lecture 10: DNA topoisomerases 2
early structures of topoisomerases provided detailed structural information regarding individual parts of the topoisomerase machine. A significant challenge remains in understanding how all the molecular events are coordinated in order to bring about strand passage.
The paper by JM Berger shows the structural basis for gate-DNA recognition and bending by type 2a topoisomerases. The G segment is how the piece of DNA is bound in the protein. TOPRIM is a conserved rossman fold domain found in topoisomerase and DNA primases.. WHD is the winged helix domain which contains the active site tyrosine and tower which is a alpha helix beta sheet domain next to the WHD domain.
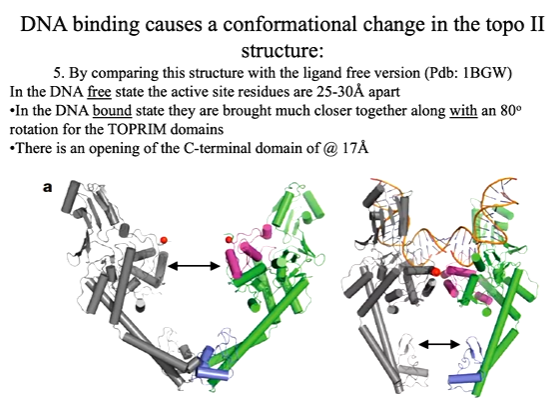
The paper expressed expressed part of yeast topoisomerase 2 which lacked the ATP domain and expressed this. They got a crystal structure in the presence of DNA at 3 angstroms. This structure shows DNA is bent at 150 degrees when bound in this complex. The protein has any alpha helices where the DNA progresses through the centre hole. They also had a structure without DNA. Electrostatic structures showed that there is positively charged residues around the DNA binding site. There is a beta hairpin loop that has an isoleucine that is specific and pokes into the minor groove of the DNA and causes displacement of the DNA. This motif is in many other proteins that bind DNA and cause bending such as IHF. The binding event also helps to induce different shapes of the DNA. On its own it is not enough to bind DNA but in the tower region there are basic residues that make charged interactions with the DNA backbone such as lysines. These residues channel the DNA upwards and hold it in place. This binding causes a conformational change in t he protein. When the DNA free structure is compared to the DNA bound orm, the binding of the DNA shows show the protein is bought together which causes opening ip of the bottom section of the topoisomerase
The DNA in this structure is not at the site where the tyrosine can help it undergo cleavage. This suggests that additional features and conformational changes are required to get the DNA strand in the vicinity of the tyrosine to make its nucleophilic attack. A close of of t he active site showed conserved 2 asp and glutamate around a magnesium atom. Tyrosine is the active site residue. The DNA is too far away to be close proximity to here.
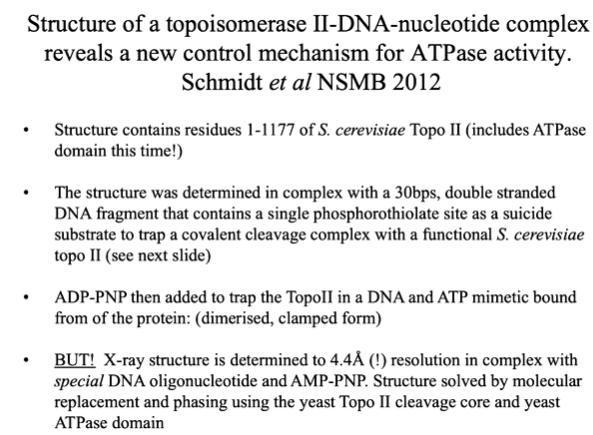
Another paper revealed how ATP hydrolysis is linked to the translocation of DNA. Here, there was more of the protein used including the ATPase domain. There is the ATPase domain and the tower domain. There are two protomers that come together with a chunk of DNA at the centre. Residues 1-1177 in the presence of shorter DNA that can trap it. There is also a non hydrolysable form of the ATP in this domain. This structure is at 4.4 angstrom resolution and so structure was solved with molecular replacement. The DNA has a bond that allows it to trap the topoisomerase when it comes to cleave this bond
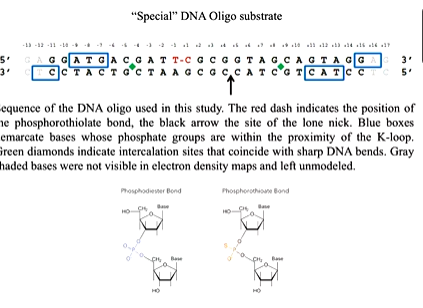
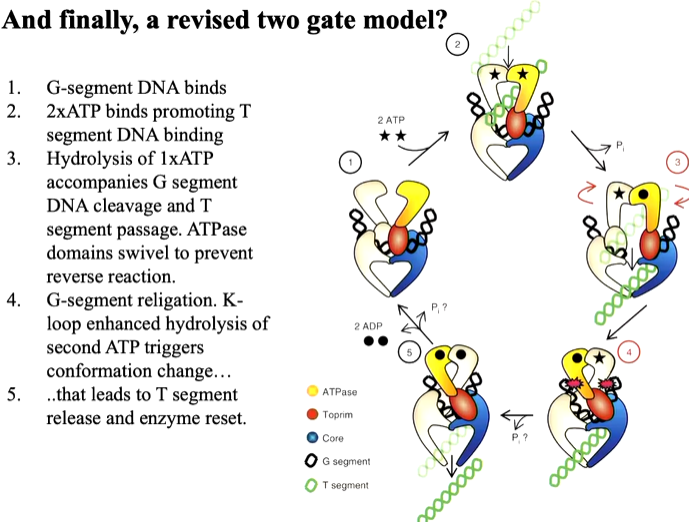
There are key observations from the moodle. There is an ATP domain that sits on top of the DNA cleavage domain. The two holes are offset by 45 degrees. This is because one polypeptide wraps around the other. This domain twist protein structure is found in other proteins such as HSP 90 which is also involved in DNA binding. One protomers N terminus reaches into the ATP site of the other and there is observation of a K loop which is not seen in other structures. The K loop has many lysines. The K loop also is highly conserved between different species and this in DNA bound structure suggests it becomes ordered when DNA binds. This is a common feature in many structures where the protein closes down upon binding of a substrate. Lysines are positively charged and so bind the DNA backbone. Using a functional assay, they tested the relationship between the protein/DNA to ATP hydrolysis. IN the gel based assay you start with supercoiled DNA and look for the ability of the protein to relax that inthe presence of ATP. You change the amount of protein in the assay and run it on the gel in a fixed time point incubation. There are different levels of relaxation reached with different amounts of the protein. Mutants of the protein with the lysines mutated to alanine shows a similar result. IN another mutant where they are mutated to alanine or glutamate (opposite charge) shows no activity until you get to high protein concentration. A time course experiment with fixed concentrations of the enzyme and samples taken out a specific time points. The wt ENZYME COMPLETES and quad ala mutation also does this but slower. The mutation with some alanine and some glutamate is much slower and so the result is that the K loop influences how the ATPase is acting processivity. The same hing is happening in other topoisomerase in different species.
The DNA binding stimulates the ATP activity and it is though that in the WT this is about 15 fold and the K loop has a direct role in influencing this.