🤐 AP BIO Chapter 15 - Regulation of Gene Expression
Overview
15.1 Bacteria often respond to environmental change by regulating transcription
15.2 Eukaryotic gene expression is regulated at many stages
15.3 Noncoding RNAs play multiple roles in controlling gene expression
15.4 Researchers can monitor the expression of specific genes
BIG IDEAS: Organisms use feedback mechanisms and transcription factors to maintain dynamic homeostasis through differential gene expression (Big Idea 2) with the interaction of key molecules (Big Idea 4), conserved core processes, and features that are shared by all organisms (Big Idea 1)
Anableps anableps

Describe the specialization of the anableps anableps
The eye’s upper half is particularly well-suited for aerial vision and the lower half for aquatic vision. The cells of the two parts of the eye express a slightly different set of genes involved in vision, even though these two groups of cells are quite similar and contain identical genomes
Both prokaryotes and eukaryotes must alter their patterns of gene expression in response to changes in ___
- environmental conditions
What challenges do multicellular eukaryotes face?
They must develop and maintain multiple cell types each expressing a different subset of genes
15.1 - Bacteria often respond to environmental change by regulating transcription
In bacteria, genes are often clustered into an operon with a single promoter. An operator site on the DNA switches the operon on or off, resulting in coordinate regulation of the genes
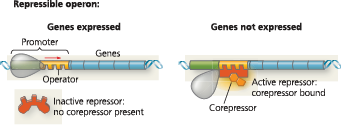
Both repressible and inducible operons are examples of negative gene regulation: Binding of a specific repressor protein to the operator shuts off transcription. (The repressor is encoded by a separate regulatory gene.) In a repressible operon (usually encoding anabolic enzymes), the repressor is active when bound to a corepressor:
In an inducible operon (usually encoding catabolic enzymes), binding of an inducer to an innately active repressor inactivates the repressor and turns on transcription
Some operons have positive gene regulation. A stimulatory activator protein (such as CRP, when activated by cyclic AMP), binds to a site within the promoter and stimulates transcription
Natural selection has favored bacteria that express only the genes whose products are ___ to conserve resources and energy
- needed by the cell
How can metabolic pathways be controlled?
Cells can adjust enzyme activity (be increased, decreased, or inhibited) and adjust the production of enzymes (regulate the expression of the genes encoding the enzymes)
Regulation of a metabolic pathway - In the pathway for tryptophan synthesis, an abundance of tryptophan can both (a) inhibit the activity of the first enzyme in the pathway (feedback inhibition), a rapid response, and (b) repress expression of the genes encoding all subunits of the enzymes in the pathway, a longer-term response. Genes trpE and trpD encode the two subunits of enzyme 1, and genes trpB and trpA encode the two subunits of enzyme 3. (The genes were named before the order in which they functioned in the pathway was determined.) The symbol stands for inhibition
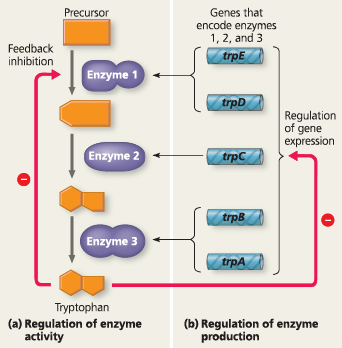
}}Feedback Inhibition:}}
A method of metabolic control in which the end product of a metabolic pathway acts as an inhibitor of an enzyme within that pathway
The trp operon in E. coli: regulated synthesis of repressible enzymes - Tryptophan is an amino acid, produced by an anabolic pathway catalyzed by three enzymes. (a) The five genes encoding the polypeptide subunits of the enzymes in this pathway are grouped, along with a promoter, into the trp operon. The trp operator (the repressor binding site) is located within the trp promoter (the RNA polymerase binding site) (b) Accumulation of tryptophan, the end product of the pathway, represses transcription of the trp operon, thus blocking synthesis of all the enzymes in the pathway and shutting down tryptophan production
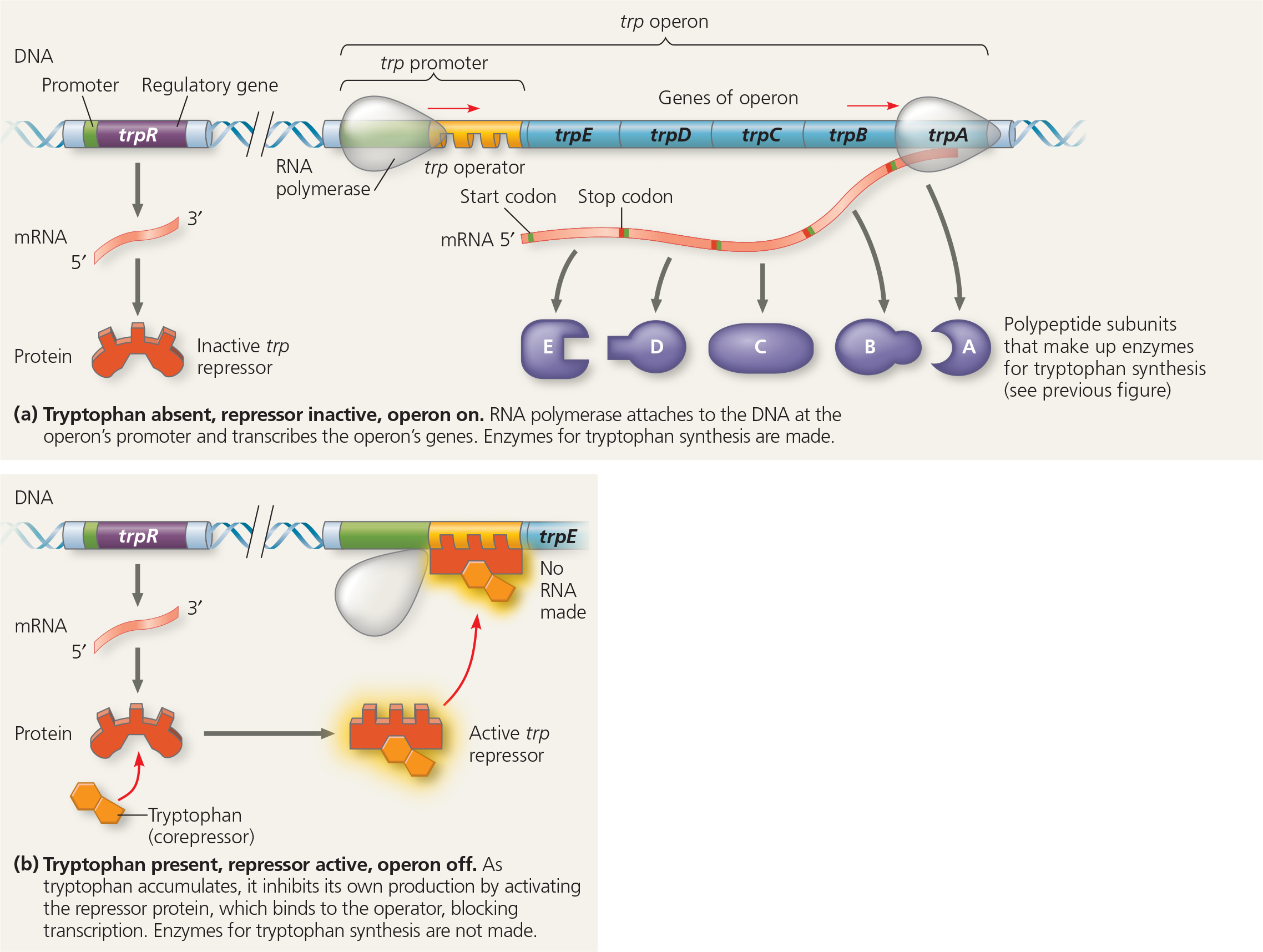
Describe what happens to the trp operon as the cell uses up its store of tryptophan
As the concentration of tryptophan in the cell falls, eventually there will be none bound to trp repressor molecules. These will then change into their inactive shapes and dissociate from the operator, allowing transcription of the operon to resume. The enzymes for tryptophan synthesis will be made, and they will again synthesize tryptophan in the cell
A key advantage of grouping genes of related function into one transcription unit is that a single ___ can control the whole cluster of functionally related genes
- “on-off switch“
}}Coordinately Controlled:}}
When genes on the same chromosome are activated by the same chemical cues
}}Operator:}}
In bacterial and phage DNA, a sequence of nucleotides near the start of an operon to which an active repressor can attach. The binding of the repressor prevents RNA polymerase from attaching to the promoter and transcribing the genes of the operon
}}Operon:}}
A unit of genetic function found in bacteria and phages, consisting of a promoter, an operator, and a coordinately regulated cluster of genes whose products function in a common pathway
}}Repressor:}}
A protein that inhibits gene transcription. In prokaryotes, repressors bind to the DNA in or near the promoter. In eukaryotes, repressors may bind to control elements within enhancers, to activators from binding to DNA
A repressor protein is ___ for the operator of a particular operon
- specific
}}Regulatory Gene:}}
A gene that codes for a protein, such as a repressor, that controls the transcription of another gene or group of genes
Regulatory genes are expressed ___ at a _ rate
- continuously; low
The binding of repressors to operators is ___
- reversible
}}Allosteric Protein:}}
A protein whose shape is changed when it binds a particular molecule; oscillates between active and inactive
}}Corepressor:}}
A small molecule that binds to a bacterial repressor protein and changes the protein’s shape, allowing it to bind to the operator and switch an operon off
}}Repressible Operon:}}
Transcription is usually on but can be inhibited (repressed) when a specific small molecule binds allosterically to a regulatory protein (trp operon)
}}Inducible Operon:}}
Usually off but can be stimulated (induced) when a specific small molecule interacts with a regulatory protein (lac operon)
}}Inducer:}}
A specific small molecule that binds to a bacterial repressor protein and changes the repressor’s shape so that it cannot bind to an operator, thus switching an operon on
The lac operon in E. coli: regulated synthesis of inducible enzymes - E. coli uses three enzymes to take up and metabolize lactose, the genes for which are clustered in the lac operon. The first gene, lacZ, codes for β-galactosidase, which hydrolyzes lactose to glucose and galactose. The second gene, lacY, codes for a permease, a membrane protein that transports lactose into the cell. The third gene, lacA, codes for an enzyme that detoxifies other molecules entering the cell via the permease. Unusually, the gene for the lac repressor, lacI, is adjacent to the lac operon
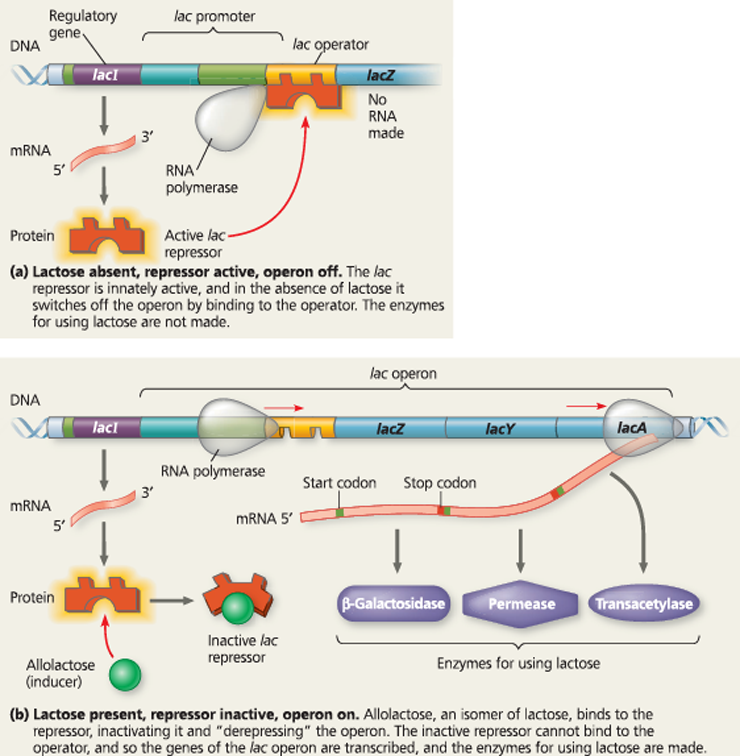
}}Inducible Enzymes:}}
Synthesis is induced by a chemical signal
}}Repressible Enzymes:}}
When a particular metabolite produced in a pathway suppresses or inhibits the production of an enzyme
}}Cyclic AMP (cAMP):}}
Cyclic adenosine monophosphate, a ring-shaped moelcule made from ATP that is a common intracellular signaling molecule (second messenger) in eukaryotic cells. It is also a regulaor of some bacterial operons
}}Activator:}}
A protein that binds to DNA and stimulates gene transcription. In prokaryotes, activators bind in or near the promoter; in eukaryotes, activators generally bind to control elements in enhancers
Positive control of the lac operon by cAMP receptor protein (CRP) - RNA polymerase has high affinity for the lac promoter only when CRP is bound to a DNA site at the upstream end of the promoter. CRP, in turn, attaches to its DNA site only when associated with cyclic AMP (cAMP), whose concentration in the cell rises when the glucose concentration falls. Thus, when glucose is present, even if lactose also is available, the cell preferentially catabolizes glucose and makes very little of the enzymes for using lactose
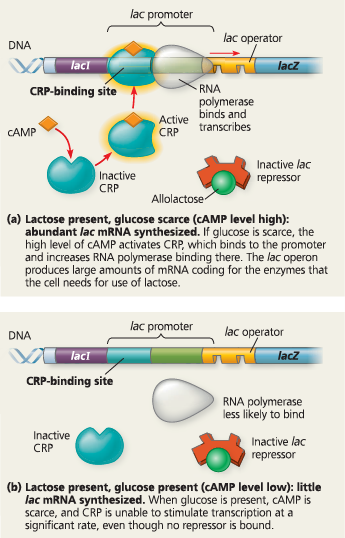
How is the lac operon under negative control by the lac repressor and positive control by CRP?
If the amount of glucose in the cell increases, the cAMP concentration falls, and without cAMP, CRP detaches from the operon. Because CRP is inactive, RNA polymerase binds less efficiently to the promoter, and transcription of the lac operon proceeds at only a low level, even in the presence of lactose
How does binding of the trp corepressor to its repressor alter repressor function and transcription? What about binding of the lac inducer to its repressor?
Binding by the trp corepressor (tryptophan) activates the trp repressor, allowing it to bind to the trp operator, shutting off transcription of the trp operon. Binding by the lac inducer (allolactose) inactivates the lac repressor so it can no longer bind to the lac operator, leading to transcription of the lac operon
Describe the binding of RNA polymerase, repressors, and activators to the lac operon when both lactose and glucose are scarce. What is the effect of these scarcities on transcription of the lac operon?
When glucose is scarce, cAMP is bound to CRP and CRP is bound to the lac promoter, favoring the binding of RNA polymerase. However, in the absence of lactose, the lac repressor is bound to the operator, blocking RNA polymerase binding to the promoter. Therefore, the lac operon genes are not transcribed
A certain mutation in E. coli changes the lac operator so that the active repressor cannot bind. How would this affect the cell’s production of β-galactosidase?
The cell would continuously produce β-galactosidase and the two other enzymes for using lactose, even in the absence of lactose, thus wasting cell resources
Compare and contrast the roles of a corepressor and an inducer in negative regulation of an operon
A corepressor and an inducer are both small molecules that bind to the repressor protein in an operon, causing the repressor to change shape. In the case of a corepressor (like tryptophan), this shape change allows the repressor to bind to the operator, blocking transcription. In contrast, an inducer causes the repressor to dissociate from the operator, allowing transcription to begin
15.2 - Eukaryotic gene expression is regulated at many stages
Both unicellular organisms and the cells of multicellular organisms continually turn genes on and off in response to ___
- signals from their external and internal environments
}}Differential Gene Expression:}}
The expression of different sets of genes by cells with the same genome
What are the differences in gene types?
It’s not due to different genes being present, but rather from uniquely expression genes leading to a specific function
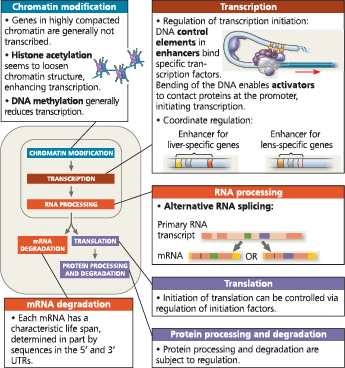
Stages in gene expression that can be regulated in eukaryotic cells - In this diagram, the colored boxes indicate the processes most often regulated; each color indicates the type of molecule that is affected (blue = DNA, red/orange = RNA, purple = protein). The nuclear envelope separating transcription from translation in eukaryotic cells offers an opportunity for post-transcriptional control in the form of RNA processing that is absent in prokaryotes. In addition, eukaryotes have a greater variety of control mechanisms operating before transcription and after translation. A miniature version of this figure accompanies several figures later in the chapter as an orientation diagram
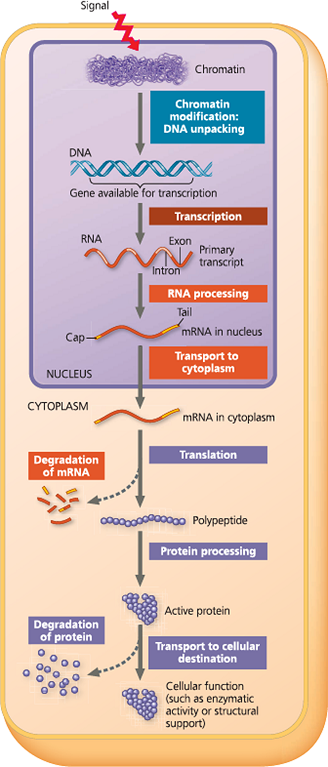
In all organisms, gene expression is commonly controlled at ___; regulation at this stage often occurs in response to signals coming from outside the cell, such as hormones or other signaling molecules
- transcription
Recall that the DNA of eukaryotic cells is ___
- packaged with proteins in an elaborate complex known as chromatin, the basic unit of which is the nucleosome. A nucleosome consists of a cluster of eight histone proteins around which the DNA double helix is wrapped
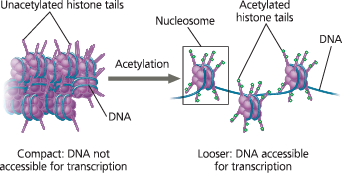
}}Histone Acetylation:}}
The attachment of acetyl groups to certain amino acids of histone proteins
}}Histone Tails:}}
The extension of the N-terminus of each histone protein in a nucleosome; accessible to various modifying enzymes that catalyze the addition or removal of specific chemical groups
A simple model of the effect of histone acetylation - Amino acids in histone tails may be chemically modified. For example, acetyl groups (green balls) may be added, causing the chromatin to be less compact and making the DNA accessible for transcription
}}DNA Methylation:}}
The presence of methyl groups on the DNA bases (usually cytosine) of plants, animals, and fungi; also refers to the process of adding methyl groups to DNA bases
When is DNA more methylated?
It is generally more methylated in long stretches of inactive DNA than in regions of actively transcribed DNA. It is generally more methylated in an individual’s genes in genes that are not expressed
}}Epigenetic Inheritance:}}
Inheritance of traits transmitted by mechanisms not directly involving the nucleotide sequence of a genome
Whereas mutations in DNA are ___, modifications to the chromatin can be _
- permanent; reversed
}}Control Element:}}
A segment of noncoding DNA that helps regulate transcription of a gene by serving as a binding site for a transcription factor. Multiple control elements are present in a eukaryotic gene’s enhancer
A eukaryotic gene and its transcript - Each eukaryotic gene has a promoter, a DNA sequence where RNA polymerase binds and starts transcription, proceeding “downstream.” A number of control elements (gold) are involved in regulating the initiation of transcription; these are DNA sequences located near (proximal to) or far from (distal to) the promoter. Distal control elements can be grouped together as enhancers, one of which is shown for this gene. At the other end of the gene, a polyadenylation (poly-A) signal sequence in the last exon of the gene is transcribed into an RNA sequence that signals where the transcript is cleaved and the poly-A tail added. Transcription may continue for hundreds of nucleotides beyond the poly-A signal before terminating. RNA processing of the primary transcript into a functional mRNA involves three steps: addition of the 5′ cap, addition of the poly-A tail, and splicing. In the cell, the 5′ cap is added soon after transcription is initiated, and splicing occurs while transcription is still under way
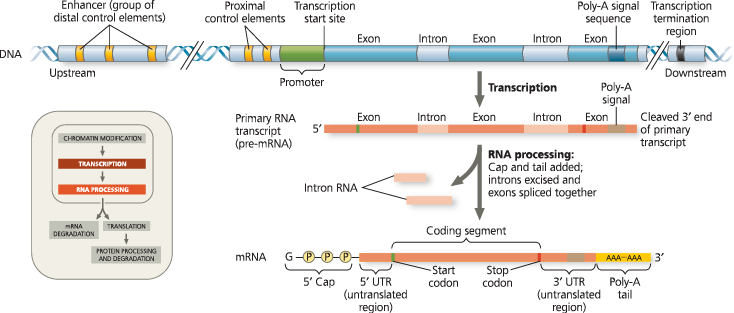
What are the types of transcription factors?
General transcription factors act at the promoter of all genes, while some genes require specific transcription factors that bind to control elements close to or further away from the promoter
}}General Transcription Factors:}}
Essential for the transcription of all protein-coding genes
}}Specific Transcription Factors:}}
In eukaryotes, high levels of transcription of these particular genes at the appropriate time and place depend on the interaction of control elements with another set of proteins
}}Proximal Control Elements:}}
Located close to the promoter
}}Distal Control Elements:}}
Located far from the promoter
}}Enhancers:}}
A segment of eukaryotic DNA containing multiple control elements, usually located far from the gene whose transcription it regulates
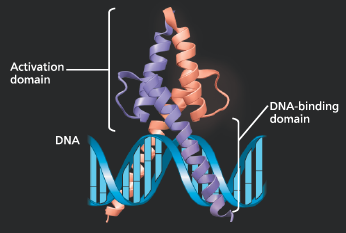
}}DNA-Binding Domain:}}
A part of the protein’s three-dimensional structure that binds to DNA
}}Activation Domains:}}
Bind other regulatory proteins or components of the transcription machinery, facilitating a series of protein-protein interactions that result in enhanced transcription of a given gene
MyoD, a transcriptional activator - The MyoD protein is made up of two subunits (purple and salmon) with extensive regions of α helix. Each subunit has one DNA-binding domain and one activation domain. The latter includes binding sites for the other subunit and for other proteins. MyoD is involved in muscle development in vertebrate embryos
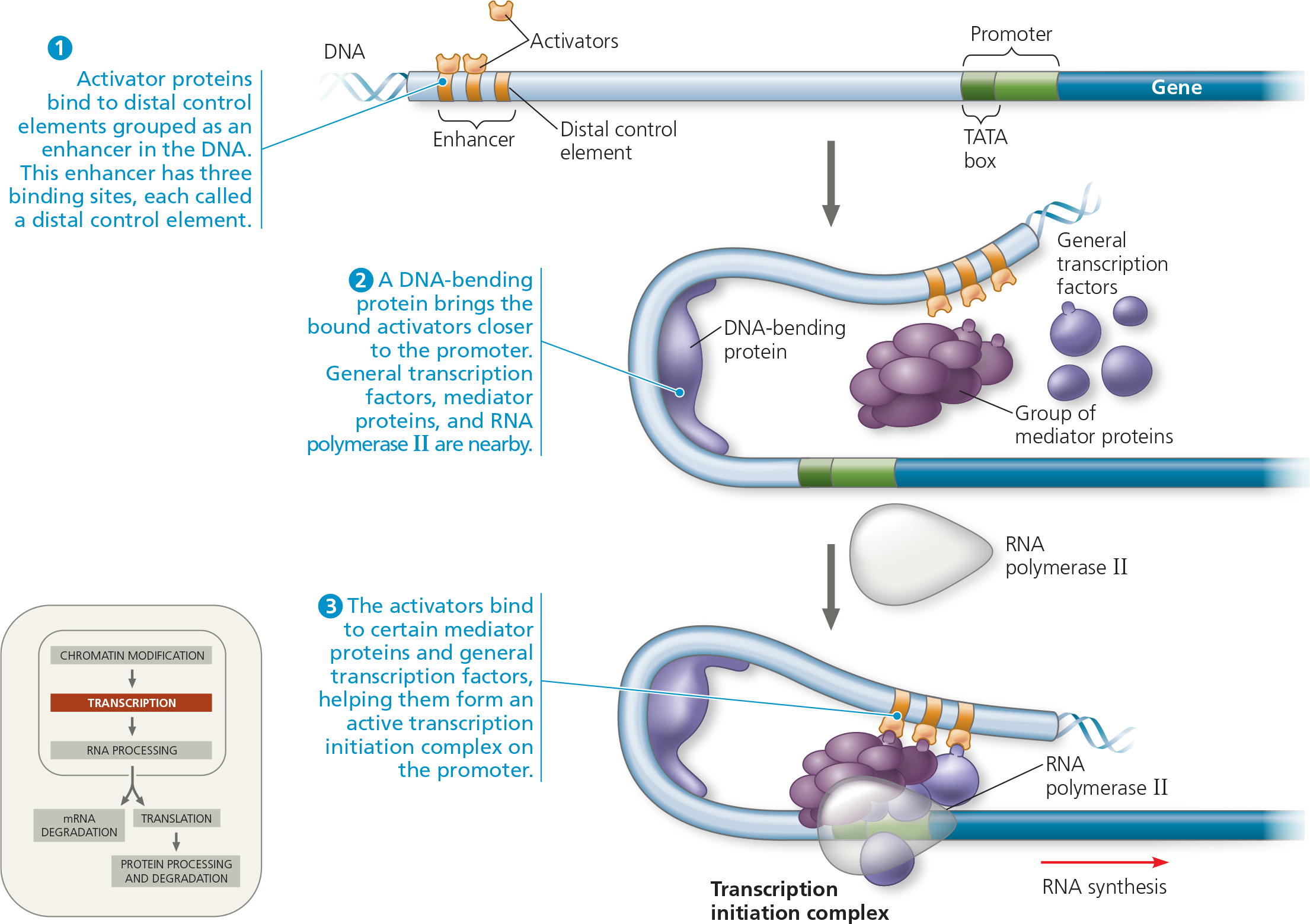
}}Mediator Proteins:}}
Interact with proteins at the promoter; brought into contact with bound activators after protein-mediated bending
A model for the action of enhancers and transcription activators - Bending of the DNA by a protein enables enhancers to influence a promoter hundreds or even thousands of nucleotides away. Specific transcription factors called activators bind to the enhancer DNA sequences and then to a group of mediator proteins, which in turn bind to general transcription factors and then RNA polymerase II, assembling the transcription initiation complex. These protein-protein interactions lead to correct positioning of the complex on the promoter and the initiation of RNA synthesis. Only one enhancer (with three gold control elements) is shown here, but a gene may have several enhancers that act at different times or in different cell types
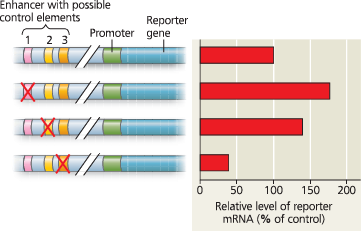
What Control Elements Regulate the Expression of the mPGES-1 Gene? - The diagrams on the left side of the figure show the intact DNA sequence (top) and the three experimental DNA constructs. A red X is located on the possible control element (1, 2, or 3) that was deleted in each experimental DNA construct. The area between the slashes represents approximately 8 kilobases of DNA located between the promoter and the enhancer region. The horizontal bar graph on the right shows the amount of reporter gene mRNA that was present in each cell culture after 48 hours relative to the amount that was in the culture containing the intact enhancer region (top bar = 100%)
}}Silencing:}}
Some repressors recruit proteins that remove acetyl groups from histones, leading to reduced transcription
It is the particular ___ of control elements in an enhancer associated with a gene, rather than a _, that is important in regulating transcription of the gene
- combination; unique control element
Cell type–specific transcription - Both liver cells and lens cells have the genes for making the proteins albumin and crystallin, but only liver cells make albumin (a blood protein) and only lens cells make crystallin (the main protein of the lens of the eye). The specific transcription factors made in a cell determine which genes are expressed. In this example, the genes for albumin and crystallin are shown at the top, each with an enhancer made up of three different control elements. Although the enhancers for the two genes both have a gray control element, each enhancer has a unique combination of elements. All the activator proteins required for high-level expression of the albumin gene are present in liver cells only (left), whereas the activators needed for expression of the crystallin gene are present in lens cells only (right). For simplicity, we consider only the role of activators here, although the presence or absence of repressors may also influence transcription in certain cell types
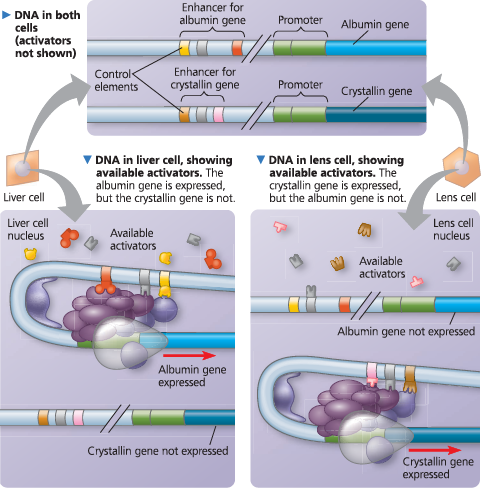
Describe the enhancer for the albumin gene in each type of cell. How would the nucleotide sequence of this enhancer in the liver cell compare with that in the lens cell?
In both types of cell, the albumin gene enhancer has the three control elements colored yellow, gray, and red. The sequences in the liver and lens cells would be identical, since the cells are in the same organism
Describe coordinated control of dispersed genes in a eukaryotic cell
They often occur in response to chemical signals from outside the cell (A steroid hormone, for example, enters a cell and binds to a receptor protein, forming a hormone-receptor complex that acts as a transcription activator)
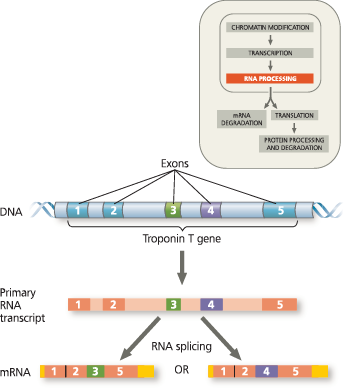
}}Alternative RNA Splicing:}}
A type of eukaryotic gene regulation at the RNA-processing level in which different mRNA molecules are produced from the same primary transcript, depending on which RNA segments are treated as exons and which as introns
Alternative RNA splicing of the troponin T gene - The primary transcript of this gene can be spliced in more than one way, generating different mRNA molecules. Notice that one mRNA molecule has ended up with exon 3 (green) and the other with exon 4 (purple). These two mRNAs are translated into different but related muscle proteins
How can gene expression be regulated at the initiation of translation?
The initiation of translation can be blocked by regulatory proteins that bind to specific sequences or structures within the untranslated region (UTR) at the 5′ or 3′ end, preventing the attachment of ribosomes
In general, what are the effects of histone acetylation and DNA methylation on gene expression?
Histone acetylation is generally associated with gene expression, while DNA methylation is generally associated with lack of expression
Compare the roles of general and specific transcription factors in regulating gene expression
General transcription factors function in assembling the transcription initiation complex at the promoters for all genes. Specific transcription factors bind to control elements associated with a particular gene and, once bound, either increase (activators) or decrease (repressors) transcription of that gene
Suppose you compared the nucleotide sequences of the distal control elements in the enhancers of three genes that are expressed only in muscle cells. What would you expect to find? Why?
The three genes should have some similar or identical sequences in the control elements of their enhancers. Because of this similarity, the same specific transcription factors in muscle cells could bind to the enhancers of all three genes and stimulate their expression coordinately
Describe what must happen in a cell before a gene specific to that type of cell can be transcribed
In that specific type of cell, the chromatin must not be tightly condensed because it must be accessible to transcription factors. The appropriate specific transcription factors (activators), which are made in that type of cell, must bind to the control elements in the enhancer of the gene, while repressors must not be bound. The DNA must be bent by a bending protein so the activators can contact the mediator proteins and form a complex with general transcription factors at the promoter. Then RNA polymerase must bind and begin transcription
15.3 - Noncoding RNAs play multiple roles in controlling gene expression
- Noncoding RNAs (for example, miRNAs and siRNAs) can block translation or cause degradation of mRNAs
}}Junk DNA:}}
DNA that does not contain meaningful genetic information, is not transcribed, and does not specify proteins or RNA (this idea was later contradicted)
}}Noncoding RNAs:}}
Genome transcribed into non-protein-coding RNAs
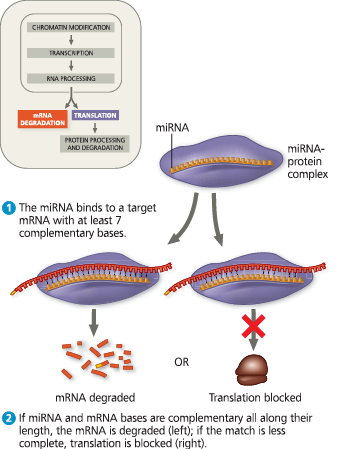
}}MicroRNA (miRNA):}}
A small, single-stranded RNA molecule, generated from a hairpin structure on a precursor RNA transcribed from a particular gene. The miRNA associates with one or more proteins in a complex that can degrade or prevent the translation of an mRNA with a complementary sequence
Regulation of gene expression by microRNAs (miRNAs) - An miRNA of about 22 nucleotides, formed by enzymatic processing of an RNA precursor, associates with one or more proteins in a complex that can degrade or block translation of target mRNAs
}}Small Interfering RNA (siRNA):}}
One of multiple small, single-stranded RNA molecules generated by cellular machinery from a long, linear, double-stranded RNA molecule. The siRNA associates with one or more proteins in a complex that can degrade or prevent the translation of an mRNA with a complementary sequence. In some cases, siRNA can also black transcription by promoting chromatin modification
}}RNA Interference (RNAi):}}
A technique used to silence the expression of selected genes. RNAi uses synthetic double-stranded RNA molecules that match the sequence of a articular gene to trigger the breakdown of the gene’s messenger RNA
What’s a possible way the RNAi pathway evolved?
Given that the cellular RNAi pathway can process double-stranded RNAs into homing devices that lead to the destruction of related RNAs, some researchers hypothesize that this pathway may have evolved as a natural defense against infection by such viruses
Many species, including mammals, apparently produce their own ___ precursors to _ such as siRNAs. Once produced, these RNAs can interfere with gene expression at stages other than translation
- long, double-stranded RNA; small RNAs
How can small RNAs impact chromatin structure?
The siRNA system interacts with other noncoding RNAs and with chromatin-modifying enzymes to condense the centromere chromatin into heterochromatin
}}Piwi-Associated RNAs (piRNAs):}}
A class of small noncoding RNAs that induces the formation of heterochromatin and blocks the expression of some parasitic DNA elements in the genome known as transposons
Suppose the mRNA being degraded in Figure 15.13 coded for a protein that promotes cell division in a multicellular organism. What would happen if a mutation disabled the gene for the miRNA that triggers this degradation?
The mRNA would persist and be translated into the cell division–promoting protein, and the cell would probably divide. If the intact miRNA is necessary for inhibition of cell division, then division of this cell might be inappropriate. Uncontrolled cell division could lead to formation of a mass of cells (tumor) that prevents proper functioning of the organism and could contribute to the development of cancer
Inactivation of one of the X chromosomes in female mammals results in a Barr body. Suggest a model of how noncoding RNA (XIST RNA) functions to cause Barr body formation
The XIST RNA is transcribed from the XIST gene on the X chromosome that will be inactivated. It then binds to that chromosome and induces heterochromatin formation. A likely model is that the XIST RNA somehow recruits chromatin modification enzymes that lead to formation of heterochromatin
Why are miRNAs called noncoding RNAs? Explain how they participate in gene regulation
miRNAs do not “code” for the amino acids of a protein—they are never translated. Each miRNA associates with a group of proteins to form a complex. Binding of the complex to an mRNA with a complementary sequence causes that mRNA to be degraded or blocks its translation. This is considered gene regulation because it controls the amount of a particular mRNA that can be translated into a functional protein
15.4 - Researchers can monitor expression of specific genes
In nucleic acid hybridization, a nucleic acid probe is used to detect the presence of a specific mRNA
In situ hybridization and RT-PCR can detect the presence of a given mRNA in a tissue or an RNA sample, respectively
DNA microarrays are used to identify sets of genes co-expressed by a group of cells. Their cDNAs can also be sequenced (RNA-seq)
L idk if i’m gonna finish these
Nucleic Acid Hybridization:
T
Nucleic Acid Probe:
I
Synthesized single-stranded DNA probe ![]()
![]()
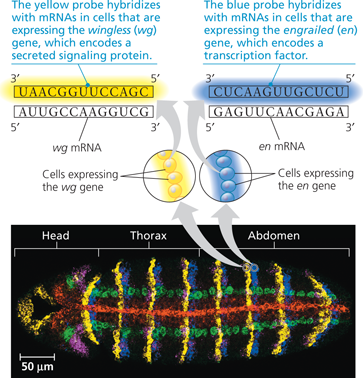
In Situ Hybridization:
A
Determining where genes are expressed by in situ hybridization analysis - This Drosophila embryo was incubated in a solution containing probes for five different mRNAs, each probe labeled with a different fluorescently colored tag. The embryo was then viewed using fluorescence microscopy. Each color marks cells in which a specific gene is expressed as mRNA. The arrows from the groups of yellow and blue cells above the micrograph point to a magnified view of nucleic acid hybridization of the appropriately colored probe to the mRNA
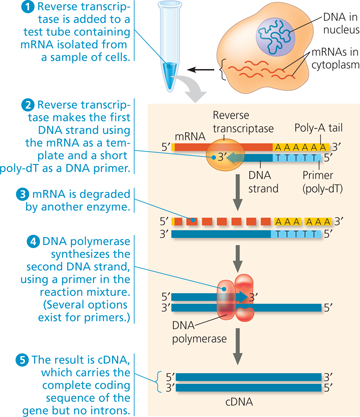
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR):
A
Complementary DNA (cDNA):
A
Making complementary DNA (cDNA) from eukaryotic genes - Complementary DNA is DNA made in a test tube using mRNA as a template for the first strand. Only one mRNA is shown here, but the final collection of cDNAs would reflect all the mRNAs that were present in the cel
DNA Microarray Assay:
A
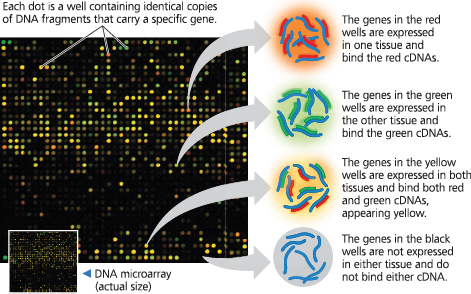
DNA microarray assay of gene expression levels - Researchers synthesized two sets of cDNAs, fluorescently labeled red or green, from mRNAs from two different human tissues. These cDNAs were hybridized with a microarray containing 5,760 human genes (about 25% of human genes), resulting in the pattern shown here. The intensity of fluorescence at each spot measures the relative expression in the two samples of the gene represented by that spot: Red indicates expression in one sample, green in the other, yellow in both, and black in neither
RNA Sequencing (RNA-seq):
A
Describe the role of complementary base pairing during RT-PCR and DNA microarray analysis
In RT-PCR, the primers must base-pair with their target sequences in the DNA mixture, locating one specific region among many. Also, the DNA polymerase (e.g., Taq polymerase) used in PCR relies on complementary base pairing to the template strand to add new nucleotides during synthesis of the fragments. In DNA microarray analysis, the labeled probe binds only to the specific target sequence due to complementary nucleic acid hybridization (DNA-DNA hybridization)
Study the microarray in Figure 15.17. If a sample from normal tissue is labeled with a green fluorescent dye, and a sample from cancerous tissue is labeled red, what color spots would represent genes that you would be interested in if you were studying cancer?
As a researcher interested in cancer, you would want to study genes represented by spots that are green or red because these are genes for which the expression level differs between the two types of tissues. Some of these genes may be expressed differently as a result of cancer, while others might play a role in causing cancer, so both would be of interest
What does detecting expression of specific genes tell a researcher?
The genes that are expressed in a given tissue or cell type determine the proteins (and noncoding RNAs) that are the basis of the structure and functions of that tissue or cell type. Understanding which groups of interacting genes establish particular structures and carry out certain functions will help us learn how the parts of an organism form and are maintained. We will also be better able to treat diseases that occur when faulty gene expression leads to malfunctioning tissues