CMMB
Midterm
Topic 1
Lecture 1
Hook is credited with ‘discovering’ (visualizing) cells for the first time
Observed cork under a microscope and compared the cells to monks’ dorm cells
Cell theory:
Schwann proposed the first two principles of cell theory:
All organisms are composed of one or more cells
The cell is a structural unit of life
Virchow proposed a third principle:
Cells can only arise by division from a preexisting cell
Since the discovery of DNA, a fourth principle has been added:
Cells contain genetic information in the form of DNA, and that information is heritable
Cells are complex and organized
Highly ordered and consistent
Cells contain genetic information
Cells store, use, and transmit genetic information
Cells acquire and use energy
Almost all energy used by life is derived from the sun
Cells carry out an array of different chemical reactions
Almost all chemical changes require enzymes to increase the reaction rate
Cells are involved in mechanical activities
Transporting materials, movement of the whole cell ex
Cells respond to stimuli
Most cells have receptors that allow them to interact with their environment
Cells are capable of self regulation
Cells maintain homeostasis using a highly complex and organized set of molecular tools
Cells evolve
All living organisms evolved from a single common ancestral cell (cells share many features including genetic code, membranes, ribosomes)
Cells reproduce by division
Prokaryote: bacteria
Eukaryote: animals, fungi, plants, protists; have membrane bound organelles**
Covalent bond: a chemical bond in which electron pairs are shared between two atoms
Number of covalent bonds that an atom can form depends on the number of electrons needed to fill its outermost (valence) shell
Polar molecules:
Molecules with an uneven distribution of charge because the component atoms have different electronegativities
Hydrophilic
Nonpolar molecules:
Molecules whose covalent bonds have a nearly symmetric distribution of charge because the component atoms have approximately the same electronegativities
Hydrophobic
Noncovalent bond:
A relatively weak chemical bond based on attractive forces between oppositely charged regions
Ionic bond (these are relatively weak in a cellular context, ex in the presence of water):
Electrostatic interaction that occurs between groups of opposite charges
Hydrogen bond:
Electrostatic interaction between H atom and a second electronegative atom (ex H bonds between strands in the DNA double helix)
Van der Waals interactions:
A weak attractive force due to temporary asymmetries of charge within adjacent atoms or molecules, distant dependent interaction (ex important when interacting proteins have complementary shapes)
Hydrophobic effect:
The tendency of nonpolar molecules to aggregate in order to minimize their collective interaction with surrounding polar water molecules
Basis for the formation of lipid bilayer membrane
Lecture 2
In a eukaryotic cell, the plasma membrane (surrounding the outside of the cell), and organelle membranes are composed of a lipid bilayer
Cell membranes: phospholipid bilayer
Membrane contains a bilayer of phospholipids
Polar phosphates face the membrane surfaces
Nonpolar fatty acid tails face into the interior of the membrane
Phospholipids are amphipathic (hydrophobic component and hydrophilic component)
Lipid bilayer prevents random movement of substances in and out of the cell (selective barrier)
Fatty acids have long, unbranched hydrocarbon chains
Fatty acids in cells typically have 14-20 carbons
Fatty acids can be saturated or unsaturated
Saturated lack double bonds
Unsaturated:
One or more double bonds
Introduce a bend into the fatty acid tail
Naturally occurring fatty acids have cis double bonds (cis introduces more of a bend into the fatty acid tail compared to trans)
Membrane lipids:
Phosphoglycerides: Are phospholipids
Most membrane phospholipids are phosphoglycerides
Built on a glycerol
Glycerol + 2 fatty acid chains + phosphate group + additional group
Often contain one unsaturated and one saturated fatty acid chain
Phosphate is negatively charged
Overall charge of head groups at physiologic pH:
Phosphatidic acid (PA) <— H (Negative)
Phosphatidylcholine (PC) <— Choline (Neutral)
Phosphatidyl serine (PS) <— Serine (Negative)
Phosphatidyl ethanolamine (PE) <— Ethanolamine (Neutral)
Phosphatidylinositol (PI) <— Inositol (Negative)
Sphingolipids: Some sphingolipids are phospholipids
Less abundant membrane lipid
Built on sphingosine
Example: ceramide is sphingosine + fatty acid
Amphipathic
Additional groups can be linked to the head group, fatty acid chain can be added to R group (ex phosphorylcholine addition makes sphingomyelin which is a sphingolipid and phospholipid)
Tend to have longer and more highly saturated fatty acid chains than phosphoglycerides
Roles in signal transduction, membrane structure
Cholesterol:
Makes up part of the plasma membrane lipids in some animal cells
Amphipathic
Oriented with the small hydrophilic group facing the membrane surfaces
Remainder is embedded in the fatty acid tails of the phospholipids
Impairs the movement of the fatty acid tails of the phospholipids
Membrane Lipid Asymmetry:
Asymmetry affects membrane permeability, surface charge, membrane shape and stability
Ex PE promotes curvature of the membrane (is ‘cone-shaped’ due to its small head group), PS negative charge interacts with transmembrane proteins, PI has roles in signal transduction
Membrane Carbohydrates:
10% of membrane carbohydrates are covalently linked to lipids (glycolipids)
90% of membrane carbohydrates are covalently linked to proteins (glycoproteins)
All membrane carbohydrates in the plasma membrane face the extracellular space
Carbohydrates attached to glycolipids and glycoproteins can have very diverse structures
Carbohydrates play important roles as receptors, in sorting membrane proteins and in cell recognition (ex blood group antigens)
Membrane function:
Myelin sheath is composed of multiple layers of plasma membrane, with very little protein
Lipid composition can determine the physical state of the membrane, facilitate protein interactions, roles in signal transduction
Overview of major membrane functions:
Compartmentalization: Membrane compartments in the cell allow specialized activities to occur without impacting one another
Scaffold for biochemical activities: Membrane helps keeps proteins organized and in the right spot so the right reactions can occur in the correct order
Selectively permeable barrier: Prevent the random movement of substances in and out of the cell, but do allow select substances in and out
Solute transport: Specialized machinery, protein channels, pumps that allow solutes to transport in and out
Response to external stimuli: Outer membrane interacts with the environment using receptors that allow it to interact
Cell-cell communication: Cells can recognize and communicate with each other, exchange substances with each other
Energy transduction: Light energy is transformed into chemical energy, mitochondria and ATP ex
Lecture 3
Increasing the percentage of saturated fatty acids in a membrane decreases the membrane’s fluidity
Fluidity (viscosity) determines the physical state of the membrane
Influenced by temperature
Transition temperature/Melting temperature: Below this temperature the membrane is in a crystalline gel (more solid state), above this temperature the membrane is in a liquid crystalline phase (relatively fluid state)
Our cells exist slightly above the transition temperature
Transition temperature (and thus fluidity) is affected by:
Fatty acid chain saturation:
Saturated fatty acids: straight, flexible rod
Cis-unsaturated fatty acids: bends at the sites of double bond = increase membrane fluidity
Cholesterol content:
Flat, rigid, hydrophobic rings impair the movement of the phospholipid fatty acid tails
Eliminates a sharp transition temperature: creates intermediate fluidity
Refer to lecture 3 pg 8 graph on D2L: cholesterol acts as a buffer
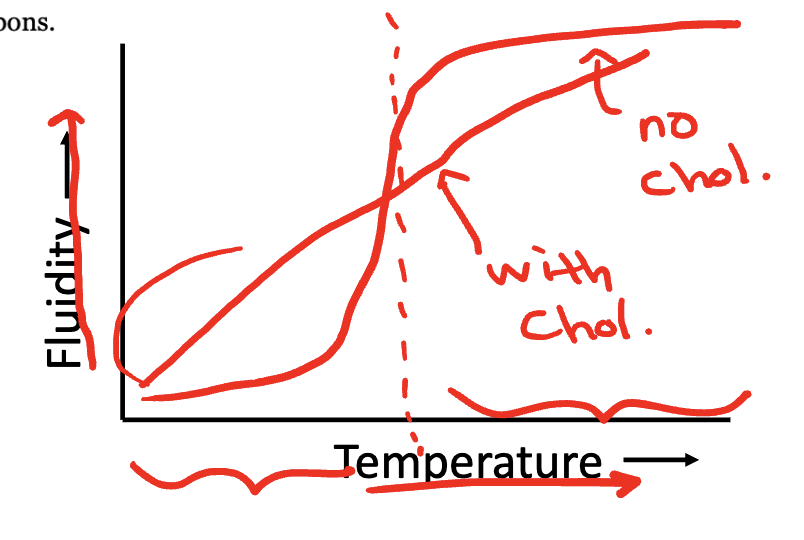
Fatty acid chain length:
Shorter fatty acid chains: fewer interactions (van der Waals) = less energy required to break them apart
A balance of membrane fluidity/rigidity is important for:
Maintaining structural organization and mechanical support
Enabling interactions (clusters of proteins)
Membrane assembly/cell growth/cell division
Cell movement, secretion, and endocytosis
Some cells (ex some fish, plants, bacteria) can alter their lipid composition in response to changing environmental conditions
In response to colder temperatures:
Desaturated single bonds in fatty acid chains to double bonds
Enzyme: desaturase
Change the types of phospholipids that it synthesizes
Synthesize more fatty acids with unsaturated bonds and shorter chain lengths
These two steps decrease transition temperature
In response to higher temperatures, the opposite would be true (in order to maintain a balance of membrane fluidity and rigidity)
Semi-aquatic mammals living in colder latitudes have increased desaturation of their fatty acids
A membrane may contain hundreds of different proteins
Proteins are distributed asymmetrically across the two leaflets of the membrane bilayer
Three classes of membrane proteins:
Integral (transmembrane) proteins
Peripheral proteins
Lipid-anchored proteins
Integral membrane proteins are usually transmembrane proteins:
Contain transmembrane domains
Pass through the lipid bilayer once (bitopic) or multiple times (polytopic)
Act as receptors, channels, or roles in electron transport
Transmembrane proteins are amphipathic
Transmembrane domains tend to be hydrophobic (form van der Waals interactions with the fatty acids in the bilayer)
Portions of the protein at the surfaces tend to be hydrophilic
Glycophorin A: bitopic membrane protein found in the red blood cells
Identifying transmembrane domains:
Peripheral membrane proteins:
Associated to the membrane by weak non-covalent bonds ex ionic interactions, hydrogen bonds
Some peripheral membrane proteins interact with integral membrane proteins
Dynamic: can be recruited to/released from the membrane
Roles in signal transduction, mechanical support for the membrane, enzymes
Mostly hydrophilic
Ex: Red blood cell peripheral membrane proteins:
Network of proteins that give the cell its shape
Major component: spectrin
On the internal surface of the membrane
Gives the cell shape, flexibility
Lipid anchored proteins:
Covalently linked to a lipid molecule in the bilayer
GPI anchored proteins:
Proteins attached to the membrane by a small, complex oligosaccharide linked to PI in the membrane
Glycosyl-Phosphatidyllnositol linkage
Outer leaflet
Roles in cell adhesion, and as receptors
Hydrocarbon chains embedded in the lipid bilayer
Usually on the cytoplasmic leaflet
Roles in signal transduction
Lecture 4
Phospholipid dynamics:
Phospholipids can easily move laterally within the same leaflet
Phospholipids ‘flip-flopping’ to the other leaflet is restricted (transverse diffusion)
Why is this thermodynamically unfavourable? Polar head groups of the phospholipid need to pass through the non-polar fatty acid tails
Enzyme flippase can play a role in establishing membrane asymmetry
Membrane protein dynamics:
Random diffusion
Immobilized (no movement)
Particular direction (motor proteins)
Restricted by other integral membrane proteins
Restricted by membrane skeleton proteins
Restrained by extracellular materials
Membranes are selectively permeable barriers: Allow the passage of some substances but inhibit the passage of others
Passive transport:
Does not require energy input from the cell
Occurs by diffusion (movement from a region of high concentration to low concentration)
Active transport: Does require energy input, can move substances against a concentration gradient
Diffusion: The spontaneous process in which a substance moves from an area of higher concentration to one of lower concentration, eventually reaching the same concentration in all areas (equilibrium)
The difference in the concentration of a substance between two areas is called the concentration gradient
Can penetrate the lipid bilayer:
Small inorganic solutes, such as O2/CO2/H2O
Solutes with high lipid solubility
Cannot penetrate the lipid bilayer:
Ions and polar organic solutes (ex sugars and amino acids)
Anything too large
Osmosis (passive transport): Water moves through a membrane from region of lower solute concentration to a region of higher solute concentration
Hypertonic solution: Higher solute concentration outside of the cell
Hypotonic solution: Lower solute concentration outside the cell
Isotonic solution: Equal solute concentration
Aquaporin (passive transport):
Channel proteins that facilitate the transport of water
Allows cells to be more permeable to water than is possible by diffusion through the bilayer
Ion channels:
A transmembrane structure permeable to a specific ion or ions (ex Na, K, Ca, Cl)
Most are highly selective
Most ion channels are gated: change conformation to be open or closed
Simple diffusion through a channel — types of ion channels:
Voltage-gated channel:
Open/closed depends on the difference in ionic charge on either side of the membrane
Voltage: difference in charge between two compartments
Ligand-gated channel:
Open/closed depends on the binding of a specific molecule (a ligand)
The ligand is usually not that solute that is passing through the channel
Ex neurotransmitter (like acetylcholine) is a ligand that binds to ion channels
Mechano-gated channel:
Open/closed depends on mechanical forces
Ex stretching
Voltage-gated K+ ion channels:
Significant impacts on electrical properties of the membrane:
Important for transmitting electrical impulses along axons
More than 10 million K+ ions can pass through the channel per second
Regulation: Different K+ channels open and close in response to different voltages
Case study: Use of the illegal drug: 3,4-methylenedioxymethamphetamine (MDMA, Ecstasy), has been associated with many serious medical complications including brain edema (water leaves the blood and enters
brain cells, causing brain swelling). MDMA has two effects on the body that together can lead to brain edema. Which two?Effect on blood sodium: Decreases blood sodium concentration
Effect on hydration: Increases thirst
Treatment: Increasing the solute concentration of the blood
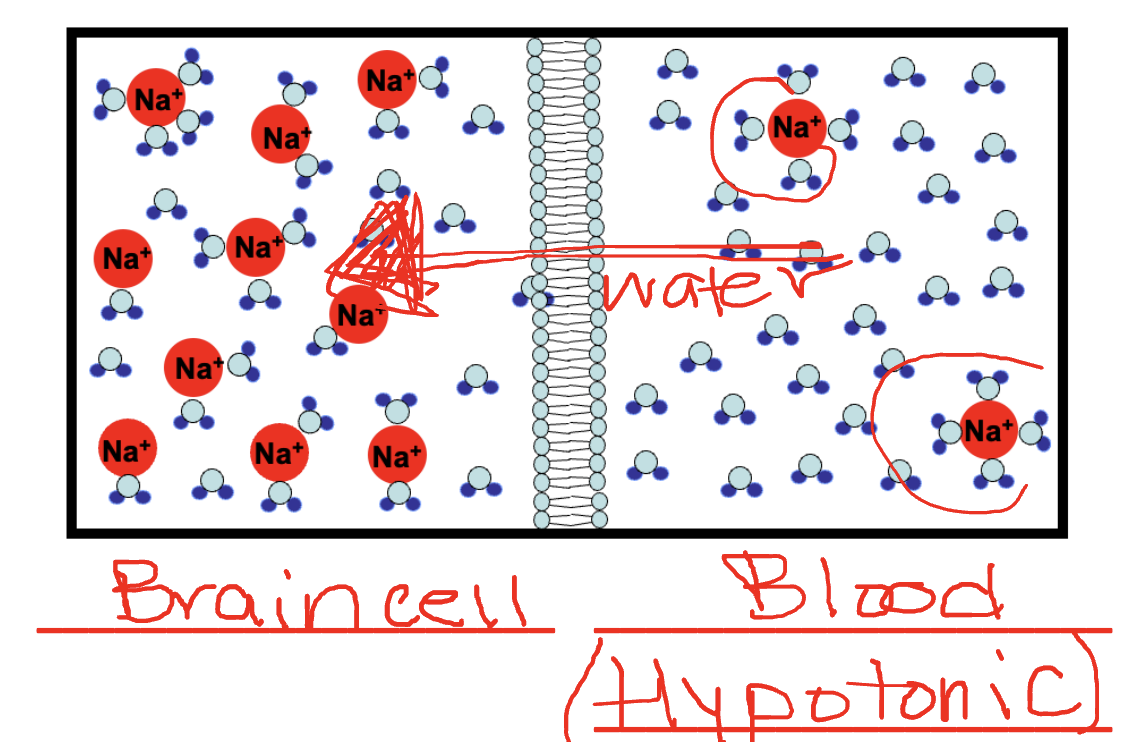
Facilitative transporter:
Binding of the solute triggers a conformational change in the transmembrane protein that exposes the solute to the other side
Exhibit saturation-type kinetics: when the concentration is high, the rate of transport levels off. This is a difference from ion channels:
Ion channels: millions of ions/sec
Facilitative transporters: 100-1000s of molecules/sec
Glucose transporter GLUT4 is an example of a facilitative transporter
Continued diffusion of glucose in the cell is possible because it becomes phosphorylated and metabolized in the cell
Lecture 5
Channel vs facilitative transporter:
Channel: Smaller conformational change, open/closed in response to ligand/voltage/mechano
Facilitative transporter: Larger conformational change, binding of the solute
Active transport:
Active transport is required to create these steep concentration gradients across the plasma membrane
Selective transmembrane protein
Protein undergoes a change in conformation
Requires energy input
Ex hydrolysis of ATP (primary active transport)
Ex flow of other substances down their concentration gradient (secondary active transport)
Primary active transport:
Three types: P-type pump, V-type pump, ABC transporter
P-type pump:
Na+/K+-ATPase is a P-type ion pump
ATPase, during active transport becomes phosphorylated
Contributes to maintaining the membrane potential (voltage) in cells
Per ATP: 3 Na+ pumped OUT, 2K+ pumped INTO the cell
Defects in the N+/K+ pump can cause impacts on the endocrine system, hypertension, neuromuscular disorders, seizures, others
Step 1:
E1 conformation: ion binding sites are accessible on the inside of the cell
High affinity for sodium ions
ATP is bound
Step 2:
When ions are bound, the protein closes (occluded E1 state)
Step 2-3:
Hydrolysis of ATP
Pump is phosphorylated
Step 3-4:
Release of ADP and conformation change to E2
Ion binding sites are accessible to the extracellular component
Loses affinity for Na+ ions, high affinity for K+ ions
Step 5-6:
When ions are bound, the protein closes (occluded E2 state)
Dephosphorylation
Step 7-8:
Dephosphorylation returns the protein to E1 conformation
Low affinity for K+ ions
Due to complex conformational changes, rate of transport is much slower than transport through ion channels (by several orders of magnitude)
V-type ion pumps:
Utilize ATP energy without becoming phosphorylated themselves
Transport H ions across organelles and vacuoles (ex maintain the low pH of lysosomes)
Also found in the plasma membrane of some cells (ex roles in maintaining acid-base balance in kidney tubules)
ABC transporters:
ATP-binding cassette transporters
Share a similar structure of ATP-binding domain
Mammalian ABC transporters transport ions, lipid, peptides, nucleosides, drugs ex
Ion gradients are a way to store energy in a cell
A concentration gradient is a form of stored (potential) energy
Symporters:
Transports two substances in the same direction
Also called cotransporter
Antiporters:
Transports two substances in the opposite directions
Also called exchanger
One of the substances is moving along (with) its concentration gradient, providing the energy to move the other substance against its concentration gradient
Na+/glucose cotransporter:
Transport glucose from the intestinal lumen into epithelial cells
Na+ ions concentration is low inside cells
Na+ ions moving down their concentration gradient is used to drive the cotransport of glucose
Transporting glucose against its concentration gradient
Primary active transport—Na+/K+ pump establishes the Na+ concentration gradient
Lecture 6
Study protein movements and dynamics within membranes:
Ex: fluorescence recovery after photobleaching (FRAP)
Can be used to study membrane dynamics in vivo within the living
Study isolated membrane proteins:
Ex examine protein size or expression levels (gel electrophoresis)
In vitro studies within the glass
Performed outside of their normal biological context
Need to isolate the protein from the membrane before you can study it
Fluorescence recovery after photobleaching (FRAP):
Technique to study movement of membrane components (proteins or lipids)
Step 1: Label membrane component with a fluorescent dye
Ex fluorescent antibody that recognizes a particular protein
Step 2: Photobleach (remove fluorescence) from a portion of the cell
~1um diameter
Step 3: Monitor reappearance of fluorescence in the previous bleached portion
Rate of recovery of fluorescence is a measure of the rate of diffusion of the fluorescently-labeled protein
Isolating membrane proteins - Lyse the cells:
Step 1: Lyse the cells and collect the plasma membrane
Mechanical disruption, freeze/thawing or hypotonic solution
Centrifuge the sample to separate into two fractions:
Pellet 1: insoluble
Supernatant 1: soluble
Membranes and membrane-associated proteins are in the pellet
Step 2: Isolate peripheral membrane proteins using high salt
Remember: Peripheral membrane proteins are associated with the membrane through hydrogen bonds and ionic interactions
Ions from salt will compete with the charged amino acids of peripheral membrane proteins to disrupt the noncovalent interactions with the membrane
Peripheral proteins are released from the membrane
Pellet 2: insoluble
Supernatant 2: soluble
Peripheral membrane proteins are in the supernatant 2
Transmembrane proteins are in the pellet 2
Step 3: Isolate transmembrane proteins with strong detergents
Amphipathic: polar end and nonpolar hydrocarbon chain (detergent)
Ionic detergents are harsher than non-ionic
Remember: transmembrane proteins are embedded in the membrane and interact with lipids by van der Waals interactions
Detergents can substitute for phospholipids to stabilize transmembrane proteins and make them soluble in aqueous solution
Pellet 3: insoluble
Supernatant 3: soluble
Transmembrane proteins are in the supernatant 3
GPI-anchored lipid proteins are in the pellet 3
Step 4: Isolate GPI-anchored proteins by treatment with phosphatidylinositol-specific phospholipase C (PI-PLC)
GPI-anchored proteins are linked to phosphatidylinositol in the membrane
GPI-anchored proteins are usually found in detergent-resistant portions of the membranes that are rich in cholesterol and sphingolipids
Pellet 4: insoluble materials
Supernatant 4: soluble materials
GPI-anchored proteins are found in the supernatant 4
Overview: A small aliquot of each pellet and supernatant is kept for separate analysis
Electrophoresis: Separation of charged molecules by migration through an electric field
Polyacrylamide gel electrophoresis (PAGE): Proteins migrate through a gel matrix made of cross-linked acrylamide polymers
Before we analyze the proteins’ migration through an acrylamide gel, we need to denature the proteins
Characterization of proteins by SDS-page:
In order to separate proteins based only on mass # of amino acids, we add SDS to the samples
SDS is a negatively charged amphipathic detergent:
Gives proteins a uniform negative charge
Denatures the protein (disrupts protein folding)
Repulsion between bound SDS molecules breaks noncovalent bonds (hydrogen, ionic)
Protein is unfolded from its native 3D structure
Protein is mixed with tracking dye and loaded on the polyacrylamide gel
Electrical current is applied and proteins migrate through the gel to the positive end
SDS-PAGE: polyacrylamide gel electrophoresis (using SDS to denature the proteins so that they are separated by mass)
To visualize all proteins in the gel, we can stain with Coomassie blue dye
To help estimate the size of proteins, we load a protein mixture called a molecular weight marker into the first lane (kDa: kilodalton; a unit often used to measure protein mass, 1kda is ~9 amino acids)
We can determine: approximate size of a protein and protein concentration (expression) by analyzing band intensity
Thicker bands indicate there is more protein present
Lecture 7
Structure of a neuron:
All organisms respond to external stimulation
Neurons (nerve cells) are specialized for communication with other cells in the form of electrical impulses
In vertebrates, most neurons are part of the central nervous system
Dendrites receive information
Axon conducts outgoing information
Terminal knobs are where impulses are transmitted to the target cell
Myelin sheath wraps most vertebrate axons
Nucleus is found in the cell body
Resting potential:
Membrane potential (membrane voltage): Difference in charge across a membrane
Resting potential: The membrane potential when a nerve cell is in an unexcited state
Resting potential of neuron is -70mV
Negative voltage: inside of cell is negative compared to the outside
What contributes to the difference in charge across the membrane?
Na+/K+ ATPase pump pumps 3Na+ ions out per 2 K+ ions pumped in
K+ ions are the charged substance with the most permeability in a resting nerve cell
Flow out through potassium leak channels (following their concentration gradient, not gated, out)
Equilibrium: balance is reached between the concentration gradient favouring K+ leaving the cell and the electrical gradient favouring K+ staying in the cell
Action potential: Changes in membrane potential after a stimulus and is the basis for neural communication
Includes depolarization and repolarization phases
Takes 5ms in a squid axon
Depolarization: A decrease in the electrical potential difference across a membrane, more positive
A stimulus (ex capsaicin in chili peppers) activates a gated channel, allowing sodium to diffuse in
If the stimulus results in depolarization above a threshold of -50mv, then voltage-gated sodium channels open
If this is reached, an action potential is triggered
The increased permeability to Na+ ions results in a membrane potential of about 40mV
Sodium channels spontaneously close after ~1ms
Repolarization: The depolarization (less negative voltage) triggers the opening of voltage-gated potassium channels
Sodium gated channels close
Membrane potential goes back to negative (-80mV)
Large negative membrane potential causes the voltage-gated potassium channels to close
Hyperpolarization: Because potassium channels are slow to close (slight dip in the graph at the end)
Propagation of an action potential:
Nerve impulse: An action potential is propagated along a neuron by triggering action potentials in adjacent portions of the membrane
Continuous conduction: Occurs in unmyelinated axons
Flow of current causes the membrane in the region just ahead to become depolarized
The action potential is propagated without any loss in intensity
Portion of the membrane that just experienced the action potential will be in a brief refractory period, can only go in one direction (Na+ channels can’t reopen for a few milliseconds after they’ve been activated)
Saltatory conduction: Occurs in myelinated axons
Impulses in myelinated axons are 20x faster than in an unmyelinated axon
Myelin prevents the passage of ions across the membrane
Most Na+ and K+ channels are found in or near unmyelinated regions called: nodes of Ranvier
Action potential at node of Ranvier triggers an action potential at the next node
Lecture 8 (Part 1)
Synaptic transmission:
Synapse: the specialized junction of a neuron with its target cell
Presynaptic cell: conducts the impulse towards a synapse (ex neuron)
Synaptic vesicles: storage for neurotransmitters in the terminal knobs of axons
Neurotransmitters: chemicals that bind to the postsynaptic cell, transmit signal across the synaptic cleft (red dots)
Synaptic cleft: space that separates the two cells
Postsynaptic cell: receives the impulse (ex another neuron, or muscle)
Turquoise channels below are ion channels
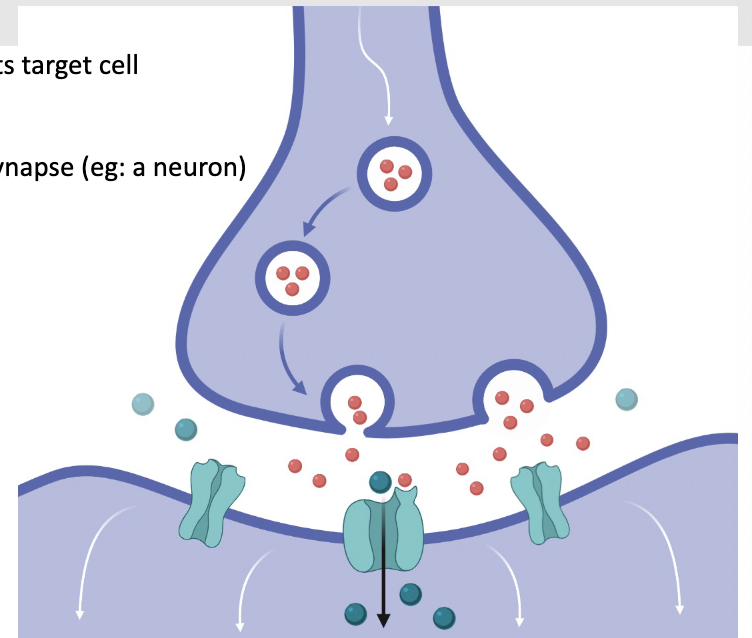
Depolarization causes voltage gated calcium channels to open in the presynaptic cell
Calcium diffuses into the cell
Increased Ca2+ in the cell triggers synaptic vesicles to fuse with the plasma membrane, releasing neurotransmitters (ex acetylcholine) which bind selectively to receptors
Neurotransmitter is acting as a ligand to open these ion channels, yellow channels below are ligand gated ion channels
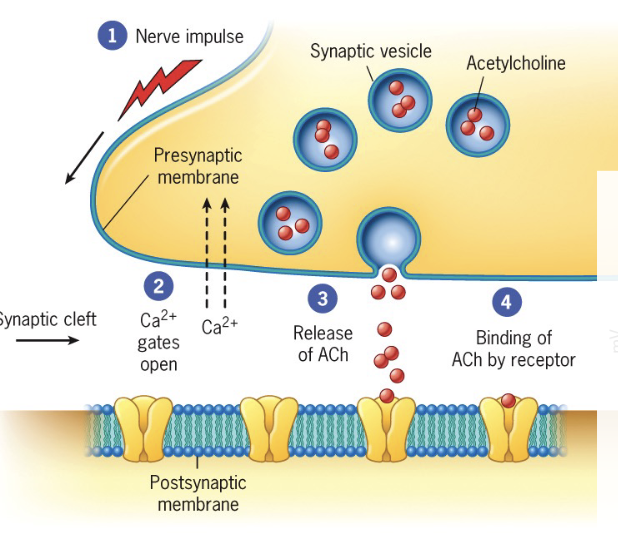
a) Influx of positive ions (ex Na+) ‘excites’ the postsynaptic cell (more likely to generate an action potential): depolarized (more positive)
Nerve impulse may be generated
b) Influx of negative cells (ex Cl-) ions: ‘inhibits’ the postsynaptic cell: hyperpolarization
Harder for a nerve impulse to be generated
After being released, neurotransmitters have a very short half life
Enzymes destroy the neurotransmitter in the synaptic cleft (ex acetylcholinesterase hydrolyzes acetylcholine)
Reuptake of neurotransmitter into the presynaptic cell
Drugs that interfere with neurotransmitters can have physiological and behavioural effects
Ex antidepressants inhibit reuptake of serotonin, cocaine interferes with reuptake of dopamine
Topic 2
Lecture 8 (Part 2)
Endomembrane system
Cytoplasmic membrane system
Composed of the cytoplasmic membranes (eukaryotic cells)
Functionally and structurally interrelated group of membranous cytoplasmic organelles including:
Endoplasmic reticulum (ER)
Golgi complex
Endosomes
Lysosomes
Vacuoles
Endomembrane system is a dynamic, integrated network
Transport materials from donor compartment to recipient compartment
Membrane-bound vesicles shuttle materials between organelles
Vesicles bud from the donor compartment
Transport in a directional manner with the help of motor proteins and the cytoskeleton network
Vesicles fuse with the membrane of the recipient compartment
Cargo is released in the destination compartment
Vesicle membrane becomes a part of the recipient compartment’s membrane
‘Escaped"‘ resident proteins of the donor compartment can be returned
Proteins (secreted proteins, lysosomal enzymes, membrane proteins ex) are directed to the correct destination with sorting signals:
Amino acid sequence (that makes up part of the protein)
Attached oligosaccharides
Signals are recognized by receptors in the membranes of budding vesicles
Transport materials out of the cell (Secretory pathway)
Examples of biomolecules synthesized in the ER (smooth or rough)
Lipids/cholesterol
Steroid hormones
Secreted proteins
Integral membrane proteins
Initial glycosolyation of proteins
Further modifications occur in the Golgi complex
Constitutive or regulated secretion
Constitutive secretion:
Most cells
Materials are continually transported in secretory vesicles from their site of synthesis and secreted
Contributes to the formation of the plasma membrane
Regulated secretion:
Materials are stored in membrane-bound compartments and only released in response to particular stimuli
Ex: endocrine cells that release hormones, pancreatic acinar cells that release digestive enzymes, nerve cells that release neurotransmitters
Transport materials into the cell (Endocytic pathway)
Materials move from the outer surface of the cell to compartments within the cell (endosomes and lysosomes)
Endosome:
Materials that are taken up are transported to early endosomes for sorting
Late endosomes are more acidic than early endosomes
Fuse with lysosomes to deliver cargo for degradation
Lysosome:
Hydrolytic (digestive) enzymes and acidic pH
Roles in breakdown of material and organelle turnover
Lecture 9
Autoradiography:
Following the location of radioactively-labeled materials in a cell
In particular: pulse-chase experiment can be used to examine a process that takes place over time
Step 1 Pulse:
Radio-labelled amino acids are incorporated in the digestive enzymes being synthesized
Exposed to the radio-labelled amino acids for only a short time
Step 2 Chase:
Transfer cells to media with only unlabelled amino acids
Enzymes synthesized during this time will not be radio-labeled
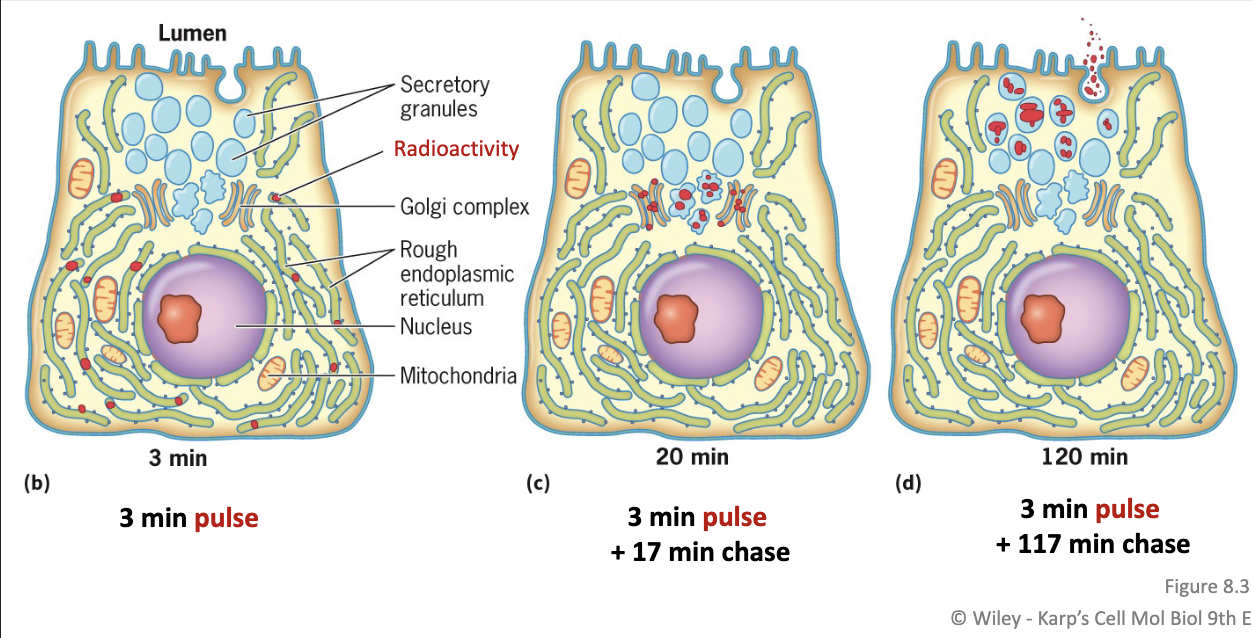
Endoplasmic reticulum (ER): a system of membranes and vesicles that encloses the ER lumen (separated from the cytosol)
Divided into smooth and rough
Rough ER:
Has ribosomes bound on the cytosolic membrane surface
Composed of a network of cisternae
Continuous with the outer membrane of the nuclear envelope
Extensive in cells with a role in protein secretion
Ex:
Pancreatic acinar cells that secrete hydrolytic enzymes
Intestinal cells that secrete mucuproteins
Endocrine cells that secrete polypeptide hormones
Functions:
Protein synthesis
Addition of sugars is initiated
Smooth ER:
Lacks ribosomes
Composed of interconnected curved, tubular membranes
Continuous with the RER
Extensive in cell types such as skeletal muscles, kidney tubules, and steroid producing endocrine glands
Functions include:
Synthesis of steroid hormones
Synthesis of membrane lipids
Detoxification of organic compounds in the liver
Sequestering calcium ions in skeletal and cardiac muscle — role in muscle contraction (sarcoplasmic reticulum)
Sites of protein synthesis:
Free ribosomes:
2/3 of proteins
Ribosomes that are not attached to the ER
Proteins are released into the cytosol
Proteins that remain in the cytosol
Peripheral proteins of the cytosolic surface of membranes
Proteins transported to the nucleus, mitochondria and chloroplast
RER ribosomes:
1/3 of proteins
Co-translational translocation: Peptides move into the lumen of the ER as it is being synthesized by the ribosome
Secreted proteins
Integral membrane proteins and soluble proteins that reside in the compartments of the endomembrane system
Integral membrane proteins in the plasma membrane
Both sets of ribosomes are structurally and functionally identical. Their location differs
Co-translation translocation - Synthesis of secreted proteins and soluble proteins that reside in the endomembrane compartments:
All protein synthesis begins on a free ribosome
Signal sequence at N-terminal end: 6-15 hydrophobic amino acids
Signal recognition particle (SRP) binds to the signal sequence and the ribosome
Polypeptide synthesis is halted temporarily
SRP directs this complex to the ER membrane by interaction with the SRP receptor
Ribosome/polypeptide are transferred from the SRP to the translocon: protein pore in the ER membrane
Contact with the signal sequence displaces the plug
Translocation through the pore: polypeptide enters the ER lumen
Upon termination, ribosome is released
Signal sequence is removed by an enzyme: signal peptidase
Protein chaperones (ex BiP) aid in protein folding
Co-translation translocation - Synthesis of integral membrane proteins:
Synthesized by co-translational translocation using the same machinery as secreted proteins (SRP, receptor ex)
SRP recognizes the hydrophobic transmembrane domain as the signal sequence
Transmembrane domains do not pass through the pore - instead, they directly enter the lipid bilayer
As polypeptides pass through the translocon, a gate in the pore opens and allows proteins to partition themselves according to their solubility properties
Either in the aqueous pore in the hydrophobic lipid bilayer
Arginine (R, arg), Lysine (K, lys), Histidine (H, his) are the positively charged amino acids*** (must know)
Direction of insertion into the bilayer is dependent on the location of the positively charged amino acids relative to the transmembrane domain
Cytoplasmic leaflet is more abundant with PS and PI phospholipids: negatively charged
The protein will orient in the membrane such that the positively charged amino acids interact with the relatively negatively charged cytosolic leaflet
If the positive charges are on the N-terminal side of the transmembrane domain, the translocon will reorient the transmembrane domain
Lecture 10
Glycosylation:
Majority of proteins produced at the RER become glycosylated (glycoproteins)
Carbohydrate groups have roles as binding sites
Aid in proper folding and stabilization
Sorting/directing proteins to different cellular compartments
N-linked glycosylation (common)
Linkage to asparagine
Is initiated in the RER
O-linked glycosylation
Linkage to serine or threonine
Occurs in the golgi complex
N-linked glycosylation in the rough ER:
First seven sugars are transferred one at a time to a lipid: dolichol pyrophosphate, embedded in the ER membrane
Initial assembly is on the cytosolic side
Sugars are added by glycosyltransferases
Dolichol and attached oligosaccharide is flipped across the membrane
Remaining sugars are attached to dolichol on the cytosolic side
Flipped across the membrane and attached to the growing oligosaccharide chain
Completed oligosaccharide is transferred to an asparagine residue of the polypeptide being translated
Transfer by the enzyme oligosaccharyltransferase to an Asn within the sequence: Asn-X-Ser/Thr (X is not proline)
Quality control for misfolded proteins:
Glucosidase I and II remove two glucoses
(and 5). Glycoprotein with one glucose is recognized by calpexin (chaperone protein in the ER)
Removal of glucose releases protein from chaperon (calpexin releases)
Incompletely folded proteins are recognized by UGGT (a conformation sensing enzyme): detects exposed hydrophobic residues. Adds glucose molecule
Properly folded proteins exit (step 6 not 5)
Improperly folded proteins are degraded in a proteosome in the cytosol (steps 7-8 not 6)
Exiting the ER:
Membrane vesicles with enclosed cargo (protein) bud from the ER and travel in the direction of the Golgi
Transport vesicles fuse with one another to form larger vesicles in a region called the ERGIC: Endoplasmic Reticulum Golgi Intermediate Compartment
Golgi Complex:
Golgi compled is composed of cisternae arranged in a stack
Distinct compartments arranged from the cis face (closest to the ER) to the trans face (exit, furthest from the ER)
Trans Golgi network (TGN): network of tubules and vesicles
Sorting station where proteins are segregated into different types of vesicles (heading to the plasma membrane or other)
Cis Golgi network (CGN): Interconnected network of tubules
Sorted station that distinguishes between proteins that need to be returned to the ER and those that should proceed through the Golgi
Protein modification with the Golgi complex:
Newly synthesized proteins leaving the ER are sequentially modified
Ex: modification of N-linked carbohydrate chains
Order that sugars are incorporated depends on the location of specific glycosyltransferases (integral membrane proteins in the membrane of the Golgi complex)
Glycosylation in the golgi complex can be quite varied
O-linked carbohydrates are entirely assembled within the golgi
Lecture 11
Movement of materials through the Golgi complex:
Model 1: vesicular transport model
Golgi cisternae are stable compartments
Vesicles carrying cargo bud from one compartment and fuse with the next
Evidence:
Golgi cisternae have different enzymes
Lots of vesicles bud from the edges of Golgi cisternae
Model 2: cisternal maturation model
Cisternae form at the cis face and move towards the trans face, ‘maturing’ as they move
Evidence:
Drugs blocking vesicle formation at the ER leads to the Golgi complex disappearing
Certain large materials (ex collagen) move from cis to trans without ever appearing in smaller vesicles
Current model of vesicle transport:
Cisternal maturation model of transport through the Golgi complex
Anterograde transport (forward): from cis to trans
Retrograde transport (backward): from trans to cis, resident Golgi and ER enzymes
Types of coated vesicles:
COPII-coated vesicles: Move cargo forward (ER to Golgi complex)
These are not required for movement from the cis to trans Golgi
COPI-coated vesicles: Move cargo backward, from ERGIC/Golgi to ER, from trans to cis Golgi
Clathrin-coated vesicles: Move materials from the TGN to endosomes, lysosomes, plant vacuoles; also endocytosis
COPII-coated vesicles:
COPII select and concentrate certain proteins for transport in vesicles: (by interacting with transmembrane proteins that have ‘ER export signals’)
Enzymes destined for the Golgi complex (ex glycosyltransferase)
Proteins involved in vesicle docking and fusion
Protein receptors that bind soluble cargo
Sar1 is a COPII coat protein. G protein (molecular switch).
Sar1-GDP is recruited by GEF (guanine exchange factor)
Sar1-GTP undergoes conformational change so that it inserts into cytoplasmic leaflet (this starts to bend the membrane)
Sec23/Sec24 dimer further bends the membrane
Sec24 is the primary adaptor protein that interacts with membrane proteins (that have ER export signals)
Sec13/Sec31 form an outer structural cage
Disassembly is triggered by hydrolysis of GTP bound to Sar1
Know the names of Sar1 and Sec24***
COPI-coated vesicles:
COPI coat is made up of a protein complex called coatamer, which forms a thick protein coat directly on the membrane
Membrane-bending G protein: Arf1 (GTP form bends membrane)
Retrograde transport of proteins:
Golgi resident enzymes
ER resident proteins (escaped)
Proteins that reside in the ER contain a retrieval signal
Soluble ER proteins usually contain the signal: lys-asp-glu-leu (KDEL)
Recognized by a KDEL receptor (shuttle between cis Golgi and ER compartments)
Membrane ER proteins also have a retrieval signal, usually: lys-lys-X-X (X is any amino acid KKXX)
KKXX retrieval signal is located on the cytosolic side so it can interact with COPI-coated recycling vesicle
Each compartment in the endomembrane system may have its own retrieval signal
Vesicle fusion:
Specific interactions between different membranes
Movement of the vesicle toward the specific target compartment
Movement mediated by microtubules and motor proteins
Tethering vesicles to the target compartment
Two types of tethering proteins:
Rod-shaped/fibrous (longer)
Multiprotein complex (closer)
G proteins called Rabs (60+ in humans) help to determine specificity
Rabs recruit specific tethering proteins
Rabs also interact with motor proteins
Docking vesicles to the target compartment SNARE proteins form complexes with another SNARE protein
Integral membrane proteins
35+ different proteins in specific compartments
v-SNARE: put into transport vesicles during budding
t-SNARE: located in the target membrane
Form four-stranded bundles
Fusion between vesicle and target membrane
Interactions between t-SNAREs and v-SNAREs pull lipid bilayers together with enough force to cause fusion
The ability of a vesicle to fuse to a specific membrane is determined by the specific combination of: Rabs, SNARES, and tethering proteins
Rab proteins are master regulators of vesicle transport between compartments within cells
Lecture 12
Lysosomes contain hydrolytic enzymes:
Contains at least 50 hydrolytic enzymes
Enzymes here have an optimal activity in acidic pH
Acid hydrolases
pH of lysosome is ~4.6
pH of lysosome is maintained by a proton pump
Roles of lysosomes:
Breakdown of material brought into the cell by endocytosis
Ex phagocytic cells in mammals ingest pathogenic microbes
Organelle turnover (autophagy)
Regulated destruction and replacement of the cell’s organelles
Organelle is surrounded by a double-membrane structure: autophagosome
Autophagosome fuses with a lysosome: autolysosome
Starved cells exhibit increased autophagy
Organelle is surrounded by a double-membrane structure
Inner autophagosomal membrane: cargo sequestration
Outer autophagosomal membrane: fusion with the lysosomal membrane
Sorting and transport of lysosomal enzymes:
Soluble lysosomal enzymes are recognized by enzymes that add phosphate groups to mannose sugars of N-linked carbohydrate chains
The phosphorylated mannose (mannose 6-phosphate) residues act as a sorting signal, directing proteins to the lysosome
Targeting lysosomal enzymes to lysosomes:
Mannose residues are phosphorylated in Golgi (mannose 6-phosphate)
Lysosomal enzymes are incorporated into a clathrin-coated vesicle
Clathrin: coat protein that forms structural scaffold
GGA Adaptor: connects clathrin to MPRs
Mannose 6-phosphate receptor (MPR): transmembrane protein that recognizes and captures proteins with the mannose 6 phosphate signal
G-protein: Arf1-GTP, binds to the membrane and initiates formation of the budding vesicle and binding of the other coat proteins
Induces membrane curvature when bound to GTP
Adaptor: physically links two or more components
GGA adaptor has multiple domains:
Binds Arf1-GTP
Binds clathrin
Binds to the cytosolic tails of the MPRs
Results in concentrating lysosomal enzymes into clathrin-coated vesicles
(Formation of the clathrin-coated vesicle)
MPRs separate from the lysosomal enzymes and are returned to the Golgi (step 5)
Clathrin coat is disassembled and lysosomal enzymes are delivered to a sorting endosome and on to a lysosome (step 6)
Simplified Steps:
Mannose residues are phosphorylated in Golgi (mannose 6-phosphate)
Lysosomal enzymes are incorporated into a clathrin-coated vesicle
Vesicle formation is complete
Clathrin coat is disassembled
Vesicle fuses with endosome for sorting
6a. MPR (receptors) are returned to the Golgi
6b. Lysosomal enzymes are delivered to the lysosome
Transport of secreted proteins:
Golgi cisternae move continually toward the TGN, which fragments into vesicles and tubules
Constitutive secretion may be the ‘default’
Endocytosis:
Bulk-phase endocytosis:
Pinocytosis
Non-specific: uptake of extracellular fluids (and any molecules that happen to be present)
Receptor-mediated endocytosis:
Clathrin-mediated
Specific molecules binding to receptors on the extracellular surface of the plasma membrane
Ex hormones, growth factors, certain nutrients
Focus in this course
Clathrin organization:
Each clathrin molecule (triskelion) is composed of three heavy chains and three light chains
AP2 complex (adaptor) links cytoplasmic tails of plasma membrane receptors with clathrin
Dynamin is a G-protein required for the clathrin-coated vesicle to bud from the membrane
Dynamin subunits polymerize to form a ring (step 3)
GTP hydrolysis induces a movement in the dynamin ring
Vesicle is cleaved and dynamin disassembles
Lecture 13 (Part 1)
Recycling pathway:
Housekeeping receptors mediate uptake of materials that will be used by the cell (cholesterol, iron, etc)
Receptors are first transported to an early endosome for sorting
Ligands dissociate due to acidic pH
Receptors are concentrated into a recycling compartment of the early endosome
Vesicles return receptors to the cell surface to be used again
Degradation pathway:
Signalling receptors bind ligands that affect cellular activities (hormones, growth factors ex)
First transported to early endosome for sorting, early endosome matures into late endosome
Late endosome fuses with lysosome for receptor degradation
Receptor degradation prevents the cell from being further stimulated by the hormone/growth factor
Topic 3
Lecture 13 (Part 2)
Cytoskeleton:
Network composed of three well-defined filamentous structures:
Microtubules
Microfilaments (actin filaments)
Intermediate filaments
General functions:
Structural support
Transport of materials (also organelles)
Contraction and motility
Spatial organization
Role in cell division
Microtubule structure and function:
Each type of cytoskeleton filament is made of protein subunits held together by weak non-covalent bonds
Allows rapid assembly and disassembly
Microtubules: hollow, unbranched, tubular structures made of tubulin
Roles in cell support and movement of materials within a cell
Can extend across the length or breadth of a cell
The microtubule is composed of 13 protofilaments aligned side by side to form a tube
Protofilaments are assembled from dimers of one ⍺-tubulin
and one β-tubulinProtofilament is asymmetric, the microtubule itself has polarity
⍺-tubulin end: negative end
β-tubulin: positive end
Assembly of microtubules:
Centrosome:
A type of microtubule-organizing centre which initiates microtubule formation
Composed of two centrioles surrounded by pericentriolar material (PCM)
PCM: loosely organized fibrous lattice
Centrioles: cylinders composed of microtubules
When centrosomes replicate, centrioles recruit PCM to form a new centrosome
Centrosomes often remain at the centre of the cell’s microtubular network
Centrosomes are microtubule organizing centers. They dictate:
the number of microtubules
their polarity
the number of protofilaments
the time and location of microtubule assembly
Not: microtubule stability nor rate of assembly
New microtubules do not make contact with the centrioles, instead they are initiated in the PCM
PCM contains ɣ-TuRC (tubulin ring complex):
ɣ-tubulin (gamma)
Non-tubulin proteins in a ring
⍺β-tubulin dimers assemble on the ɣ-tubulin, where only ⍺-tubulin can bind to the ring of ɣ-tubulin
Microtubule dynamics:
Microtubules in some structures are sensitive to disassembly: mitotic spindle
Microtubules in some structures are very stable: neurons, cilia, flagella
Stability is determined by:
MAPs: microtubule associated proteins
+TIPS, which bind at the + end of growing microtubules
Temperature: cold=disassembly
(Stabilizing) MAPS
Increase stability and promotes assembly by linking tubulin dimers together
Activity of some MAPs is controlled by the presence of phosphate groups
Ex high level of a phosphorylated MAP (called tau) has been associated with the development of Alzheimer’s disease
GTP is an energy source
analogous to ATP
β-tubulin is a G protein: hydrolyzes GTP to GDP after the dimer is added to the microtubule
GTP bound to the β-tubulin subunit is required for microtubule assembly
GTP hydrolysis affects microtubule structure
GTP is not hydrolyzed by ⍺-tubulin
Lecture 14
In a growing microtubule, the top consists of tubulin-GTP dimers in an open sheet
Tube closure is associated with hydrolysis of GTP
GDP tubulin has a different conformation, introducing mechanical strain
MAP stabilize microtubule
In the absence of stabilization, protofilaments curl outward and undergo catastrophic shrinkage
+TIPs:
Bind to the positive end of the microtubule and regulate the rate of growth or shrinkage
Mediate the attachment to subcellular structures (ex kinetochore of the mitotic chromosome)
Microtubule polymerization/disassembly can effectively ‘push’ and ‘pull’ material within a cell
Microtubules as structural supports:
Microtubules provide mechanical support: are stuff enough to resist compression or bending forces
Help determine the shape of a cell
Maintains intracellular location of organelles
Microtubules as agents of intracellular motility:
Transport of membranous vesicles from one membrane compartment to another
Transport of nonmembrane bound cargo (RNAs, ribosomes, cytoskeletal elements)
Refer to diagram on pg 14
Microtubule motor proteins:
Motor proteins: Utilize ATP hydrolysis to generate mechanical forces that move the motor protein and attached cargo along the cytoskeleton
Cargo examples: membranous vesicles, nonmembrane bound (ribosomes, RNA), organelles (lysosomes, mitochondria), chromosomes, other cytoskeletal filaments
Three types of motor proteins: microtubule motor proteins (kinesins and dyneins), actin motor proteins (myosins)
Each type of motor protein moves unidirectionally in a stepwise manner
Kinesin Structure
Kinesin-related proteins superfamily: Kinesin-1 family
Tetramer: two heavy chains and two light chains
Globular head:
Binds microtubules
ATP hydrolysis
Conserved sequences
Kinesin movement:
Kinesin moves along the microtubule towards the positive end
Leading head binds one ATP: hydrolysis and release of ADP + Pi = power stroke that swings the trailing head forward
Moves the motor 8nm (length of one tubulin dimer)
Kinesin moves in a hand-over-hand mechanism: at least one head is attached to the microtubule at all times
Highly processive: capable of moving consiserable distances without falling off
Speed is proportional to the ATP concentration (to a max speed of 1um/sec)
Dynein structure:
Dynein is much larger than kinesin
Dyein head is ~10x larger than a kinesin head (also faster than kinesin)
Two heavy chains + multiple intermediate and light chains
Bind to cargo via an adaptor protein (dynactin)
Globular head: force generation, ATP binding and hydrolysis
Dynein movement:
Dynein moves progressively along the microtubule towards minus end
Roles in:
Positioning the spindle and moving chromosomes during mitosis
Positioning organelles and moving vesicles
Structure and function of cilia and flagella:
Cilia and flagella are hairlike organelles that project from various eukaryotic cells
Often motile
Same structure, different contexts
Beware: microvilli are not the same as cilia
Lecture 15
Motile cilia in multicellular organisms move fluid
Ex cilia lining the respiratory tract sweet mucus away from lungs
Usually found in large numbers on a cell’s surface
Coordinated beating
ES: Effective power stroke
RS: Recovery stroke
Flagella have the same structure as cilia, but found in fewer numbers
Unicellular alga (eukaryote) moves by an asymmetric waveform
Cilia/flagella is covered in a membrane that is continuous with the cell’s plasma membrane
Microtubule organizing center: basal body (ɣ-Turc)
Axoneme: Core contains microtubules oriented longitudinally
All microtubules oriented:
+ at the distal end
- near the basal body
Structure of the axoneme:
9 peripheral doublet microtubules around a central pair of single microtubules (“9+2 array”)
Centriole has 9 microtubule triplets, axoneme has 9 microtubule doublets
Doublets are connected to each other via nexin
Dynein tails are anchored to one of the tubules in each pair (the ‘A tubule’)
Reminder: move towards the - end of the microtubule
Movement of cilia/flagella:
Dynenin tails attached to the A tubule and dynein stalks bind to the B tubules
Power stroke (conformational change upon ATP hydrolysis)
Dynein stalks detach
Dyenin stalks reattach (cycle begins again)
Nexin link limits the extent of movement/sliding
Actin filaments (F-actin) (microfilaments)
Actin is the most abundant protein in most cells
Filaments are composed to globular subunits (G-actin)
Involved in cellular motile processes
Ex: Movement of vesicles, phagocytosis, cytokinesis
Provides structural support: shape of cells, support for cellular projections
Structure of actin filaments:
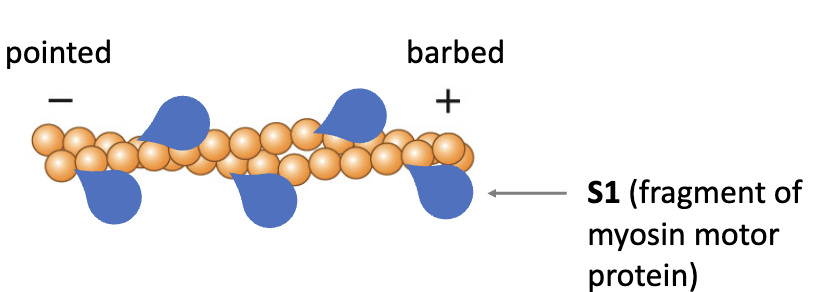
Actin filaments have polarity:
+ end barbed
- end pointed
Individual G-actin monomers have directionality and are added to the filament in a particular orientation
Filament also has directionality (polarity)
Filament is a double-stranded helix (both strands are oriented in the same direction)
Ends are named based on binding of a fragment of the myosin motor protein (S1)
Actin filament assembly and disassembly:
ATP-actin is incorporated into the filament
After incorporation, actin hydrolyzes it to ADP
ATP-actin is added to both ends
Faster addition at the barbed end
Barbed and pointed ends have different critical concentrations:
Minimal concentration of available ATP-actin required to elongate
Critical concentration of the barbed end (+) is much lower
Preformed actin filament (seed) in the presence of ATP-actin
At high ATP-actin concentrations, it will be added to both ends
Concentration reaches the critical concentration of the pointed end; addition stops at the pointed end
Loss of subunits occurs at the pointed end because ADP-actin dissociates more readily than ATP-actin, but addition continues at the barbed end
Relative position of subunits is continually moving: treadmilling
Concrete numbers:
Assume we’re starting with a high available concentration of actin-ATP, which is decreasing as the subunits get incorporated into the filament
Initial concentration of actin-ATP (higher than 1.5um)
Pointed end (-) critical concentration: 1.5um
Barbed end (+) critical concentration: 0.5um
As available actin-ATP decreases, the critical concentration of which end will be reached first? Pointed end
Treadmilling happens when the cell’s available actin-ATP is between 0.1um and 1.5um
Steady state: when the rate of addition at one end is the same as rate of loss at the other end
Occurs at approx 0.3um available actin-ATP
Lecture 16
Actin motor protein - Conventional Myosin (Type II):
Myosin superfamily:
Conventional (type II)
Unconventional (type I, types III-XVII)
Conventional Myosin Type II:
Motor (head)
Binds the actin filament
Binds and hydrolyzes ATP
Conserved sequences
Neck
(or lever arm)
Moves during the power stroke
Tail
Intertwining of the two heavy chains
Allows the formation of filaments of myosin
All myosins (except type VI) move towards the + end (barbed end)
Actin motor protein - Unconventional Myosin (Type V):
Moves processively along actin filaments
Moves in a hand-over-hand movement
Long necks act as swinging arms
Can take very large steps (~36nm)
Some myosins (types I, V, VI) can associate with vesicles and organelles (ex myosin type V tail bound to a vesicle via adaptors (including Rab 27a)
Transport by Unconventional Myosins:
Some vesicles contain both microtubule motors and actin filament motors
Movement over long distances occurs mostly on microtubules
Local movement in the outskirts of the cell: actin filaments
Myosin type 2 filaments:
Myosin II tails allow the protein to form filaments
In the myosin II filament, tails point towards the center and heads points towards the outside
Myosin filament:
Bipolar: Motor domains are oriented at opposite filament ends
Thick: Composed of myosin (in contrast to ‘thin’ filaments that are composed of actin)
Skeletal muscle organization:
Skeletal muscles are usually anchored to bones
Muscle fiber: a skeletal muscle cell. Contains multiple nuclei and hundreds of myofibrils
Myofibrils: composed of repeating contractile units called sarcomeres
Sarcomeres: contractile unit with a characteristic banding pattern
Sarcomere organization:
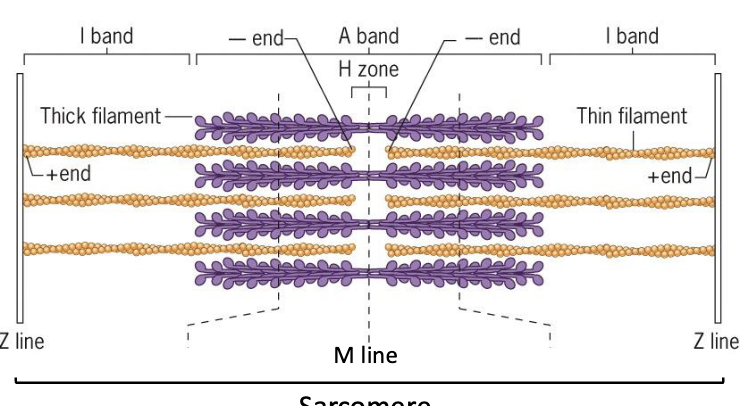
Thick filament (purple): myosin filament
Thin filament (orange): actin filament
Z-line: contains proteins important for sarcomere structure stability (one sarcomere is from Z-line to Z-line)
M-line: dark staining in the center of the sarcomere, contains anchoring proteins
I bands (light staining) - contain only thin filaments
H zone: contains only thick filaments
A band: dark staining (overlap of thick and thin, also includes the H zone)
Each thick filament is surrounded by 6 thin filaments
I band and H band decreases in length when the muscle contracts
A band length does not change when the muscle contracts
Model: thin filaments slide towards the center of the sarcomere
Molecular basis of contraction:
Myosin II heads in a thick filament binds to six surrounding actin filaments
Myosin II is nonprocessive:
Only in contact with actin for a fraction of the time
Myosin heads are not synchronized
Actin-myosin contraction cycle:
ATP binds to myosin head and myosin dissociates from actin
ATP hydrolysis, ADP and Pi remain bound to myosin
Energized myosin binds actin
Release of phosphate triggers conformational change: power stroke - actin moves towards the center of the sarcomere
ADP is released
Neuromuscular junction:
Muscle fibers (cells) within a motor unit are stimulated simultaneously by a single motor neuron
Neuromuscular junction: Point of contact between motor neuron and muscle fiber; site of transmission of the nerve impulse
Acetylcholine stimulates an action potential in the muscle cell
Lecture 17
Excitation-contraction coupling:
Transverse tubules (T-tubules): Membrane folds that propagate an impulse to the interior of a muscle cell
Sarcoplasmic reticulum: Special smooth ER in muscle cells, stores Ca2+ in lumen (pumped in from the cytosol)
Arrival of action potential at the SR opens Ca2+ channels, release Ca2+ into the cytoplasm
Thin filaments contain actin and:
Tropomyosin: rod shaped
Troponin: globular
Absence of Ca2+: tropomyosin blocks myosin-binding sites on actin
Presence of Ca2+: Ca2+ binds troponin which moves tropomyosin exposing the myosin-binding site on actin
Actin organization:
Cell cortex: Actin network on the inner face of the plasma membrane, capable of dynamic remodelling
Enable cells to crawl/move
Enable phagocytosis
Cellular constriction during cell division
Actin-binding proteins: regulate the assembly, disassembly, and rearrangements of actin networks (more than 100 different proteins)
Actin-binding proteins:
Filament nucleating:
Slowest step in the formation of an actin filament
Proteins can enhance the rate at which actin filaments are formed
Arp2/3 complex: Binds to the side of an existing filament (creates branches), remains at the pointed end of the new branch, similar structure to actin monomers
Formins: Generate unbranched filaments, stay associated with the barbed end, promote rapid elongation of filaments
Monomer-sequestering:
Bind to actin-ATP monomers to prevent them from being added to the elongating filament
Able to modulate the available monomer pool in certain regions at certain times
Ex thymosins
End-blocking (capping):
Regulate the length of actin filaments
Bind at either end
Monomer polymerizing:
Binds to actin monomers to promote growth of actin filaments
Promotes replacement of ADP with ATP on the actin monomers
Ex: profilin
Depolymerizing:
Bind to actin-ADP at the pointed end to promote depolarization
Ex: cofilin
Cross-linking and bundling:
Multiple actin-binding sites, allowing them to alter the 3D organization of the actin filament network
Ex: filamin (cross-linking), villin and fimbrin (bundling)
Filament-severing:
Break an existing filament in two
Ex: gelsolin and cofilin
Membrane-binding:
Actin filaments linked to the plasma membrane
Enabling the plasma membrane to protrude outward (cell locomotion) or inwards (phagocytosis)
Ex: spectrin family of proteins
Specific proteins/complexes to know:
Arp2/3 (branched filament nucleation)
Profilin (monomer polymerizing)
Cofilin (depolymerizing)
Cell motility (step 1):
Movement is initiated by a protrusion of the cell in the direction of movement (Lamellipodium)
A portion of the protusion anchors to the surface below
The bulk of the cell is pulled toward the front, over the adhesive contacts
Adhesive contacts break, causing retraction of the trailing edge (tail)
Lamellipodium: the leading edge of a moving cell that extends over the surface, broad and flat
Dynamic actin network at the site of lamellipodium formationL
A stimulus is received at the cell surface (ex neutrophil receiving a signal from an infected tissue)
Arp2/3 complex at the site of stimulation gets activated
Arp2/3 binds the side of an existing filament
ATP-actin monomers bind to the Arp2/3 complex, forming a new actin branch
Polymerization is promoted by profilin
Additional Arp2/3 complexes can bind to the sides of the new filaments, forming additional branches:
Older filaments are capped at their barbed ends
Newer filaments continue to grow at the barbed end, pushing the membrane of the lamellipodium outward
Older capped filaments undergo disassembly promoted by cofilin
Cell motility (step 2):
Traction forces: When the cell grips the surface (at adhesion points called focal adhesion)
Focal adhesion: structures in the cell membrane where integrin proteins connect to actin
Integrin proteins: Transmembrane proteins that mediate the interaction between actin and extracellular components
Cell motility (step 3):
Contraction forces pull the bulk of the cell forward
Myosin found near the rear of the lamellipodium
Lecture 18
Intermediate filaments:
Strong, flexible, stretchy, unbranched fibers
Only found in animal cells
Provide mechanical strength to cells
Neurons
Muscle cells
Epithelial cells
Chemically heterogeneous
Encoded by ~70 different genes in humans
Five classes (I-V)
Intermediate filaments don’t have polarity** (both ends are identical)
New units are added into the middle of an existing filament**
Bridging (ex: via the protein plectin) to intermediate filaments stabilizes other cytoskeletal elements, increasing cell stability
Intermediate filaments in neurons: neurofilaments
Have sidearms that help to maintain proper spacing
Important for determining the axon’s diameter
Final
Lecture 19
Section I: The Plant Cell
Plant and animal cells diverge from a common unicellular ancestor
Plant cells have chloroplasts
Plant cell structure and function is conserved in many ways
Conserved organelles, structures, metabolic processes, genes
Many differences between plant, animal, fungi cells
Plant cells are glued to their neighbours
~50 different plant cell types: xylem, phloem, mesophyll (leaf) actively involved in photosynthesis
Differentiated plant cells: can de-differentiate and form another cell type —> whole plant
Differentiated cell —> undifferentiated cell —> new cell type (ex lead mesophyll cell) —> whole plant
Individual plant cells can de-differentiate, divide, and form a complete plant
Totipotency: ability of a cell to divide and form any other cell type, sometimes a complete organism (ex zygote, spore)
Important in biotechnology
Genetically modified (transgenic) plant
All cells in transgenic plant have the transgene (ex canola — most of it is herbicide resistant)
After bombardment, cells that contain the transgene are selected and induced to form complete plants with each of their cells containing the transgene
Movement within plant cells: Lots of cytoplasmic streaming in plant cells
Ex root hair
Organelle movement drives cytoplasmic streaming
Ex golgi stacks (100’s in each cell), the plant Golgi stacks move along actin filaments that are associated with ER
Move on actin filaments using myosin motors
Golgi bodies - punctate structures
ER - reticulate structure
Plant myosin XI — the fastest myosin - takes 35nm steps (one helical rotation)
Myosin-mediated vesicle movement along actin filaments in Arabidopsis root hair cells
Chloroplasts move in response to light in leaf cells
Dim light: Chloroplasts align perpendicular to the direction of light
Bright light: Chloroplasts align parallel to the direction of light
Movement is triggered by blue wavelengths of light
Lecture 20
Section II: Plant Cytoskeleton
Plant cells have microtubules (MT) and actin filaments
No intermediate filaments
Motor proteins: myosin and kinesin
No dynein in plant cells
No centrioles (or centrosomes) in higher plant cells
No flagellated or ciliated plant cells
4 MT arrays in plant cells
Cortical array - interphase: on inner side of plasma membrane
Pre-prophase band of MTs
Mitotic spindle
Phragmoplast
These 4 are found during cell division
Section III: Cell Wall
All plant cells have a cell wall
~15-30% dry weight of a herbaceous plant
All have a primary cell wall: extracellular - outside plasma membrane
Many others also have a secondary cell wall: made after the primary wall (also extracellular)
Lies inside of the primary wall (ex xylem cell - dead at maturity)
Lignin: a compound that gives wall strength
Middle lamella: Glues plant cells together
Contains pectin
Plant cells have high internal pressure (turgor pressure)
Primary wall prevents cell from bursting
Vacuole: high solute concentration (sugar, salts, amino acids ex); osmosis—water moves in
Leaf: solar panel
Cellular turgor pressure is important in maintaining leaf shape
Components of the Cell Wall:
Highly organized structure composed of polysaccharides and proteins
Cellulose: bundles of long glucose polymers, glucose subunits bond by B(1,4) linkages
Hemicellulose: crosslinks adjacent cellulose microfibrils
using H-bonds - weak
polymer of glucose and another sugar (ex xyloglucan)
Pectins: heterogeneous, branched carbohydrate
Proteins: involved in wall stability (ex extensin locks wall into place, expansin loosens wall)
Synthesis of Wall Compounds:
Cellulose: made by cellulose structure
Embedded in plasma membrane
Rosettes synthesize cellulose microfibrils
Cortical MTs guide cellulose synthase movement
Non-cellulose: hemicellulose and pectin, made in Golgi stacks, protein are made on RER
Lecture 21
Direction of cellulose microfibrils - determines the direction of cell growth
Expansin loosens hemicellulose-cellulose interaction
Ex root cell
Coordinated cell elongation can result in complex plant movements
Cytokinesis of plant cell: moves centrifugally outward
Mitosis:
Interphase - Non-dividing cell
Prophase - Chromosomes begin to condense
Metaphase - Condensed chromosomes line up at equator
Anaphase - Sister chromatids separate
Telophase - New nuclear envelopes begin to form
Pre-prophase band of MTs:
Transient band of MTs - predicts plane and position of new cell plate
Forms prior to prophase —> gone by metaphase
Against plasma membrane —> leaves a “footprint”
Phragmoplast:
Forms after anaphase
Helps cell plate development
Double band of MTs - deliver Golgi derived vesicles to developing cell plate
Anti-parallel MTs - plus-ends pointing toward each other
Uses kinesin motor
Moves laterally
A GFP-MT labelled cell demonstrates the dynamics of three MT arrays during plant cell division - PPB, spindle, and phragmoplast
Plasmodesma(ta):
Cytoplasmic connections between adjacent cells
Can move small (ions, sugars) molecules and large (proteins, RNA)
Connect most cells — plants are supracellular organisms: one big cell
Protein spokes: regulate large molecule movement
Microinjection Studies:
Fluorescent dextrans — known molecular weights (0.5kDalton, 5kDalton ex)
Size exclusion limit (SEL) of PD ~1kDa ~ sugar
Proteins ~30kDa
Some proteins have a signal in peptide sequence which can open up a gate and selectively move that protein in
Ex transcription factor: cell-cell movement controls gene expression, move into phloem (long distance transport)
Actin in plasmodesmata (PD): regulate movement between cells
Section VI - Vacuole:
Fluid filled compartment bounded by the tonoplast membrane
Usually 30% of cell volume but can be up to 90%
Functions:
Storage: ions, organics, sugars, proteins
Digestion: nucleases, proteases
pH and ion homeostasis - decrease pH ex lemon
Defense: accumulate toxic compounds
2 types: lytic vacuole and protein storage vacuole
Module 1
Lecture 22
Extracellular Interactions:
Materials present outside the plasma membrane play an important role in the life of a cell
Most cells in a multicellular plant or animal are organized into clearly defined tissues
There are many diverse activities that are regulated by this:
Tissue development
Wound healing
Fighting infection
Receptor and ligand interactions are common methods of cell interactions
Direct cell-cell interactions such as in cell-cell contacts
Cells interact with their extracellular environment
The epidermis has closely packed cells of epithelial tissue
The dermis is a type of connective tissue
Fibroblasts of the dermis have receptors that mediate interactions and transmit message
Cellular interactions are required for:
Intercellular communication
Survival
Tissue strength
Organ function
Immune system function
Embryonic development
4 different families of integral membrane proteins mediate cell-cell adhesion:
Selectins
Immunoglobulin super family
Members of the Integrin family
Cadherins
Protein interactions involving the cell surface:
Homotypic interactions of two L1 molecules through immunoglobulin (Ig) domains
Heterotypical interactions of IG super family (IGSF) protein with integrin
Cadherins:
Calcium dependent
adhesion or transmit signals
bind a similar cadherin on a neighbouring cell
Possibly the single most important factor in molding cells into cohesive tissues in the embryo and holding them together in the adult
Cadherin loss associated with malignancy
Distributed along cell surfaces or part of intracellular junctions:
synapses
Adherens junctions
Desmosomes
During embryogenesis:
Cells from different ‘germ’ layers display distinctive adhesive properties (ectoderm: outside skin, mesoderm: middle)
Selective cell affinities help establish the spatial order of different tissues in the embryo
Requires specific molecular interactions: cadherin-cadherin
Experiments demonstrated that separated cells redistribute themselves, so each cell adhered to cells of the same type
Stem cells were induced from differentiated human cells and injected into a developing pig embryo, the human cells successfully integrated into the tissues of the developing pig
The human and pig cells must have been able to interact appropriately with their cadherins initially
Immunoglobulin super family (IgSF):
contain Ig domains that can connect to the integrin family, or connect to another IgSF
mediate calcium independent adhesion
many IgSF proteins are ICAMs
ICAMs - intracellular adhesion molecules
Integrins are some of the proteins that acts as receptors for ICAMs
Selectins:
E-selectin, present on endothelial cells (blood vessels)
P-selectin, present on platelets and endothelial cells
L-selectin, present on all types of leukocytes (white blood cells)
Calcium dependent
Selectins are a family of membrane glycoproteins that bind to specific oligosaccharide (carbohydrate moiety)
“Lectin” is a term for a compound that binds to specific carbohydrate group
Selectins have a small cytoplasmic segment, a single membrane-spanning domain and a large extracellular portion
Lecture 23
Movement of neutrophils from the bloodstream during inflammation:
Inflammation activates endothelial cells, which upregulates the selectins and they ___
Selectins bind to the carbohydrate residues (PsgI-1) on neutrophil, a phagocytic leukocyte
Platelet activating factor on IL-8 on the surface of endothelial cells activates G-protein coupled receptors on the neutrophil and this leads to _____
Integrins bind to ICAMs on endothelial surface and a cascade of events results in cytoskeletal rearrangement such that the cell can ___
Transendothelial migration
Cancel calls escape the normal growth control mechanisms and proliferate in an unregulated manner
Metastasis is the spread of cancer
One of the most important proteins that reduces metastasis is the presence of ____
How can we visualize cell junctions?
Electron microscopy uses electrons rather than light
Very high resolution (visualization) of cellular structures
Samples are imaged under a vacuum so live cells can’t be imaged
The Junctional Complex:
Tight junctions (zonula occluden)
Adherens junctions
Desmosomes (macula adherens)
Tight junctions:
At the top of the cell
Occur between neighbouring epithelial cells
Prevent solute distribution where different solute concentrations are in adjacent compartments
They can control:
Gate function - controls the passage of the following between cells (paracellular pathway):
Ions
proteins
blood brain barrier - ions or water can’t pass, but cells of the immune system can pass
water - mutations in claudin 1 cause death due to dehydration
Fence function - block diffusion of integral membrane proteins between apical and basolateral membranes of one cell, connect to the actin cytoskeleton and microtubules
Tight junctions regulate the passage of solutes between cells
Form close contacts between cells
TJ restrict plasma membrane proteins to a particular domain of the membrane
Contribute to cell polarity - forming a barrier that blocks proteins
Adherens junctions and desmosomes increase tissue strength
Adherens junctions:
Connect the external environment to the actin cytoskeleton
Provide a pathway for signals to be transmitted from the exterior to the cytoplasm and nucleus
AJ form a belt (zonula adherens) that encircles the cells near their apical surface in epithelial cells
Desomosomes - primarily adhesive:
Also contain cadherins = desmosomal cadherins
The cadherins interact with multiple proteins to form a cytoplasmic plaque on the inner surface of the plasma membranes
The intermediate filament cytoskeleton anchors two cells together, this provides strength to a sheet of cells
Gap junctions are communication channels:
Form intercellular channels
Transmit small soluble signaling molecules directly through the membrane
They are made of connexin proteins: 6 identical connexins from each cell form a transmembrane channel with a central pore called a connexon
Two connexons form a gap junction**
Gap junctions are molecular “pipelines” that pass through the adjoining plasma membranes and open into the cytoplasm of the adjoining cells
Hemidesmosomes:
Cell matrix attachment in vivo is seen at the basal surface of epithelial cells, anchored to the underlying basement membrane
Hemidesmosomes contain a dense cytoplasmic plaque with keratin filaments
Keratin filaments are linked to ECM by integrins
Focal adhesions are discrete sites of cell attachment and are dynamic
Cultured cells are anchored to the surface of the dish only at scattered, discrete sites, called focal adhesions
Focal adhesions play a key role in cell locomotion
Focal adhesions are dynamic structures
 Knowt
Knowt