Chapter 11: Serology Techniques: Past, Current, and Future
11.1: Introduction to Forensic Serology
- Forensic serology is the component of forensic biology that deals with the examination and identification of biological evidence.
- It focuses on determining the presence and identification of various bodily fluids such as blood, semen, and saliva in a questioned sample.
Class Characteristics and Individual Characteristics of Biological Evidence
- The identification of an unknown fluid sample is based on a comparison of the class characteristics of a sample with known standards of its class.
- Forensic identification typically involves bodily fluids, such as blood, semen, and saliva.
- If the presence of the bodily fluid is confirmed, the individual characteristics of the biological evidence are then determined to find out whether or not a bodily fluid sample has come from a particular individual.
- Short Tandem Repeats (STR): The most commonly utilized forensic DNA analysis used for human identification.
- Y-chromosomal STR analysis is often utilized for the investigation of sexual assault crimes.
- Mitochondrial DNA analysis is used for the identification of human remains.
- Establishing the probative value of a sample requires both the identification of its class characteristics and the individualization of its contributor.
Presumptive and Confirmatory Assays
The identification of bodily fluids can be carried out using presumptive and confirmatory assays to identify the type of bodily fluid in question.
Presumptive Assays
- Positive assay indicates the possibility of the presence of the bodily fluid in question.
- Negative assay indicates that the questioned bodily fluid is absent.
Confirmatory Assays
These assays are utilized to identify bodily fluids with higher certainty than presumptive assays.
These are performed when a sample has to be identified as blood.
Primary and Secondary Binding Assays
- Primary Binding Assays
- It involves the initial binding between a single epitope of an antigen and a single binding site of an antibody.
- They are very sensitive assays.
- Secondary Binding Assays
- They are less sensitive and easier to perform than primary binding assays.
- Precipitation-based assays have been used for species identification.
- Agglutination-based assays are more sensitive. It detects antigens located on the surface of cells or carriers, which are normally applied to blood group typing.
11.2: Primary Binding Assays
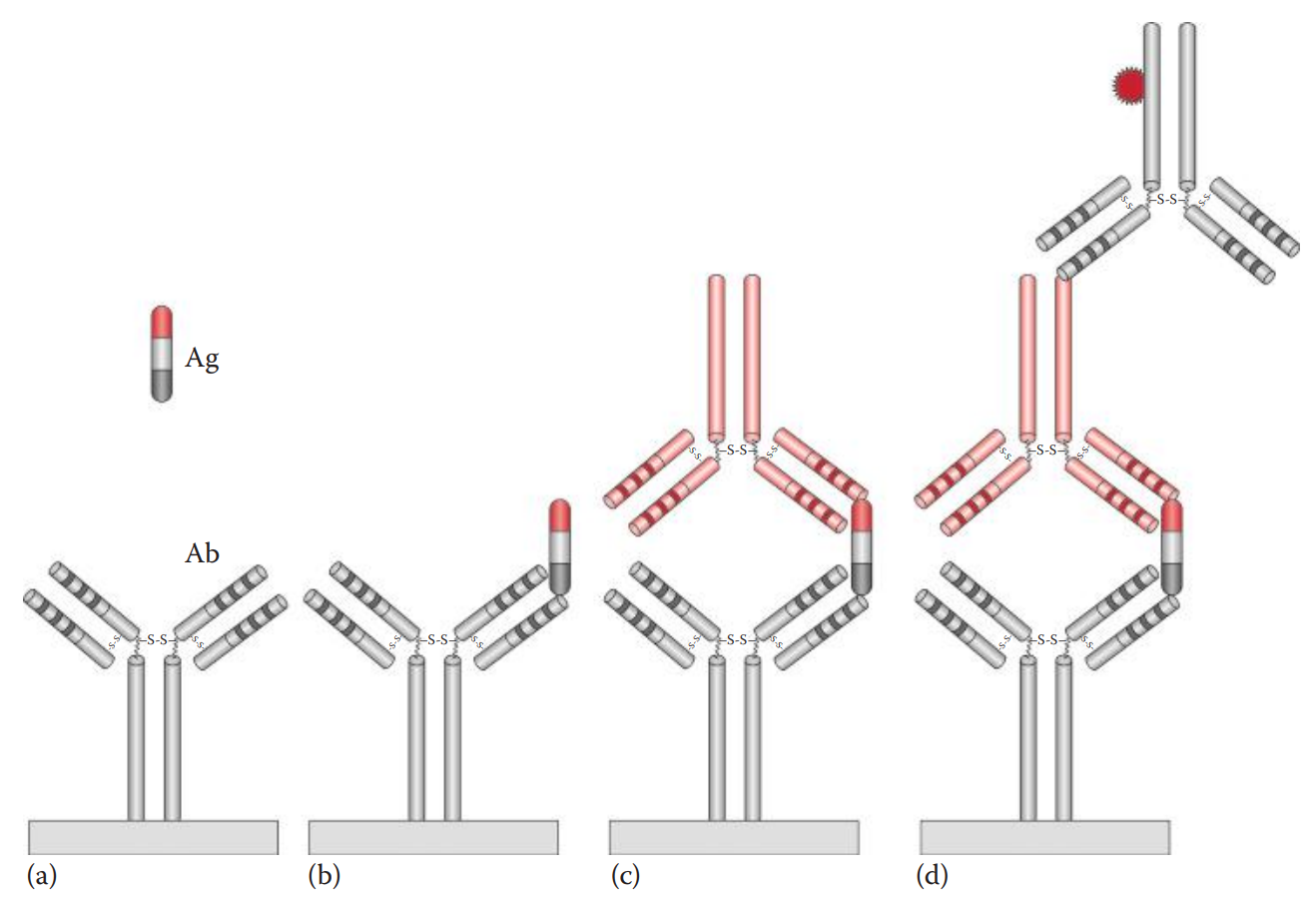
Enzyme-Linked Immunosorbent Assay (ELISA)
It is an immuno-enzyme assay that can be used to detect and measure the antibody or antigen in question.
Antibody-sandwich ELISA: The most common ELISA that is used in forensic serology.
- It is utilized to detect the prostate-specific antigen (PSA) to identify seminal stains and amylase for the identification of saliva.
An antibody coating is formed by nonspecific adsorption onto a solid phase such as the wells of a polystyrene plate.
Several enzymes such as alkaline phosphatase and horseradish peroxidase have been used as reporting enzymes to label the antiglobulin for ELISA.
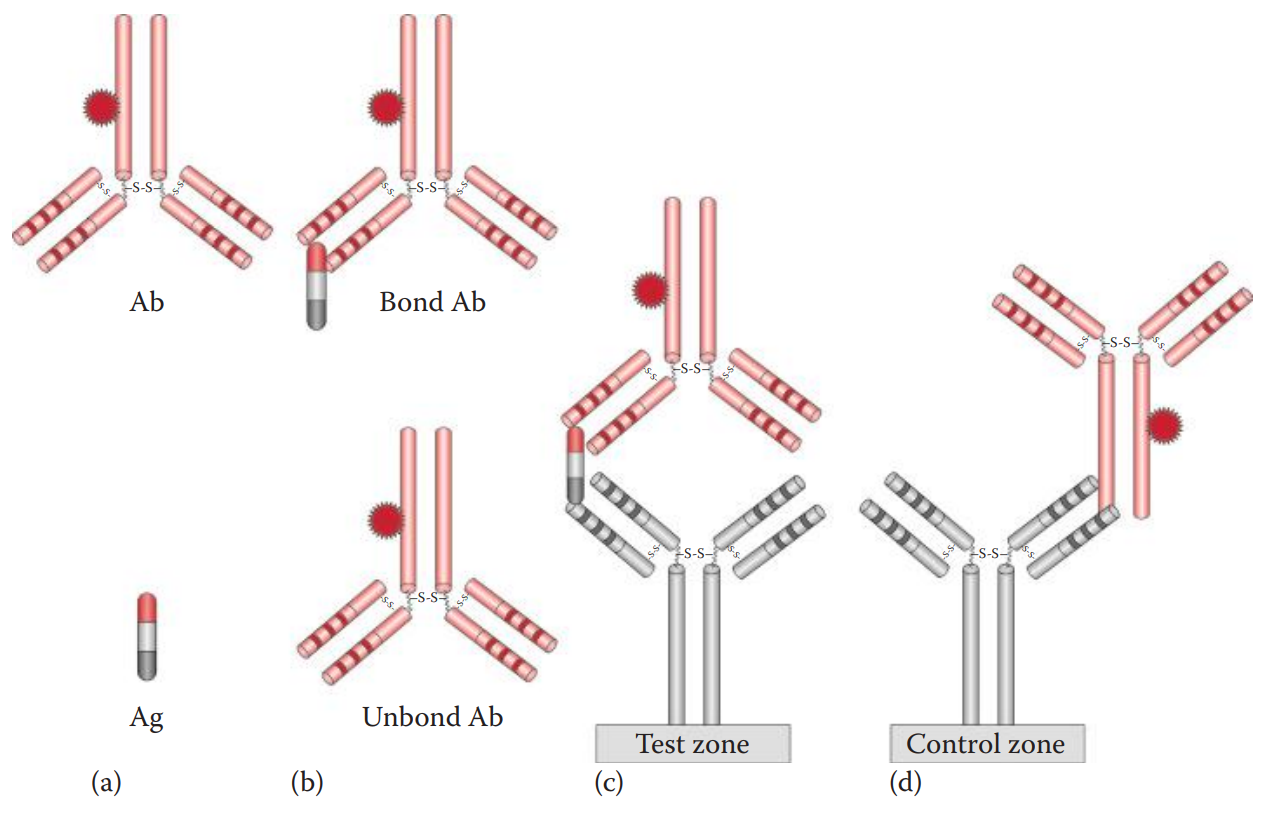
Immunochromatographic Assays
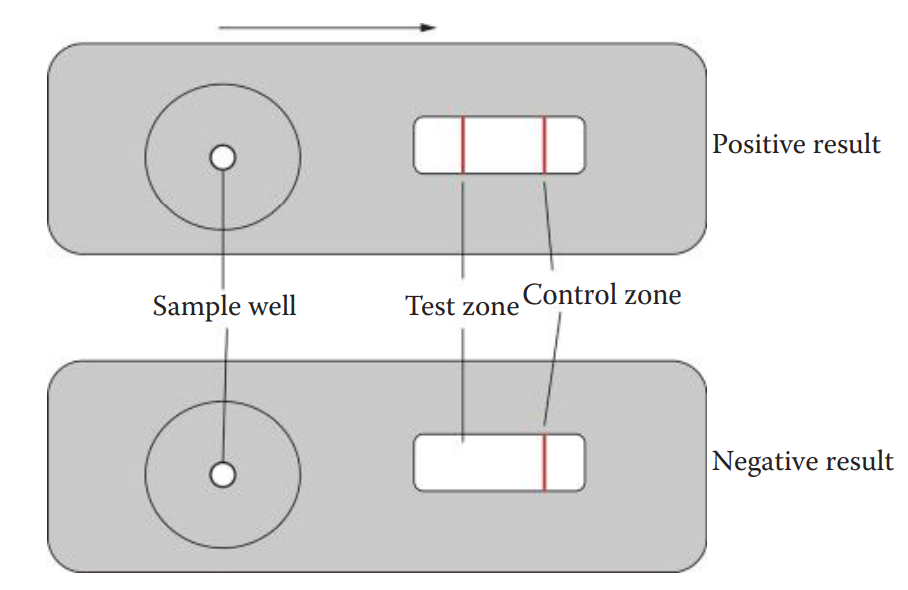
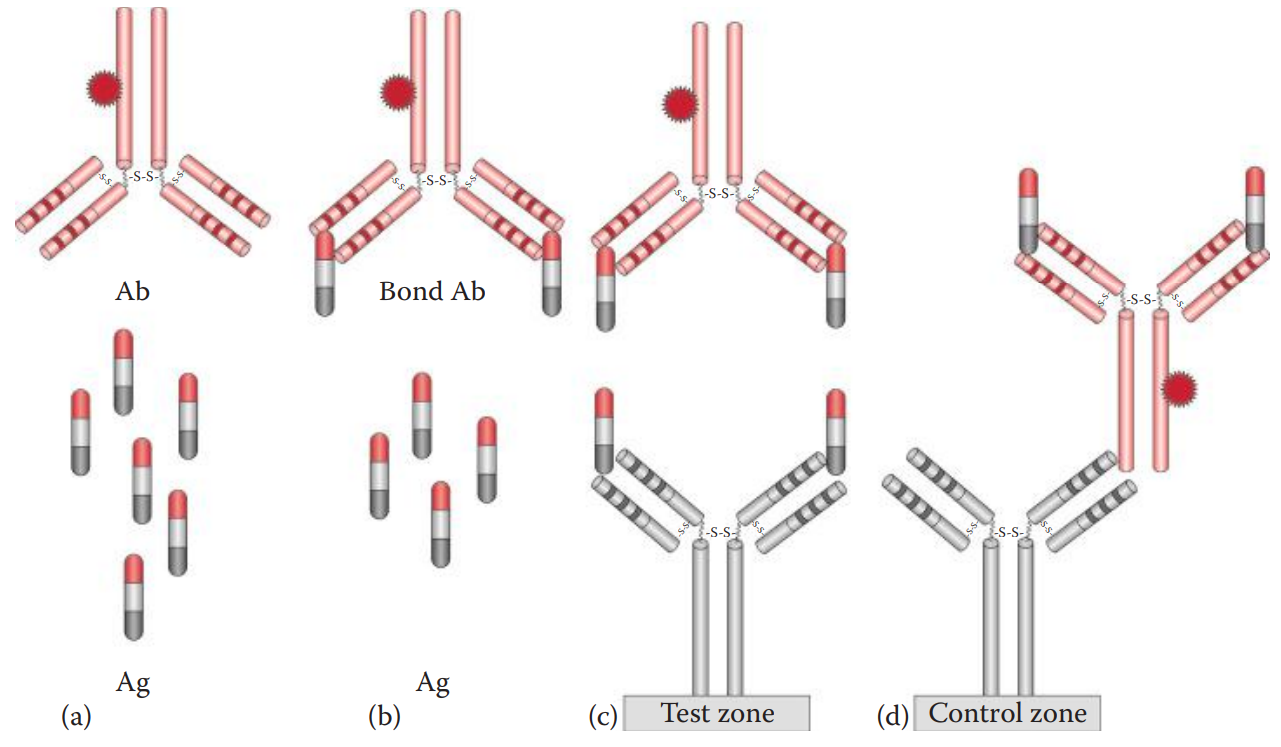
The assay is carried out by loading a sample into the sample well.
The antigen in the sample binds to the dye-labeled antibody already in the sample well to form an antigen–antibody complex.
The complex then diffuses across the nitrocellulose membrane until it reaches the test zone.
The antibody immobilized at the test zone traps the antigen–antibody complex to form an antibody–antigen–antibody sandwich.
The presence of the antigen in the sample results in a colored vertical line at the test zone.
The immunochromatographic device also utilizes a control zone to ensure that the device works properly and that the sample has diffused completely along the test strip.
Common Immunochromatographic Assays for Forensic Applications
| Assay | Antigen | Labeled Antibody | Immobilized Antibody | Forensic Application |
|---|---|---|---|---|
| ABAcard®; HemaTrace® | Hemoglobin (Hb) | Monoclonal anti-human Hb antibody | Polyclonal anti-human Hb antibody | Blood and species identification |
| RSID-Blood | Glycophorin A (GPA) | Monoclonal antihuman GPA antibody | Monoclonal antihuman GPA antibody | Blood and species identification |
| RSID-Saliva | Human salivary α-amylase (HAS) | Monoclonal antihuman HAS antibody | Monoclonal antihuman HAS antibody | Saliva identification |
| One-Step ABAcard PSA® | Prostate-specific antigen (PSA) | Monoclonal antihuman PSA antibody | Polyclonal antihuman PSA antibody | Semen identification |
| RSID-Semen | Semenogelin (Sg) | Monoclonal antihuman Sg antibody | Monoclonal antihuman Sg antibody | Semen identification |
11.3: Secondary Binding Assays
Precipitation-Based Assays
Immunodiffusion
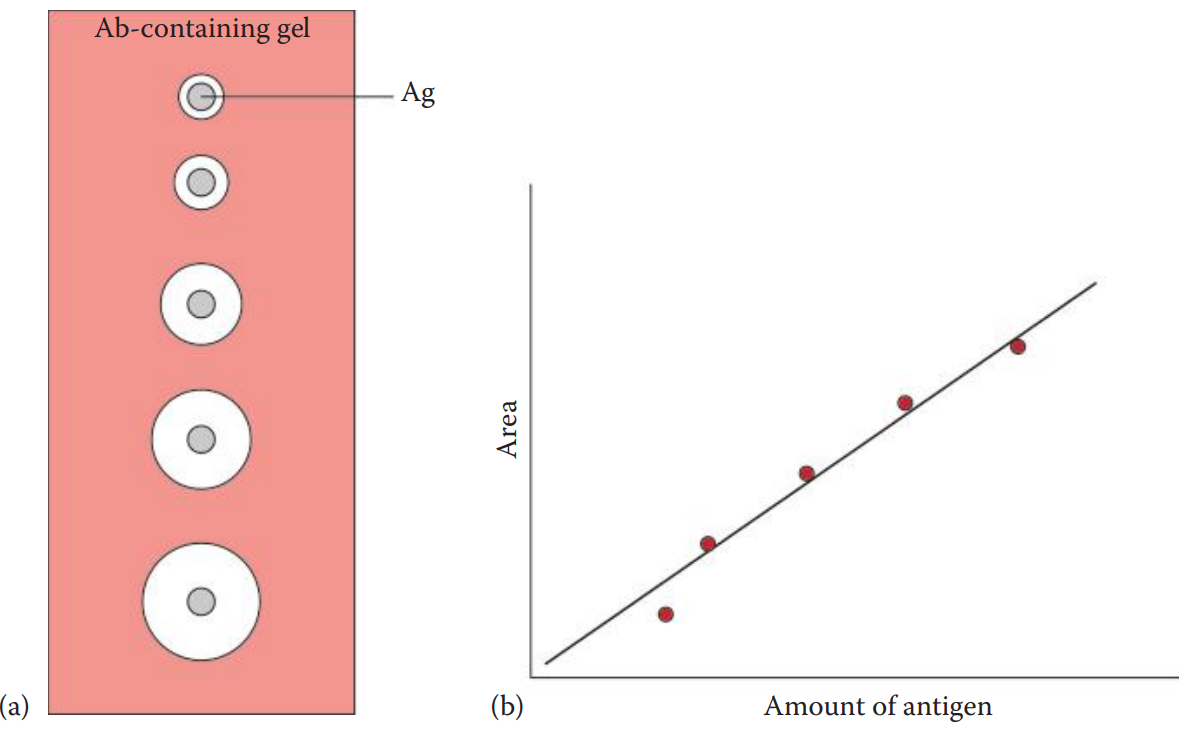
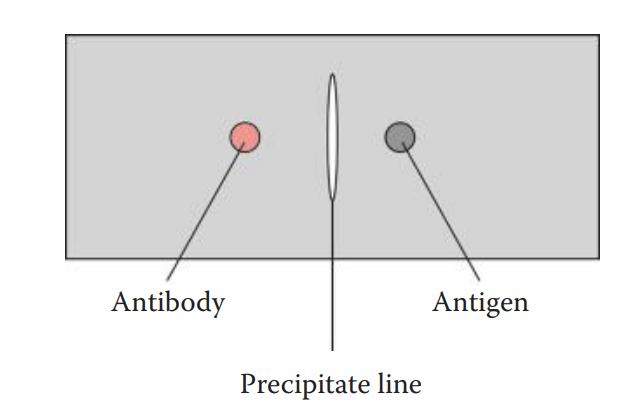
A passive method in which an antigen or an antibody or both are allowed to diffuse and therefore a gradient, from low to high concentration, is established for an antigen or an antibody or both.
Single Immunodiffusion: A concentration gradient is established for either an antigen or an antibody.
Double Immunodiffusion: A concentration gradient is established for both an antigen and an antibody.
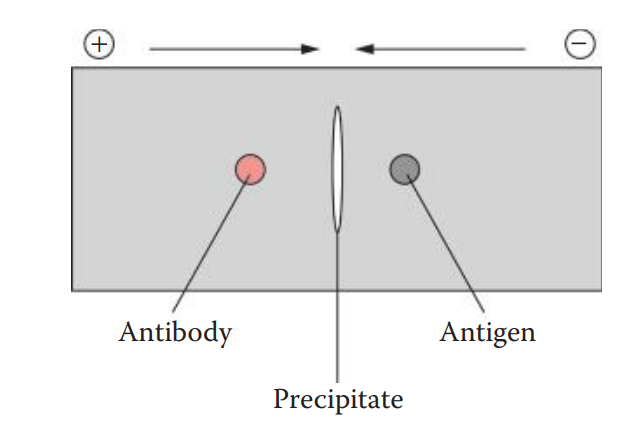
The Ouchterlony assay is named after the Swedish immunologist, Örjan Ouchterlony, who developed it. The assay can be performed in an agarose gel supported by a glass slide or polyester film
Immunoelectrophoretic Methods
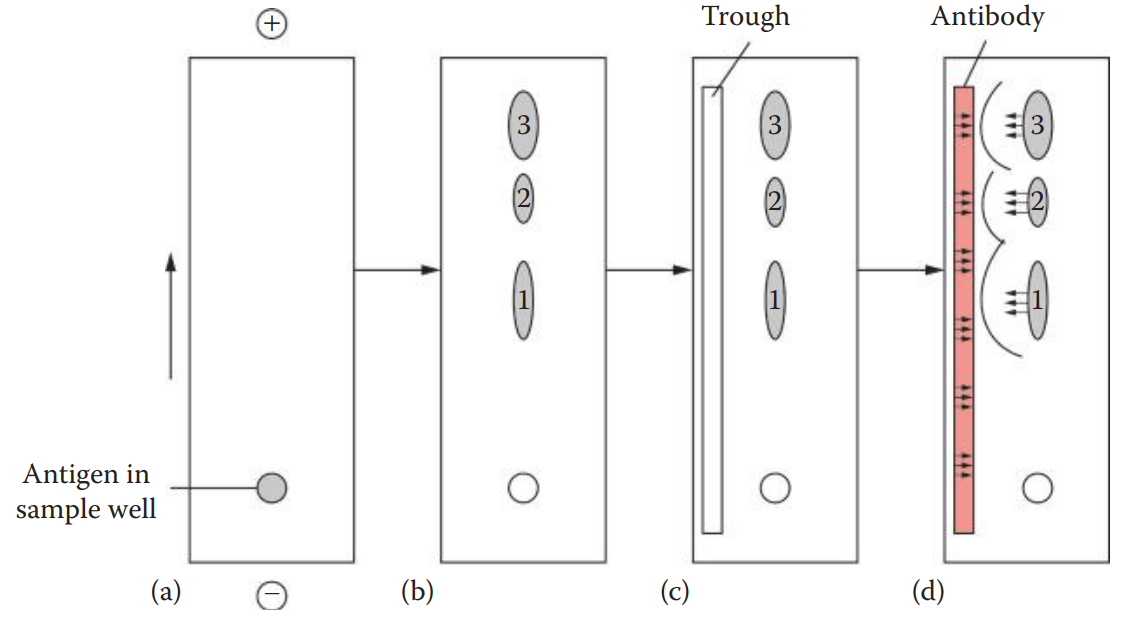
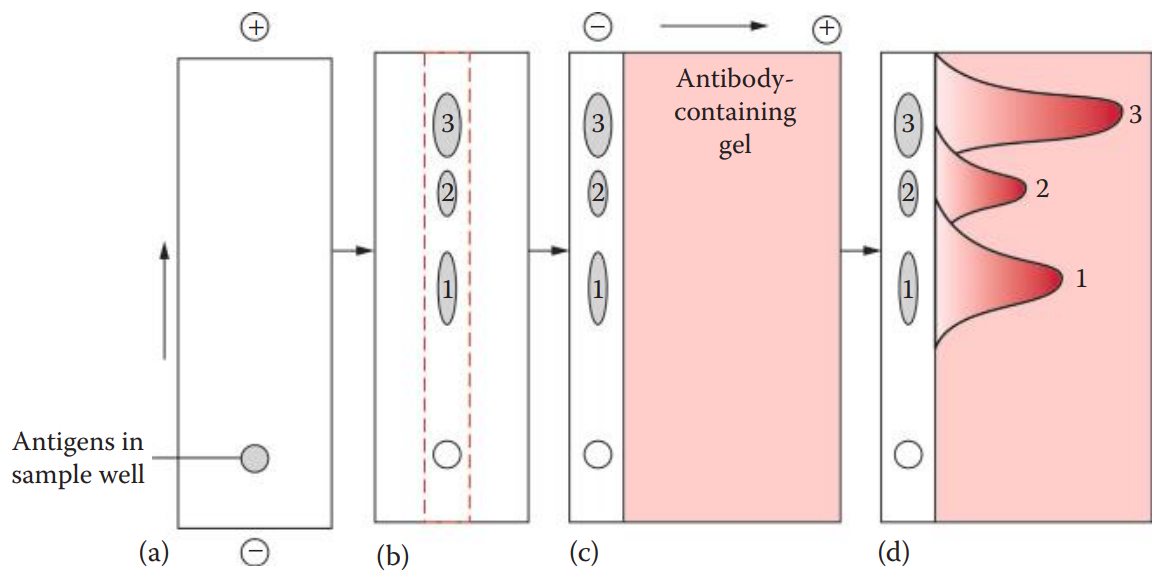
Immunoelectrophoresis (IEP): This technique uses electrophoresis to separate the antigen mixture before immunodiffusion.
Crossed Immunoelectrophoresis (CRIE): Also known as two-dimensional IEP, is a modification of IEP.
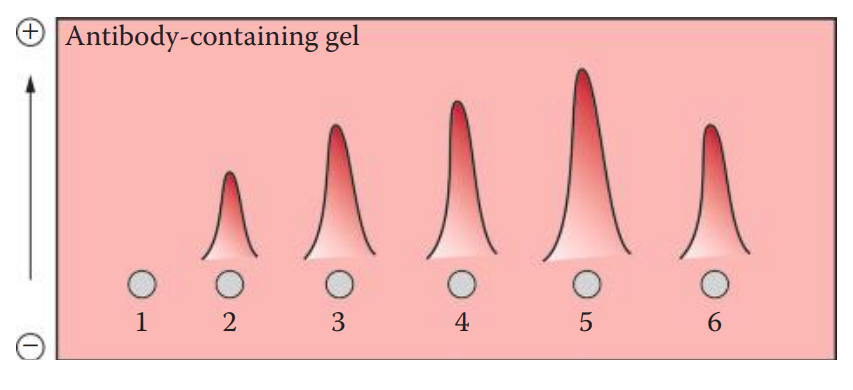
Rocket Immunoelectrophoresis: An antibody-containing agarose gel is used. The antigen is loaded into the well.
Crossed-Over Immunoelectrophoresis: This technique is also known as counterimmunoelectrophoresis (CIE).
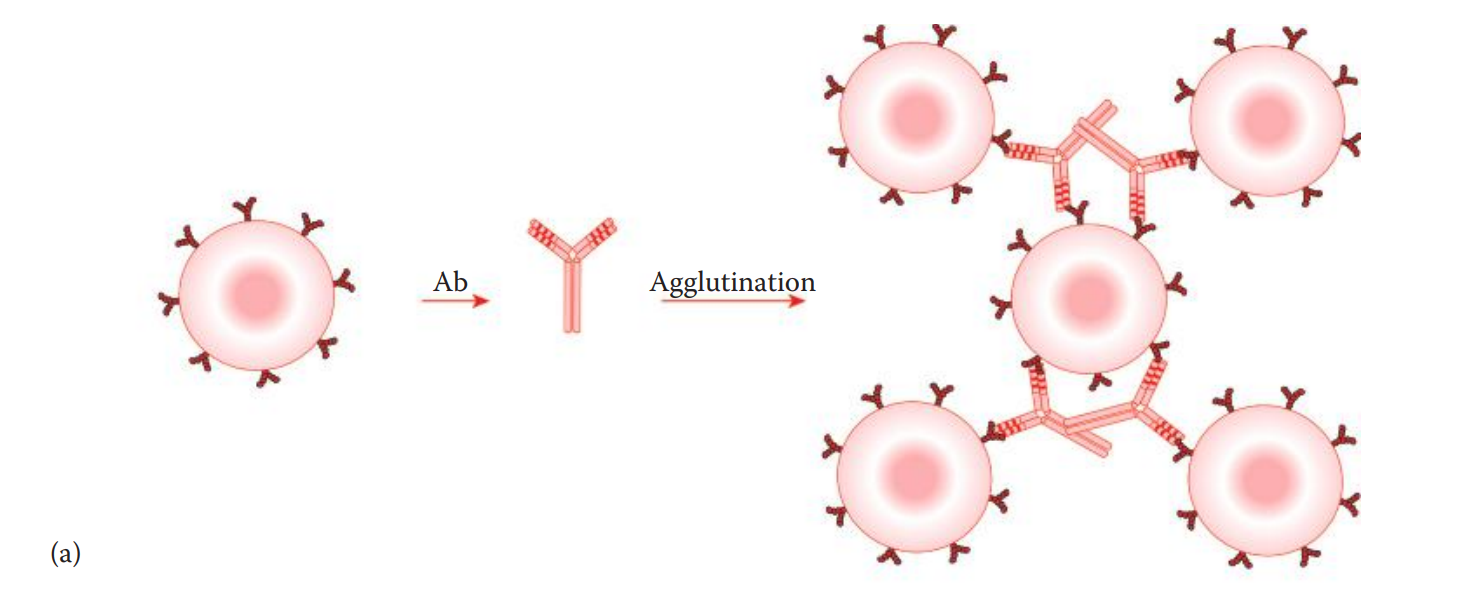
Agglutination-Based Assays
Agglutination reactions can be used as forensic serological assays such as for blood group typing and menstrual blood identification.
Agglutination assays are qualitative, indicating the absence or presence of antigens or antibodies.
Semi-quantitative assays results can be obtained by titration.
Direct agglutination assays involve reactions in which an antibody interacts with antigens originally located on cell surfaces.
In a hemagglutination reaction, an antibody binds to the antigens located on erythrocytes. This method is used for the identification of blood types.
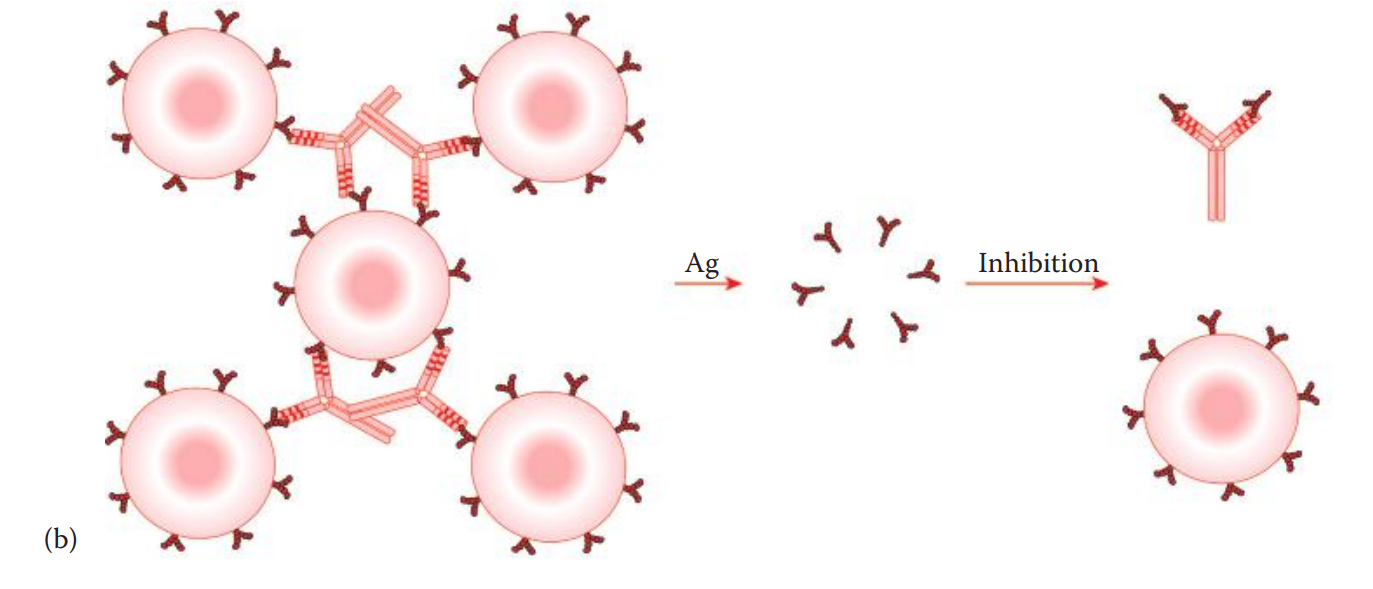
Agglutination inhibition assays: The presence of an antigen in question is indirectly detected.
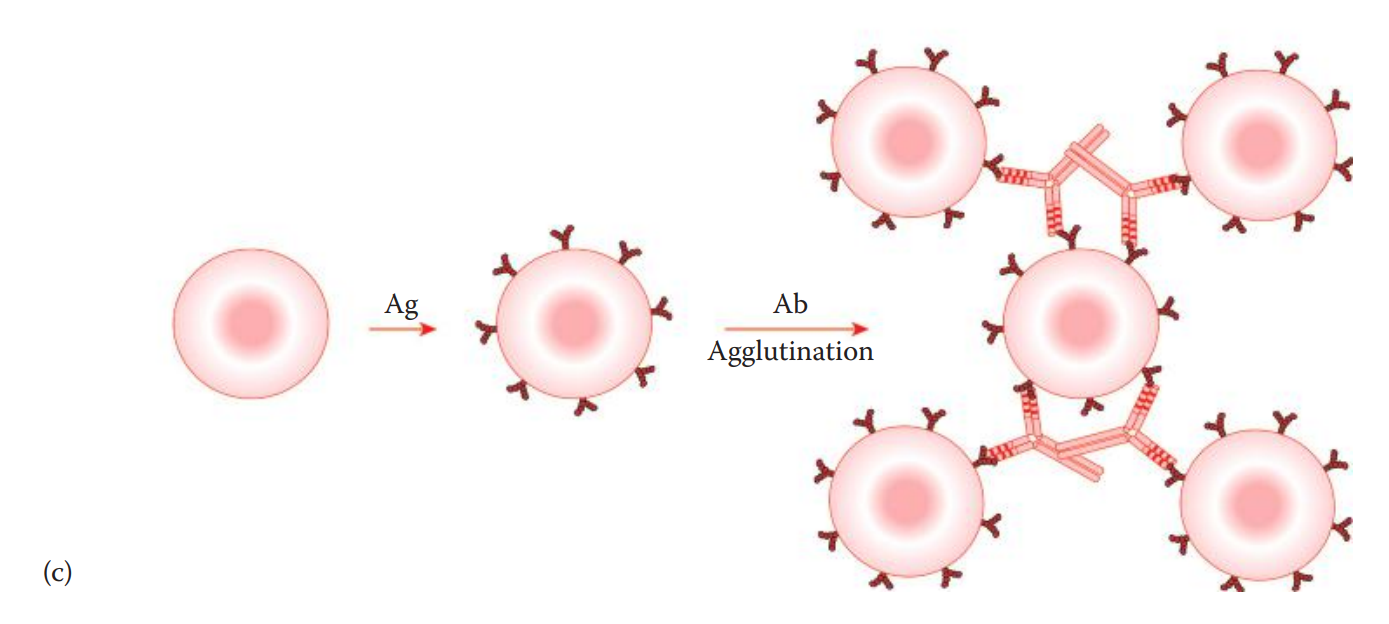
Passive agglutination assays: The antigen is coated on the surface of carrier cells such as tannic acid–treated sheep erythrocytes.
11.4: DNA Methylation Assays for Bodily Fluid Identification
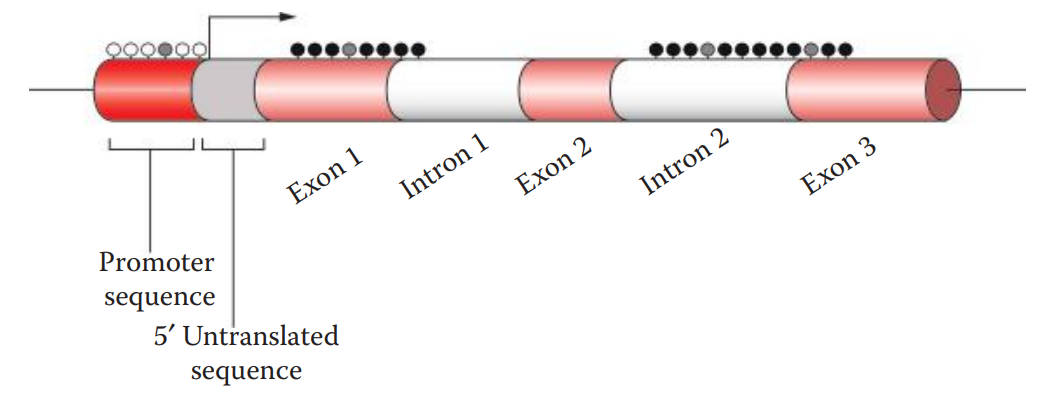
In the eukaryotic genome, methylation occurs at the cytosine residues commonly in the CpG dinucleotide sequences of both DNA strands.
The “p” in CpG refers to the phosphodiester bond between cytosine and guanine.
Cytosine methylation is carried out in vivo by a methyltransferase.
Genomic Loci: Also known as tissue-specific differentially methylated regions (TDMRs).
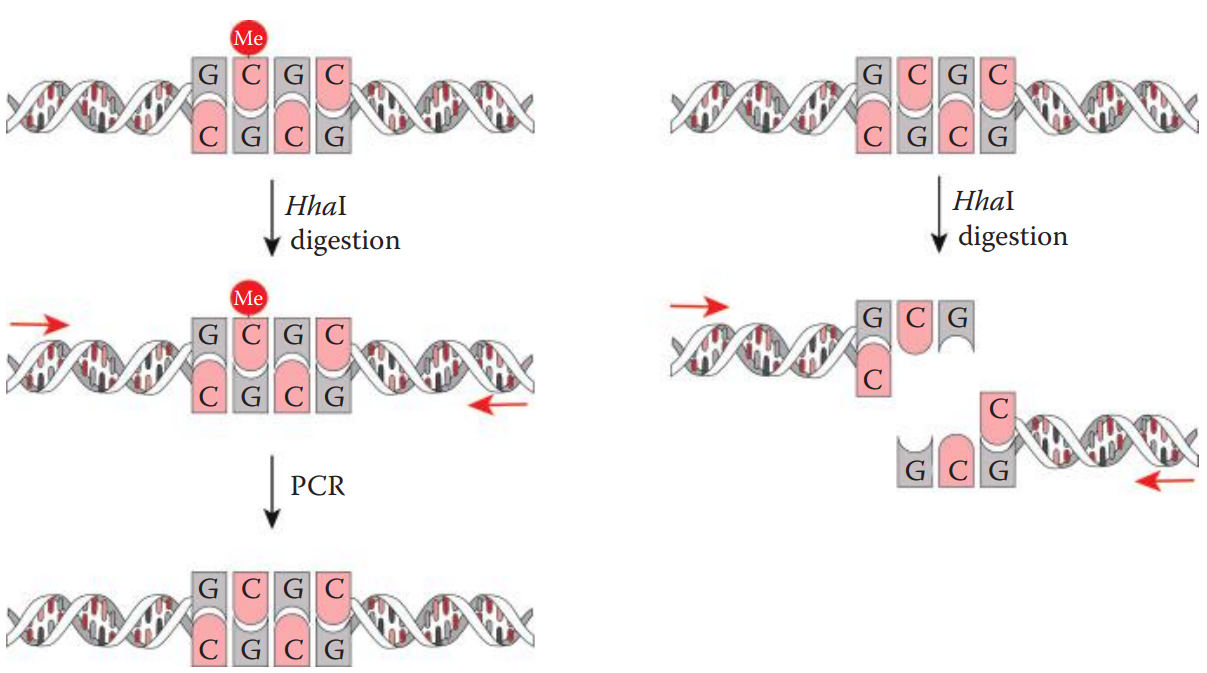
Methylation-sensitive restriction enzyme digestion polymerase chain reaction (MSRE-PCR) can be used for the rapid detection of DNA methylation.
- This method utilizes the methylation-sensitive restriction enzyme (MSRE).
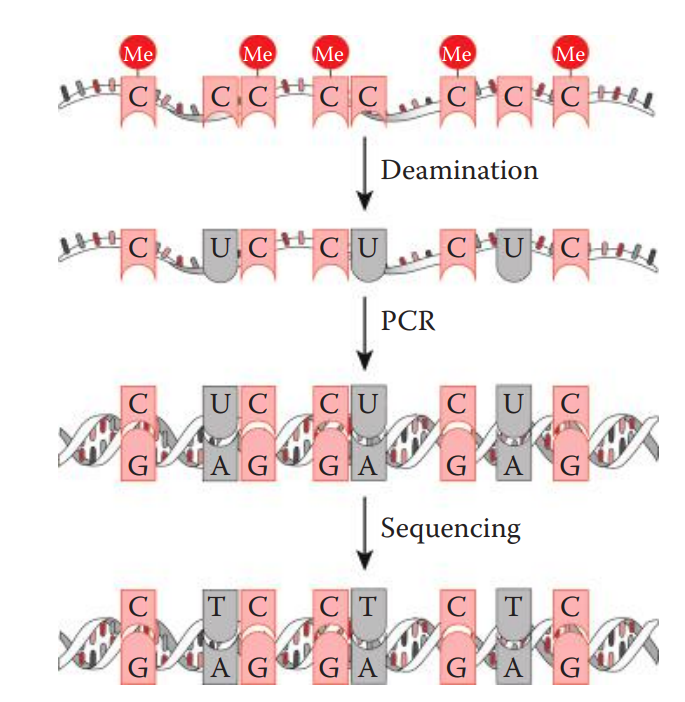
Bisulfite sequencing: The genomic DNA is treated with sodium bisulfite, which catalyzes the hydrolytic deamination of unmethylated cytosines.
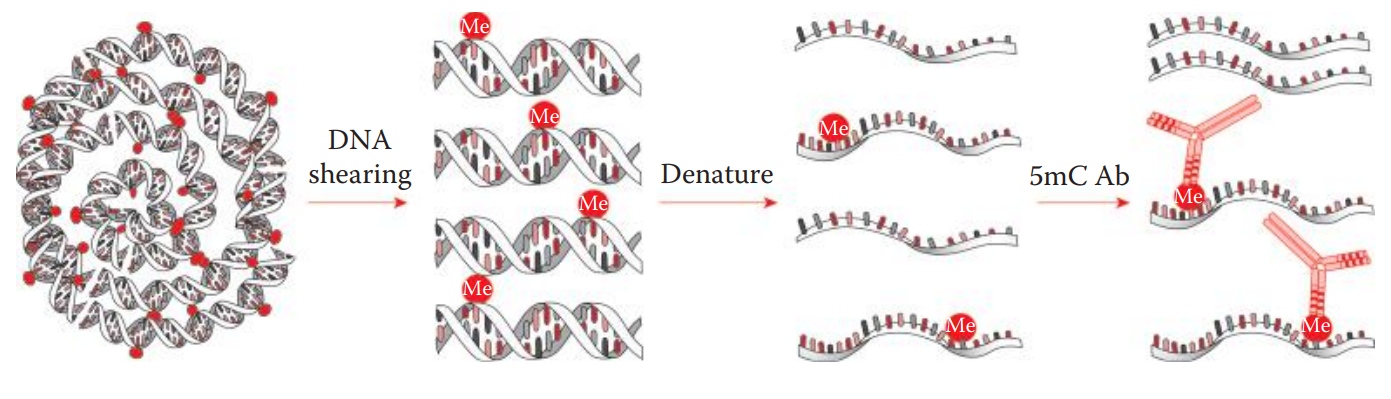
Methyl-DNA immunoprecipitation (MeDIP) is a useful method for isolating methylated DNA.
11.5: Forensic Applications of RNA-Based Assays and RNA Profiling
Messenger RNA-Based Assays
- mRNA-based Assays can potentially be used for wound age estimation and the age of biological stains.
- The tissue-specific genes that are utilized for bodily fluid identification.
- Reference genes: Constitutively expressed housekeeping genes are utilized as internal controls.
MicroRNA-Based Assays
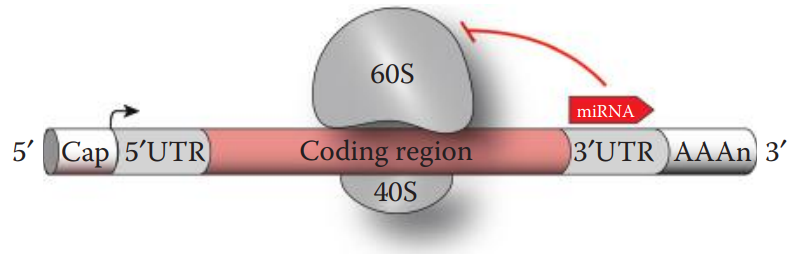
The biological function of microRNAs (miRNAs) is to regulate gene expression.
The mature miRNA strand associated with the RNA-induced silencing complex (RISC) binds to its target mRNA.
A single miRNA can bind different mRNA transcripts encoded by multiple genes.
Animal miRNAs usually form a base pairing with their target mRNAs through partial complementary sequences, which lead to translation repression to inhibit protein synthesis.
If the base pairing is exactly complementary to its target mRNA sequence, then the cleavage and the degradation of the target mRNA occur.
11.6: Proteomic Approaches Using Mass Spectrometry for Bodily Fluid Identification
- Mass spectrometry (MS) is a susceptible and rapid technique for protein identification from complex biological samples.
Mass Spectrometric Instrumentation for Protein Analysis
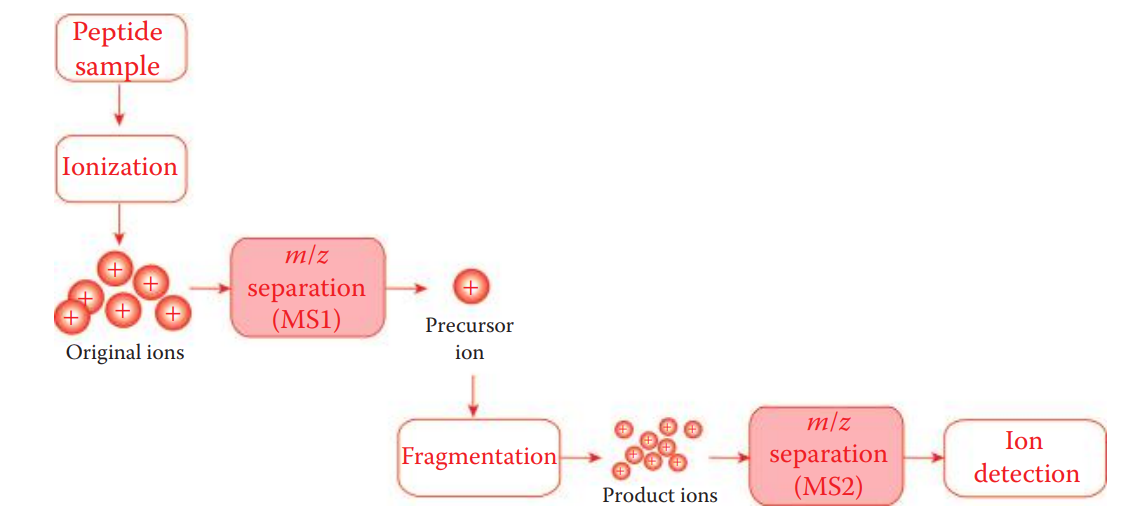
Mass spectrometer: An analytical technique to identify a molecule by measuring the ratio of the mass (m) to the charge (z) of a charged molecule.
- A typical mass spectrometer instrument analyzes ionized molecules while in their gas phase.
Ion source: Converts analytes into gaseous phase ions through ionization.
Ionization: A process used by gaining a positive or a negative charge from a neutral species.
Mass Analyzer: A part of a mass spectrometer where ions are accelerated under electric fields and separated based on their m/z ratio.
Biomarker proteins can be identified by peptide sequencing using the tandem mass spectrometry (MS/MS) mode of operation in which a mass spectrometer uses two or more mass analyzers.
After an initial mass analysis, individual peptide ions in the mass spectrum can be selected. The selected ions are known as precursor ions.
A precursor ion is then subjected to the second round of fragmentation through collision and is broken into smaller ions known as product ions.
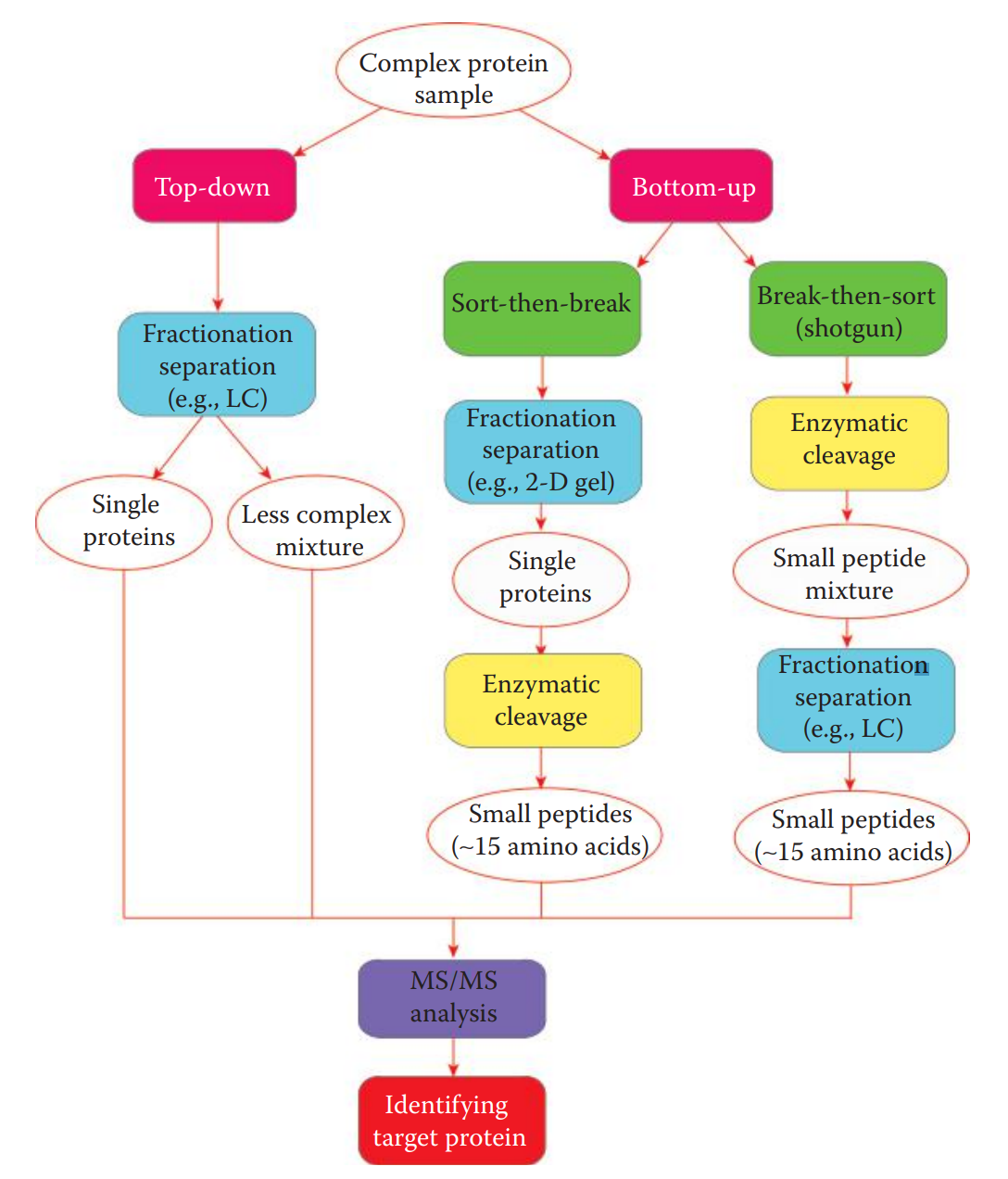
Analysis Strategies for Protein Identification
In top-down strategy, intact proteins in a complex mixture are fractionated and separated into less complex protein mixtures or single proteins.
The bottom-up strategy is commonly used for high-throughput analysis of small peptides derived from highly complex samples.
Sort-then-break approach: Proteins in a complex mixture are first isolated and then enzymatically cleaved.
Break-then-sort approach: Also known as the shotgun approach, wherein proteins are cleaved first and the resulting peptide mixture is then separated by LC and analyzed by tandem MS.
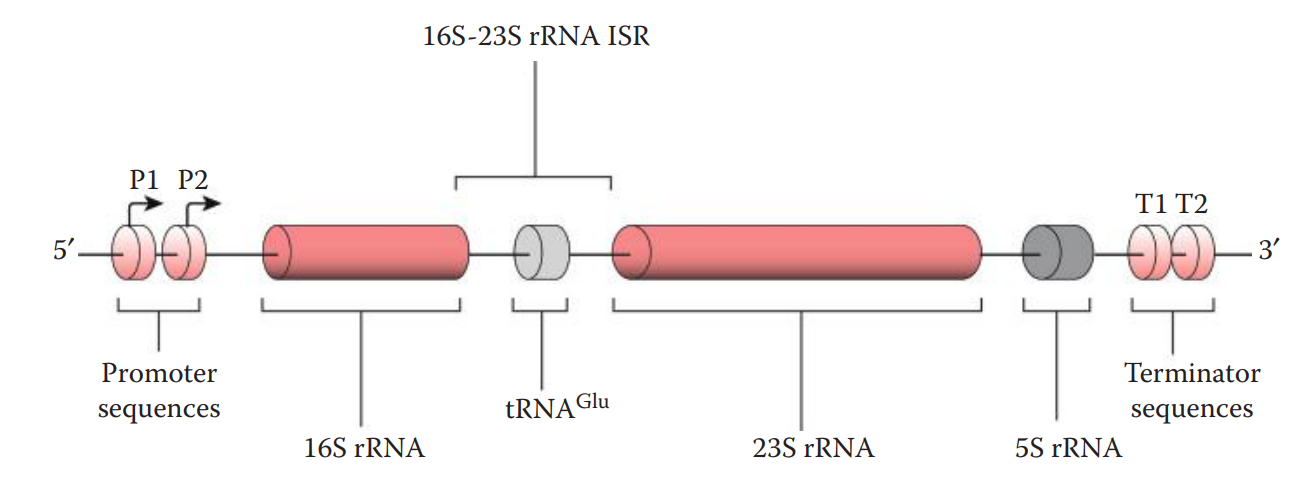
11.7: Microbial DNA Analysis for Bodily Fluid Identification
Human Microbiota: Microbial community.
Metagenomics: Allows the study of the microbial genome of complete microbial communities harvested directly from natural environments.
The bacterial rRNA operon contains three rRNA genes:
- 5S and 23S rRNA genes encode the RNA components of the large subunit of the ribosome.
- 16S rRNA gene encodes the RNA component of the small subunit of the ribosome.
The intergenic spacer region (ISR) between the 16S rRNA and 23S rRNA genes in the rRNA operon is another commonly used marker.
11.8: Nondestructive Assays for the Identification of Bodily Fluids
Fluorescence: The emission of light by a fluorophore.
Fluorophore: A moiety in a molecule that fluoresces on absorbing the energy from an excitation light source or radiation.
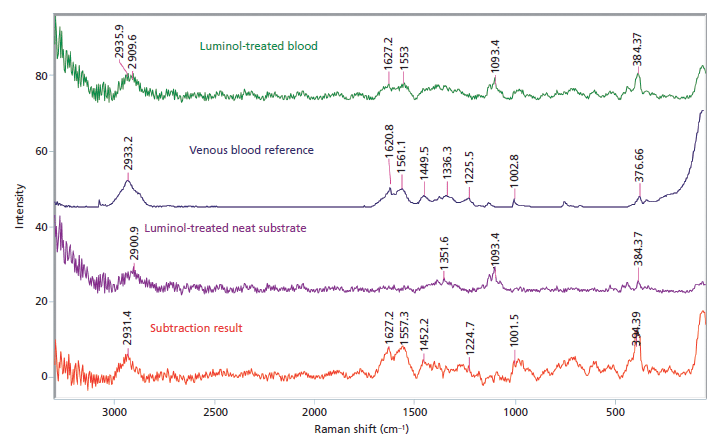
Raman spectroscopy: Utilizes a near-infrared excitation light source and measures the scattering of laser light caused by the vibrating molecules of a sample.