lecture 11: bacterial cell wall peptidoglycan biosynthesis 1
There are classical targets for antibodies such as protein synthesis machinery such as ribosomes, DNA replication and cell wall (peptidoglycan) biosynthesis. This is the target for beta lactam antibodies such as penicillin. bacterial metabolism can be other targets but are not discussed here.
peptidoglycan is a polymer that sits on outside the bacterial cell membrane to form a mesh to give strength rigidity and strength. In MRSA it is a huge polymer and is important as functionally, it can give cell shape. Bacteria can adopt a variety of different shapes such as rod shaped or rounded shapes. Cell shape is a fundamental and conserved property of all cells but especially bacteria. We know little about its origin but its a feature of a cell wall and links processes occurring across the cytoplasmic membrane that controls peptidoglycan biosynthesis. If this is disturbed, it can be recognised by the immune system and can lead to cell death.
The depth of the peptidoglycan poly determines whether it is gram positive or negative. Gram positive bacteria have a thick layer of this layer above the cell membrane but no outer membrane which gram negative have. These also have a thinner peptidoglycan layer:
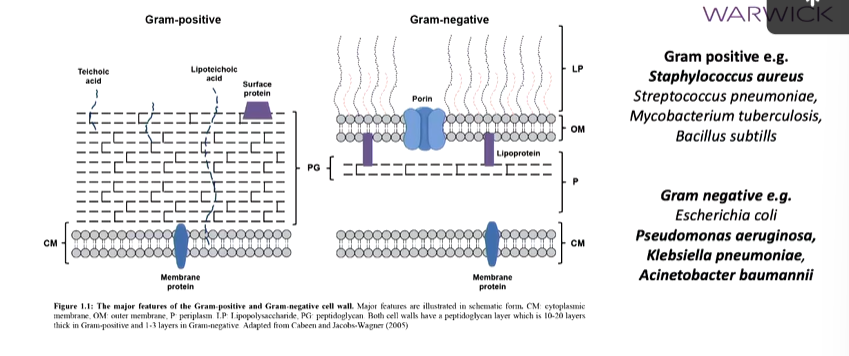
Bold bacteria are the ones that form escape pathogen list which are of most conserved from an antibiotic resistance perspective.
Peptidoglycan is an alternative polymer of NAM and NAG residues in the polymer backbone. NAM residues have a pentapeptide that becomes a tetrepeptide and this forms peptide linkages between chains that gives the peptidoglycan its 3D shape. This process is interfered by beta lactam antibiotics. This leads to weakening of the cell wall. Beta lactams look like the terminal D alanine peptide which is recognised by enzymes that do the cross linking process. Interference can either lead to bulges that make the cell lyse which is a failure in the cell division process or there can be a change in shape where rod cells go to round cells which is a failure in cell elongation and also leads to lysis
Gram negative bacteria have L alanine D glutamate mesodipolymeric lyse in the peptide stem while gram positive have L Alanine, D isoleucine, L lysine, D Alanine:

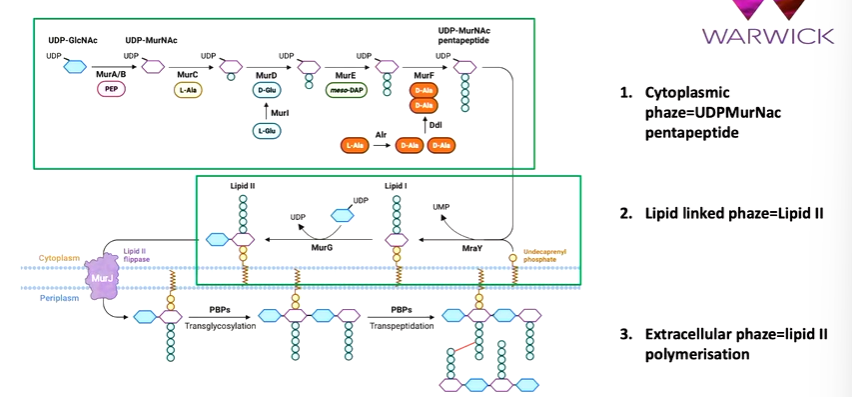
the biosynthetic pathway of peptidoglycan starts with the cytoplasmic phase that takes a nucleotide activated sugar precursor (UDP glucnac) that is converted to UDP MurNac by MurA/B. Then MurC,D,E and F sequentially add amino acid groups. The last two are a dipeptide. This makes it a pentapeptide. Next is the lipid linked phase where the UDP MurNac pentapeptide is linked to a C 55 carrier lipid called undecaprenyl phosphate The enzyme that does this is MraY. There is single sugar peptide linked to the C55 carrier peptide and MurG adds UDP glucnac to make lipid II. Next is the extracellular phase where LIpid II is polymerised. This is flipped by MurJ to the outer part of the membrane. A series of enzymes called penicillin binding proteins polymerize lipid II to a long strand and enable cross linking reactions:
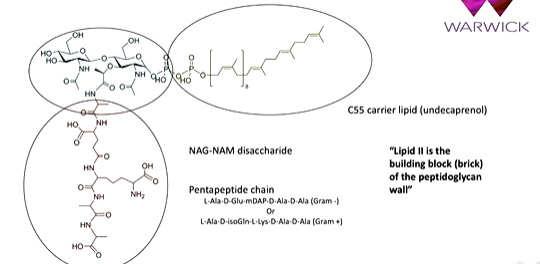
lipid II has a disaccharide NAG NAM core with a pentapeptide chain which is different is Gram Pos and Neg. There is also a C55 carrier lipid. This is used in other biosynthetic pathways
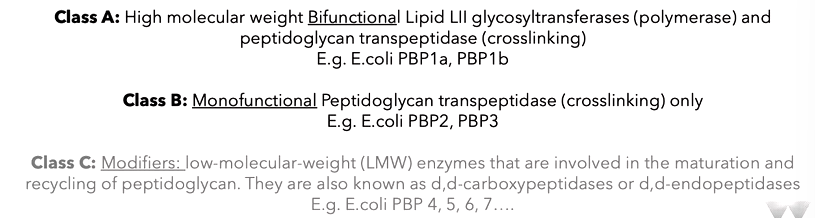
the enzymes that polymerise this are penicillin binding proteins which bind penicillin and the transpeptidase activity is in the literature. A PDP can bind penicillin but are peptidoglycan synthesis and crosslink the polymers. There are three classes:
Radioactive or fluorescent penicillin could have been used to identify PBPs which showed high molecular PBPs at the top and working its way down to type 3. In some organisms, a PBP1 could be PBP2 in another. The numbering is not systematic but class A is 1 etc.
PBP 1b has a glycosyl transferase domain and a transpeptidase domain. Lipid 2 is polymerised by the first domain and when it is long enough, it reaches into the transpeptidase domain. you cant have peptisation without the formation of a glycan strand. This formation of forming the glycan strand is not a classic pathway but instead a processive reaction. Lipid 2 forms a beta 1,4 glycosidic bond between it and the existing glycan strand in the acceptor and donor sites. This can cyle around to form the polymer. Monamycin can inhibit this in PBP1s. It bonds to the glycosyl transferase site very tightly and was used in animal feed to inhibit the growth of bacteria in the feed. It was banned as it could promote antibiotic resistance. There was no development towards monamycin in nature during this time.
It was thought that class 1s were the most important but this isn’t necessarily true. This came from a paper that showed that in some gram + bacteria, you can take away class A PBPs, the bacteria still survives. This is the first evidence that there is also something else going on. If you look at affinity profiles of the clinically used beta lactam antibiotics, you see they bond the class B PBPs. Genetically, the cluster of genes that are responsible for the biosynthesis of cell walls - MurE etc - there is a protein called FDSW which is a SEDS protein. IN the cell elongation cluster, MrdA there is MrdB which is a SEDS protein. Class B enzymes are making a complex with a SEDS protein which polymerises the lipid II for its class B partner. You always find RodA and PBP2 associated together and FtsW and PBP3 associated together. The reason that the beta lactam drugs are useful is they specifically inhibit the cell elongation and division complexes.
In any bacterial species, there are a selection of Class A PBP and SEDS associated PBPs that together are required for competent cell wall peptidoglycan biosynthesis that supports cell division and elongation.
RodA and PBP2 is the main system important for the Rod system for cell division and this is why they are targeted by beta lactams.