DTU BIO 2 - Information flow in biology
- Be able to compare combinatorial diversity of DNA/RNA and protein sequences of different lengths
- If given the table of the genetic code, you need to be able to translate an RNA sequence into an amino acid sequence and vice versa.
- Be able to determine/estimate how many (open) reading frames there are in a given piece of dsDNA or in a piece of DNA of given length (see exercises).
- Be able to understand the mathematics of the genetic code and perform simple calculations of alternative genetic codes, e.g. using doublets instead of triplets, or more or less nucleotides.
- Be able to compute the quantity of information stored in a piece of DNA (see exercises).
Other concepts you need to know about qualitatively:
- What are genotype and phenotype of an organism?
Genotype: genes, DNA
Phenoypes: apperance, analog.
- What is the principal direction of information flow in the central dogma of molecular biology?
○ Genetic information is converted from DNA to RNA to protein; never from protein to RNA or DNA
- What is the role/significance of the DNA double strand structure?
○ Its helpful to replicate DNA into two new strands
- By what mechanism is the DNA compacted such that it fits into the nucleus?
DNA is a million times smaller inside a living cell than it would be in a solution
Chromatin is what enables this compactness.
- What is the principle of the PCR reaction and what is it used for?
DNA Amplification
The polymerase chain reaction (PCR)
Uses thermostable polymerase.
Can be cycled (amplified) indefiently.
(1) denaturation of the template into single strands; (2) annealing of primers to each original strand for new strand synthesis; and (3) extension of the new DNA strands from the primers.
- What are the overall ideas behind DNA sequencing by synthesis and nanopore DNA sequencing?
○ Synthesis
2nd gen: Sequencing by synthesis.
Add fluo, mix, cleave, repeat.
Cheap, effective
○ Nanopore
§ Nanopore sequencing is a third generation approach used in the sequencing of biopolymers. Low cost
- What is the chemical difference between DNA and RNA?
○ A single extra O. Usually one stranded.
○
○
- What are the different roles of DNA and RNA in an organism?
DNA is used for long term information storage (half life of 100 or 1000s of years)
RNA is used for short term data (half life of hours)
RNA is cheaper to produce in terms of energy.
- What happens in translation? What is the ribosome?
○ Translation is the decoding of RNA into proteins
○ The ribosome is macromolecular machine found in all cells and it handles the translation.
- How many different amino acids are used to make proteins? How are they encoded by nucleotides in RNA?
○ 20 amino acids make up all proteins.
○ They are read in triplets within the open reading frame.
- What is kinetic proof reading and why does it exist?
○ Is the introduction of a high energy intermediate. So we have more places in the reaction where the wrong building block (tRNA) can detach.
○ It costs more energy but it reduces error rates in translation.
- Why does gene expression need to be regulated?
○ It can have unforseen consequences and is not well enough understood. One gene is not only responible for one thing. (eg. Height is determined by 400 genes).
- What is an operon? How does the lac operon work (schematically)?
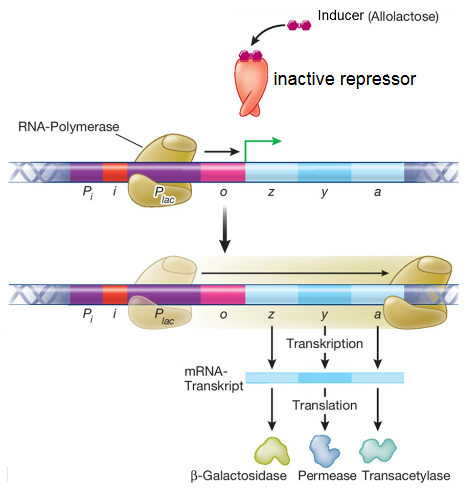
An operon is a cluster of genes under the control of a single promoter and is typically associated with a regulatory mechanism that coordinates the expression of those genes. Lac operon is regulater of lactose
- What are epigenetic modifications?
Epigenetic modifications refer to changes in gene expression that do not involve alterations to the underlying DNA sequence.
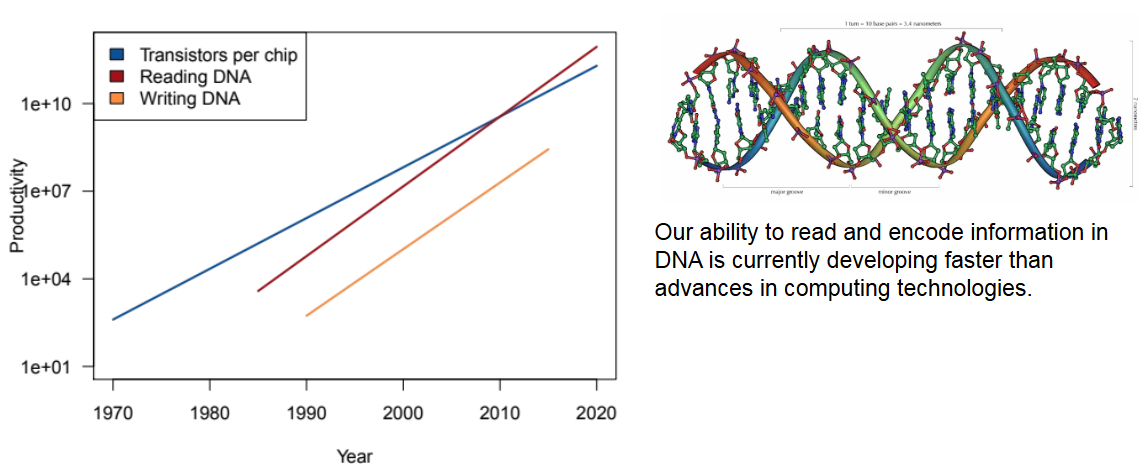
- What are advantages and disadvantages of DNA as a data storage medium?
pro: high density storage, Stability in amount of errors, energy efficiency
con: high cost of reading DND, Speed at retrival, complexity of the process
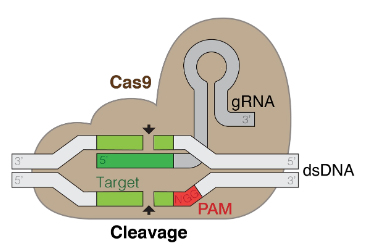
- What is CRISPR-Cas and what is it used for?
(Clustered Regularly Interspaced Short Palindromic Repeats)
cut and paste for targeted gene editing, allowing scientists to modify DNA sequences with precision and efficiency.