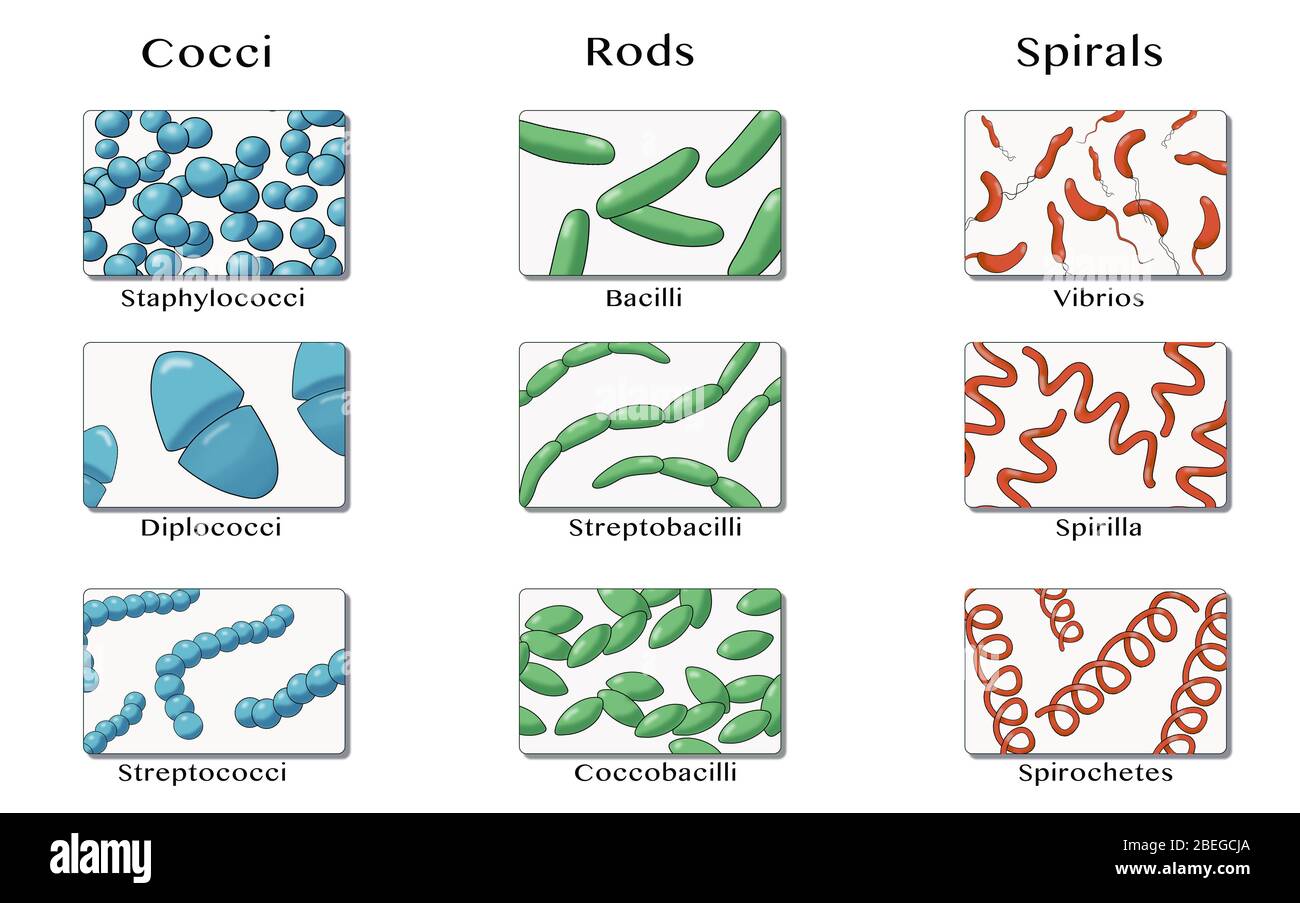
Morphological Shapes:
Cocci: Spherical bacteria may appear as singles (monococci), pairs (diplococci), or clusters (staphylococci).
Bacilli: Rod-shaped bacteria can appear as single cells (monobacilli), chains (streptobacilli), or branched forms.
Spirilla: Spiral-shaped bacteria often appear as single spirals, or may have multiple turns (spirochetes).
Vibrio: Comma-shaped.
Arrangement: Affects how the bacteria appear in lab cultures:
Streptococci: Chains of cocci, typical of certain pathogens like Streptococcus pneumoniae.
Staphylococci: Clusters of cocci, characteristic of Staphylococcus aureus.
Tetrads: Groups of four cocci, typically seen in Micrococcus species.
Sarcina: Cubic arrangement of cocci.
Negative Staining
Purpose: To visualize the shape and size of bacteria without heat fixation, preserving their natural morphology and arrangement.
Principle: Negative stains such as India ink or nigrosin do not penetrate the bacterial cell. Instead, they stain the background, leaving the cells transparent and allowing better visualization of their structure. This technique is particularly useful for observing the size and arrangement of delicate bacterial forms.
Technique:
Place a small drop of negative stain on a slide.
Using a sterile loop, obtain a sample of the bacterial culture and mix it with the stain.
Use another slide to spread the mixture across the slide.
Allow the slide to air dry; observe under oil immersion for better detail.
Acid-Fast Staining
Purpose: To detect mycobacteria, particularly Mycobacterium tuberculosis, which are resistant to ordinary staining techniques due to their waxy cell walls.
Principle:
Primary Stain: Carbol fuchsin penetrates the waxy cell wall, staining acid-fast organisms red.
Decolorization: Acid-alcohol (3% hydrochloric acid in alcohol) is used; acid-fast bacteria retain the pink stain while non-acid-fast bacteria lose it.
Counterstaining: Methylene blue is applied; non-acid-fast organisms take up the blue stain, appearing blue under the microscope.
Interpretation:
Acid-fast bacteria: Retain the red color (e.g., Mycobacterium tuberculosis).
Non-acid-fast bacteria: Take up the counterstain and appear blue.Study Guide: Anaerobic Jar and Thioglycolate Tube
Anaerobic Jar
Principle: An anaerobic jar is used to create an oxygen-free environment for the growth of anaerobic bacteria.
Gases Released from Gas Pack: The gas pack contains chemicals that release hydrogen and carbon dioxide when activated. The hydrogen combines with oxygen present in the jar, creating water and removing oxygen from the environment.
Organisms:
Facultative Anaerobes: Organisms like E. coli can grow in both aerobic and anaerobic environments. In an anaerobic jar, E. coli will still grow well since it can metabolize with or without oxygen.
Obligate Aerobes: Organisms like Pseudomonas require oxygen to grow. They will not grow in an anaerobic jar.
Obligate Anaerobes: Organisms such as Clostridium thrive in environments without oxygen and grow well in the anaerobic jar.
Comparison of Plates
Use aerobic plates (exposed to oxygen) and anaerobic plates (in anaerobic jars) to identify and observe the growth of E. coli, Pseudomonas, and Clostridium based on their growth patterns and colony morphology.
Thioglycolate Tube
Information about Thioglycolate Media:
Composition: Thioglycolate media is a complex broth that includes sodium thioglycolate, providing a reducing environment to minimize oxygen.
Type: It is a reducing medium and allows for the growth of both aerobic and anaerobic organisms.
Organisms:
Obligate Aerobes: These organisms require oxygen to grow and will grow at the top of the thioglycolate tube where oxygen is present (e.g., Pseudomonas).
Obligate Anaerobes: They grow at the bottom of the tube where there is no oxygen (e.g., Clostridium).
Facultative Anaerobes: Grow throughout the tube but are most dense at the top where oxygen is present (e.g., E. coli).
Enzymes Present:
Different bacteria have different enzymes facilitating oxygen metabolism:
Obligate Aerobes: Catalase and superoxide dismutase (to handle reactive oxygen species).
Obligate Anaerobes: Often lack these enzymes and will not survive in the presence of oxygen.
Facultative Anaerobes: Possess both aerobic and anaerobic metabolism enzymes, including the ability to produce catalase to neutralize hydrogen peroxide when oxygen is present
Comparison of Aerobic and Anaerobic Plates
Comparing the growth of E. coli, Pseudomonas, and Clostridium on aerobic and anaerobic plates helps identify their oxygen requirements and growth patterns:
E. coli (Facultative Anaerobe):
On Aerobic Plates: E. coli will grow well in the presence of oxygen, leading to colonies that are typically off-white to yellowish.
On Anaerobic Plates: E. coli can also grow in the absence of oxygen, and colonies may be similar in appearance, reflecting its capability to metabolize under both conditions.
Pseudomonas (Obligate Aerobe):
On Aerobic Plates: Pseudomonas thrives with oxygen, forming characteristic colonies that are often greenish or blue-green due to pigment production.
On Anaerobic Plates: Pseudomonas will not grow in an anaerobic environment, and the absence of growth will indicate its obligate aerobic nature.
Clostridium (Obligate Anaerobe):
On Aerobic Plates: Clostridium will typically show no growth in aerobic conditions, as it is unable to survive in the presence of oxygen, which can be toxic to it.
On Anaerobic Plates: Clostridium will flourish in anaerobic conditions, with colonies appearing white, cream, or yellow, depending on the species.
Interpretation:
The growth patterns observed on the plates allow microbiologists to determine the oxygen requirements of the bacteria. A lack of growth on aerobic plates and robust growth on anaerobic plates suggests the organism is an obligate anaerobe (like Clostridium), while significant growth on both suggests a facultative anaerobe (like E. coli). Meanwhile, growth only on aerobic plates indicates an oblig
Study Guide: Culture Media
Broth and Slant Media
Broth: A liquid medium that supports the growth of a wide range of microorganisms, allowing for the growth of both aerobic and anaerobic bacteria. It is ideal for culturing large numbers of bacteria and for various biochemical assays.
Slant: A solid medium in a test tube that provides a larger surface area for microbial growth on the slanted agar's surface. Slants are typically used for storage, isolating pure strains, and studying colony morphology.
Selective and Differential Media
EMB (Eosin Methylene Blue Agar)
Purpose: Primarily used to isolate Gram-negative bacteria and differentiate lactose fermenters from non-fermenters.
Selective: Inhibits Gram-positive organisms due to the presence of methylene blue and eosin dye, which interfere with their growth.
Differential: Lactose fermenters produce acid, leading to colony color changes:
Strong fermenters like E. coli produce a greenish metallic sheen.
Non-fermenters appear colorless or pale pink, making it easy to identify them.
Growth Observed:
Positive: E. coli (purple/greenish metallic sheen)
Negative: Salmonella, Shigella (No growth, or colorless colonies).
MacConkey Agar
Purpose: Used to isolate and differentiate enteric (intestinal) Gram-negative bacteria.
Selective: Contains bile salts and crystal violet, which inhibit Gram-positive bacteria, allowing only Gram-negative organisms to grow.
Differential: Identifies lactose fermenters, producing acid that turns the pH indicator pink:
Strong fermenters like E. coli yield pink colonies.
Non-fermenters remain colorless.
Growth Observed:
Positive: E. coli (pink colonies)
Negative: Non-lactose fermenters (colorless colonies) including Salmonella and Shigella.
MSA (Mannitol Salt Agar)
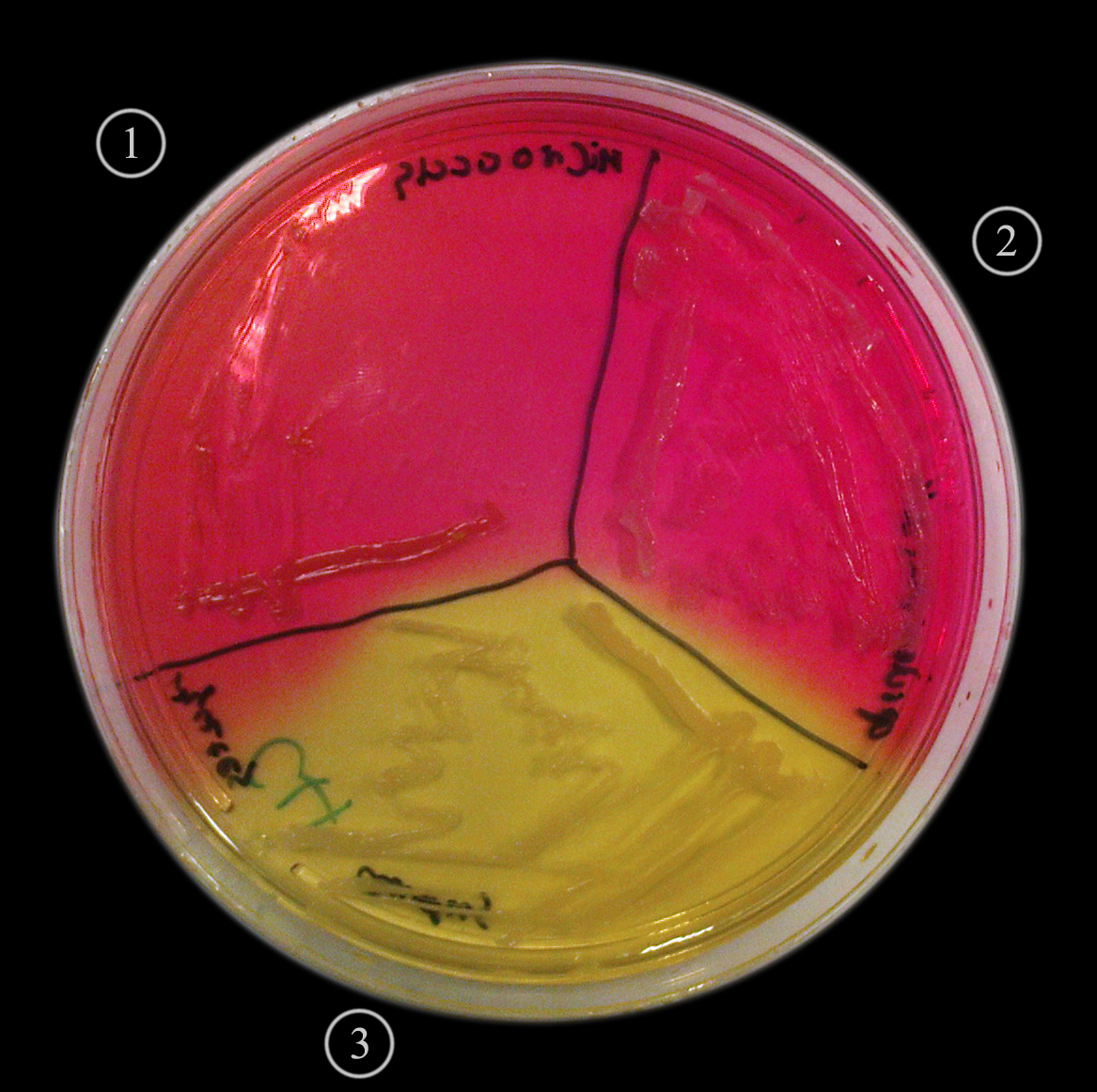
Purpose: Designed to isolate Staphylococcus species and differentiate between mannitol fermenters and non-fermenters.
Selective: High salt concentration (7.5%) inhibits the growth of most bacteria except halophilic staphylococci.
Differential: Staphylococcus aureus ferments mannitol, resulting in acid production and a color change of the phenol red indicator from red to yellow:
Other staphylococci that do not ferment mannitol will leave the medium red.
Growth Observed:
Positive: Staphylococcus aureus (yellow colonies)
Negative: Staphylococcus epidermidis (red colonies)
PEA (Phenylethyl Alcohol Agar)
Purpose: Primarily used for isolating Gram-positive organisms, especially staphylococci and streptococci.
Selective: Phenylethyl alcohol inhibits Gram-negative bacteria by disrupting their outer membrane, leading to selective growth of Gram-positive organisms.
Differential: Generally not used for differential purposes; it is primarily selective.
Growth Observed:
Positive: Staphylococcus, Streptococcus species (growth)
Negative: Enterobacteriaceae (no growth)
Blood Agar
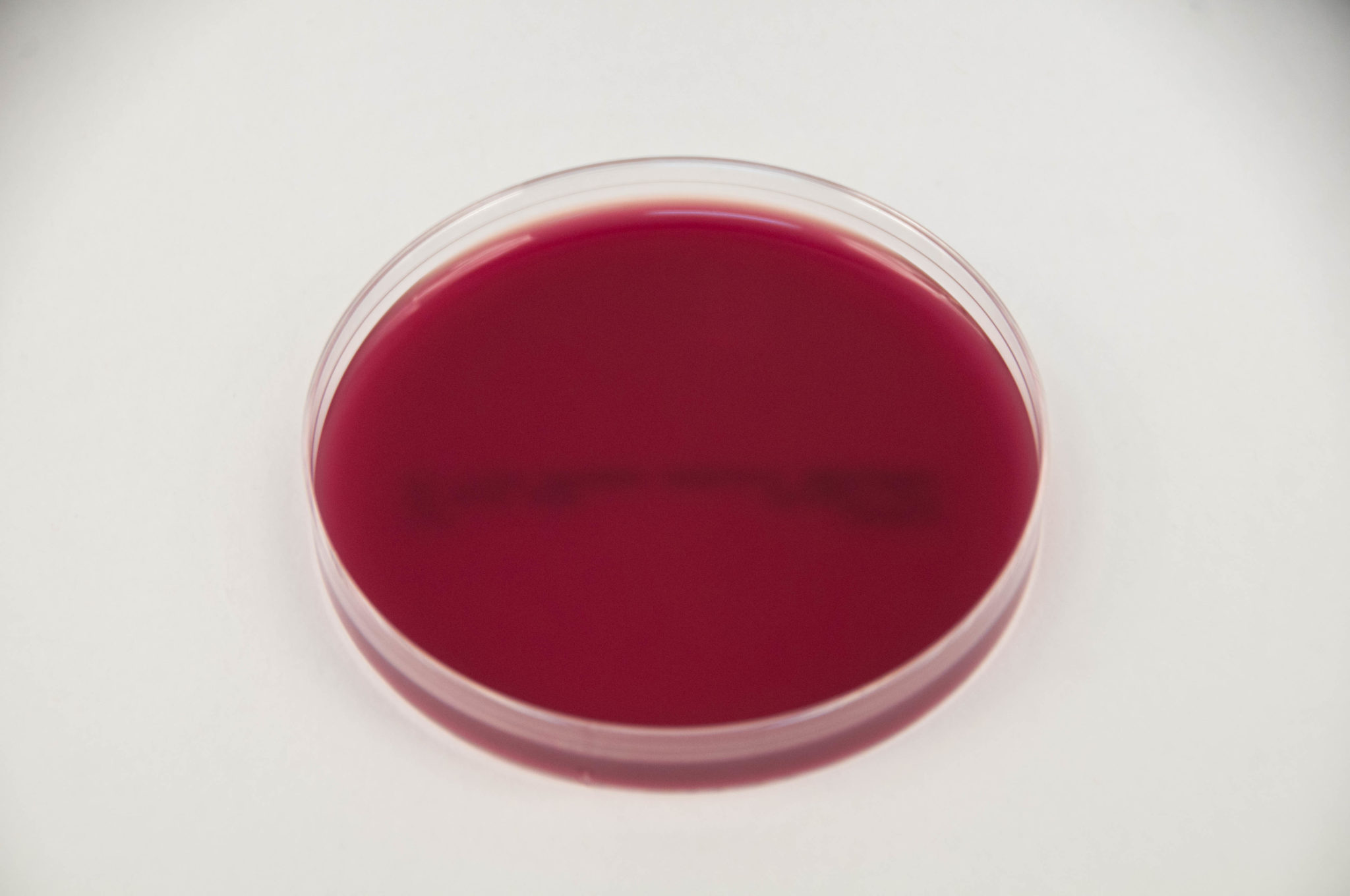
Purpose: Used for the cultivation of a wide variety of fastidious organisms and to assess hemolytic activity.
Selective: Not significantly selective; supports growth of both Gram-positive and Gram-negative organisms.
Differential: Differentiates by hemolytic activity:
Alpha-hemolysis (green): partial hemolysis by bacteria such as Streptococcus pneumoniae.
Beta-hemolysis (clear): complete lysis of red blood cells by organisms such as Streptococcus pyogenes.
Gamma-hemolysis (no change): no hemolysis, seen with species like Enterococcus faecalis.
Growth Observed:
Positive: Streptococcus pyogenes (beta-hemolysis, clear zone)
Negative: Some Staphylococcus species (gamma-hemolysis, no change)
Cetrimide Agar
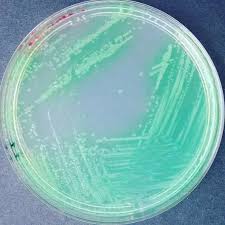
Purpose: Primarily used to isolate Pseudomonas aeruginosa, which is important in clinical and environmental microbiology.
Selective: Contains cetrimide, which inhibits the growth of most non-pseudomonad species.
Differential: While it does not exhibit pronounced differential characteristics, Pseudomonas aeruginosa typically produces a blue-green pigment due to pyocyanin, especially under aerobic conditions.
Growth Observed:
Positive: Pseudomonas aeruginosa (typically blue-green colonies)
Negative: Other Gram-positive and many Gram
Blood Agar
Purpose: Used for the cultivation of a wide variety of fastidious organisms and to assess hemolytic activity.
Selective: Not significantly selective; supports growth of both Gram-positive and Gram-negative organisms.
Differential: Differentiates based on hemolytic activity:
Alpha Hemolysis: Partial lysis of red blood cells, causing a green discoloration around colonies.
Example: Streptococcus pneumoniae. The green color results from the reduction of hemoglobin to methemoglobin.
Beta Hemolysis: Complete lysis of red blood cells, resulting in a clear zone around colonies.
Example: Streptococcus pyogenes. The hemolytic enzymes (streptolysins) lyse red blood cells completely.
Gamma Hemolysis (or non-hemolytic): No lysis of red blood cells, no change in the appearance of the agar around colonies.
Example: Enterococcus faecalis. These organisms do not produce enzymes that lyse red blood cells, leading to no discoloration of the medium.
Kirby-Bauer Test - Antibiotic Susceptibility Test
Purpose: The Kirby-Bauer test is a standardized method used to determine the susceptibility of specific bacterial strains to various antibiotics. It is an essential tool in clinical microbiology for guiding antimicrobial therapy.
Plates Used: The test is conducted using Mueller-Hinton agar plates. This particular medium is chosen because of its ability to support the growth of a wide range of bacteria and its suitable properties for the diffusion of antibiotics, providing reliable and reproducible results.
Other Names for Antibiotic Susceptibility Test:
Antibiotic sensitivity test
Antimicrobial susceptibility test
Disk diffusion test
Mechanism of Action for Common Antibiotics:
Penicillins (e.g., Penicillin G, Amoxicillin): These beta-lactam antibiotics function by inhibiting the synthesis of bacterial cell walls. They bind to and inactivate penicillin-binding proteins (PBPs) involved in cross-linking the peptidoglycan layer, leading to cell lysis and death, especially effective against gram-positive bacteria.
Tetracyclines (e.g., Doxycycline, Minocycline): These antibiotics work by targeting the bacterial ribosome. They bind to the 30S subunit, preventing the attachment of aminoacyl-tRNA and thus inhibiting protein synthesis. This makes them broad-spectrum agents effective against a variety of bacterial infections.
Macrolides (e.g., Erythromycin, Azithromycin): Macrolides inhibit protein synthesis by binding to the 50S ribosomal subunit, blocking peptide bond formation during translation. They are particularly effective against gram-positive bacteria and some gram-negative bacteria as well.
Fluoroquinolones (e.g., Ciprofloxacin, Levofloxacin): These are broad-spectrum antibiotics that inhibit bacterial DNA replication. They target two essential enzymes, DNA gyrase and topoisomerase IV, disrupting DNA supercoiling and cell division, making them effective against both gram-positive and gram-negative bacteria.
Zone of Inhibition: The zone of inhibition refers to the clear area around an antibiotic disk on the agar plate where bacterial growth has been inhibited. This area is measured in millimeters; a larger zone indicates that the bacteria are more susceptible to the antibiotic, while a smaller zone suggests resistance.
Effect of UV on Bacterial Growth
Mechanism of Action:
Ultraviolet (UV) light kills bacteria primarily through its ability to induce DNA damage. The most significant process is the formation of thymine dimers, which occur when UV light causes adjacent thymine bases in the DNA strand to bond improperly. This leads to distortion in the DNA structure, preventing proper replication and transcription.
Penetration of UV Light:
Glass: UV light, particularly UVC (200-280 nm), does not effectively penetrate glass. Most glass types, including quartz glass, can block UVC, thus limiting its bactericidal effects when bacteria are covered by glass.
Cardboard: Similar to glass, cardboard also absorbs UV light and therefore does not allow significant UV penetration, hindering its effectiveness for surface sterilization.
Plastic: The effect of UV on plastic depends on the specific type. Many plastics can absorb some UV radiation, particularly those not specifically designed for UV transmission, which limits their bactericidal capacity. However, clear or UV-resistant plastics might allow some penetration, although generally much less than desired for effective disinfection.
Thymine Dimers:
The formation of thymine dimers is a critical result of UV exposure. In typical bacteria, these dimers may occur at a significant rate, with estimates suggesting that for every 100-500 bases of exposed DNA, around 10 to 20 thymine dimers can be generated under intense UV exposure. This quantifiable alteration leads to errors in DNA replication, ultimately resulting in cellular death if not repaired by cellular mechanisms such as nucleotide excision repair (NER). Therefore, the cumulative effect of UV light on bacterial cells is lethal, particularly if they cannot effectively repair the damage.
In conclusion, while UV light can be an effective means of controlling bacterial growth, its efficiency greatly depends on the environmental medium it passes through and the biological effects on microbial DNA, particularly the formation of thymine dimers.
Study Guide: Microbiology Techniques
A. Streaking and Lab Equipment
Streaking:
A method used to isolate a pure strain from a single species of microorganism, facilitating easy growth and observation of colonies.
Technique:
Sterilize the inoculating loop by flame until red hot and let it cool.
Dip the loop into the sample and streak it across the agar surface in a zig-zag manner.
Flame the loop again, cool, and streak the plate in a different section to dilute the sample. Repeat to create isolated colonies.
Lab Equipment:
Essential items in a microbiology lab include:
Inoculating Loop: For transferring microorganisms.
Petri Dishes: Circular dishes used for culturing microorganisms.
Agar Plates: Nutrient media solidified with agar.
Incubator: Maintains an optimal temperature for microbial growth.
Main Safety Precautions:
Always wear gloves and lab coats to protect from contaminants.
Use biosafety cabinets when handling pathogenic microbes.
Properly dispose of biohazardous waste following institutional guidelines.
B. Colony Morphology:
The physical appearance of microbial colonies on solid media. Key characteristics to observe include:
Size: Range from small (1-2 mm) to large (>5 mm).
Color: Different pigments can indicate species (e.g., yellow for Staphylococcus aureus).
Shape: Circular, irregular, filamentous, or rhizoid.
Margin: Smooth, wavy, lobate, or entire.
Elevation: Raised, convex, flat, or umbonate.
C. Staining Techniques
Identifying Gram Stain:
Gram-positive (+): Retain crystal violet stain, appearing purple due to thick peptidoglycan layers. Example genera: Staphylococcus, Streptococcus.
Gram-negative (-): Do not retain crystal violet and appear pink/red after counterstaining because of a thinner peptidoglycan layer and an outer membrane. Example: Escherichia coli, Salmonella.
Principles of Staining:
Gram Staining:
Crystal Violet Stain: All bacteria initially absorb this purple dye.
Iodine Treatment: Acts as a mordant, enhancing dye retention in gram-positive bacteria.
Decolorization: Ethanol or acetone selectively washes out the dye from gram-negative bacteria.
Counterstaining with Safranin: Confirms gram-negative bacteria by coloring them pink.
Negative Staining:
Purpose: Visualizes sizes and arrangements of delicate bacteria without heat fixation.
Principle: Negative stains (e.g., India ink) stain the background, contrasting against colorless bacteria, providing clearer structure visibility.
Acid-Fast Staining:
Purpose: Detects bacteria with waxy cell walls like Mycobacterium.
Principle: Uses carbol fuchsin to stain acid-fast bacteria red; after decolorization, non-acid-fast organisms take up blue dye (Methylene blue), appearing blue under the microscope.
Gram Positive Biochemical Tests
1. Catalase Test
Principle: This test detects the presence of the enzyme catalase, which breaks down hydrogen peroxide (H₂O₂) into water and oxygen.
Positive Result: Bubbling observed when hydrogen peroxide is added (e.g., Staphylococcus aureus).
Negative Result: No bubbling (e.g., Streptococcus pyogenes).
2. Coagulase Test
Principle: This test determines the ability of the organism to clot plasma, indicating the presence of coagulase enzyme.
Positive Result: Clot formation in the plasma (e.g., Staphylococcus aureus).
Negative Result: No clotting (e.g., Staphylococcus epidermidis).
3. Latex Agglutination Test
Principle: This test utilizes latex beads coated with specific antibodies to detect the presence of antigens found in the bacterial cell wall.
Positive Result: Visible agglutination (e.g., Streptococcus pneumoniae).
Negative Result: No agglutination (e.g., Staphylococcus saprophyticus).
4. Bile Esculin Test
Principle: This test evaluates an organism's ability to hydrolyze esculin in the presence of bile salts.
Positive Result: Dark brown/black precipitate formed (e.g., Enterococcus faecalis).
Negative Result: No color change (e.g., Streptococcus).
5. Oxidase Test
Principle: This test assesses the presence of cytochrome c oxidase enzyme, which contributes to the electron transport chain.
Positive Result: Color change to dark purple (e.g., Enterobacter aerogenes).
Negative Result: No color change (e.g., Escherichia coli).
Confirmatory Tests:
MSA Plate: Used to identify Staphylococcus species based on mannitol fermentation (S. aureus gives yellow colonies).
Blood Agar Plate: Used to assess hemolytic activity (Streptococcus pyogenes shows beta hemolysis).
Gram Negative Biochemical Tests
1. Oxidase Test
Principle: Determines the presence of cytochrome oxidase, which is involved in the electron transport chain.
Positive Result: Development of a dark blue/purple color after the addition of oxidase reagent (e.g., Pseudomonas aeruginosa).
Negative Result: No color change (e.g., Escherichia coli).
2. Nitrate Reduction Test
Principle: Tests for the ability to reduce nitrate (NO₃) to nitrite (NO₂) or further to nitrogen gas (N₂).
Positive Result: Color change to red after reagent addition or the gas bubble in the Durham tube (e.g., Enterobacter cloacae).
Negative Result: No color change, remains yellow (e.g., Staphylococcus saprophyticus).
3. Citrate Test
Principle: Assesses the ability of the organism to use citrate as a sole carbon source.
Positive Result: Color change of the medium to blue (e.g., Klebsiella pneumoniae).
Negative Result: No color change, remains green (e.g., Escherichia coli).
4. Urease Test
Principle: Determines the ability to hydrolyze urea into ammonia and carbon dioxide.
Positive Result: Pink color indicates an alkaline pH (e.g., Proteus vulgaris).
Negative Result: No color change (e.g., Escherichia coli).
5. Triple Sugar Iron (TSI) Test
Principle: Assesses the ability of an organism to ferment sugars and produce gas and hydrogen sulfide.
Positive Result: Yellow slant/yellow butt for fermentation; black precipitate for H₂S (e.g., Salmonella spp.).
Negative Result: Red slant/red butt (e.g., Pseudomonas aeruginosa).
6. Sulfide Indole Motility (SIM) Test
Principle: Tests for sulfulfide production, indole production, and motility.
Positive Result: Black precipitate for H₂S, red ring for indole, diffusion from stab line for motility (e.g., Escherichia coli).
Negative Result: No black precipitate, no red ring, and single stab line (e.g., Salmonella).
7. Methyl Red and Voges-Proskauer (MRVP) Test
Principle: MR tests for stable acid production from glucose, and VP tests for acetoin production.
Positive Result: Red color for MR (e.g., Escherichia coli); pink color for VP (e.g., Enterobacter aerogenes).
Negative Result: Yellow color for MR (e.g., Enterobacter cloacae); no color change for VP (e.g., Escherichia coli).
Confirmatory Tests:
MacConkey and EMB Plates: Used to isolate and differentiate Gram-negative bacteria based on lactose fermentation (E. coli produces pink colonies).
Cetrimide Agar: Specifically used to isolate Pseudomonas aeruginosa (shows characteristic blue-green colonies).
Questions and Answers about Gram Negative Biochemical Tests
1. Oxidase Test
Question: What is the principle of the oxidase test?
Answer: It determines the presence of cytochrome c oxidase, which is involved in the electron transport chain.
Question: Which organisms give a positive result and what color indicates this result?
Answer: A positive result is indicated by the development of a dark blue/purple color, seen in organisms like Pseudomonas aeruginosa.
Question: Which organism gives a negative result?
Answer: Escherichia coli gives a negative result, showing no color change.
2. Nitrate Reduction Test
Question: What is the principle of the nitrate reduction test?
Answer: It tests for the ability to reduce nitrate (NO₃) to nitrite (NO₂) or further to nitrogen gas (N₂).
Question: What indicates a positive result, and which organisms exhibit this?
Answer: A positive result is indicated by a color change to red after reagent addition or the presence of a gas bubble in a Durham tube, seen in organisms like Enterobacter cloacae.
Question: Which organism gives a negative result?
Answer: Staphylococcus saprophyticus shows no color change, remaining yellow, which indicates a negative result.
3. Citrate Test
Question: What is the principle of the citrate test?
Answer: It assesses the ability of the organism to use citrate as a sole carbon source.
Question: Which organisms give a positive result and what color indicates this?
Answer: A positive result is indicated by a color change to blue, seen in Klebsiella pneumoniae.
Question: Which organism gives a negative result?
Answer: Escherichia coli shows no color change and remains green.
4. Urease Test
Question: What is the principle of the urease test?
Answer: It determines the ability to hydrolyze urea into ammonia and carbon dioxide.
Question: Which organisms give a positive result and what color indicates this?
Answer: A positive result is indicated by a pink color, which shows an alkaline pH, as seen in Proteus vulgaris.
Question: Which organism gives a negative result?
Answer: Escherichia coli shows no color change during this test.
5. Triple Sugar Iron (TSI) Test
Question: What is the principle of the TSI test?
Answer: It assesses the ability of an organism to ferment sugars and produce gas and hydrogen sulfide.
Question: What indicates a positive result in this test?
Answer: A yellow slant/yellow butt for fermentation and a black precipitate for hydrogen sulfide (H₂S) are indicators of a positive result, typically seen with Salmonella spp.
Question: Which organism gives a negative result?
Answer: Pseudomonas aeruginosa shows a red slant/red butt, indicating no fermentation.
6. Sulfide Indole Motility (SIM) Test
Question: What is the principle of the SIM test?
Answer: It tests for sulfide production, indole production, and motility.
Question: What indicates a positive result in this test?
Answer: A black precipitate for H₂S production, a red ring for indole production, and diffusion from the stab line for motility indicate positive results, commonly seen with Escherichia coli.
Question: Which organism gives a negative result?
Answer: Salmonella shows no black precipitate, no red ring, and maintains a single stab line, indicating negative results.
7. Methyl Red and Voges-Proskauer (MRVP) Test
Question: What is the principle of the MRVP test?
Answer: The MR test detects stable acid production from glucose, while VP tests for acetoin production.
Question: What indicates a positive result for the MR test and which organisms are involved?
Answer: A red color indicates a positive result in MR, seen in Escherichia coli. For the VP test, a pink color indicates positive results, seen in Enterobacter aerogenes.
Question: Which organism gives a negative result?
Answer: For MR, Enterobacter cloacae gives a yellow color, and Escherichia coli shows no color change for VP, indicating negative results.
Confirmatory Tests
Question: What plates are used for confirmatory tests in Gram-negative biochemical tests?
Answer: MacConkey and EMB plates are used to isolate and differentiate Gram-negative bacteria based on lactose fermentation. Cetrimide agar is specifically used to isolate Pseudomonas aeruginosa, which shows characteristic blue-green colonies.