M1L2 DNA damage and repair
DNA repair pathways
Direct reversal of damage
O6 alkyl guanine DNA alkyl transferase (ATase)
Photolyase
AlkB
Nucleotide excision repair
UV damage
Bulky adducts
Bae excision repair
Base and sugar phosphate backbone damage
DNA SSBs
DNA damage tolerance, translesion polymerases
DNA is the only biomolecule to be specifically repaired, all others are replaced
~5% of all proteins encoded are involved in DNA repair
Possible upregulation of DNA repair accounting for therapeutic resistance to cancer chemotherapy
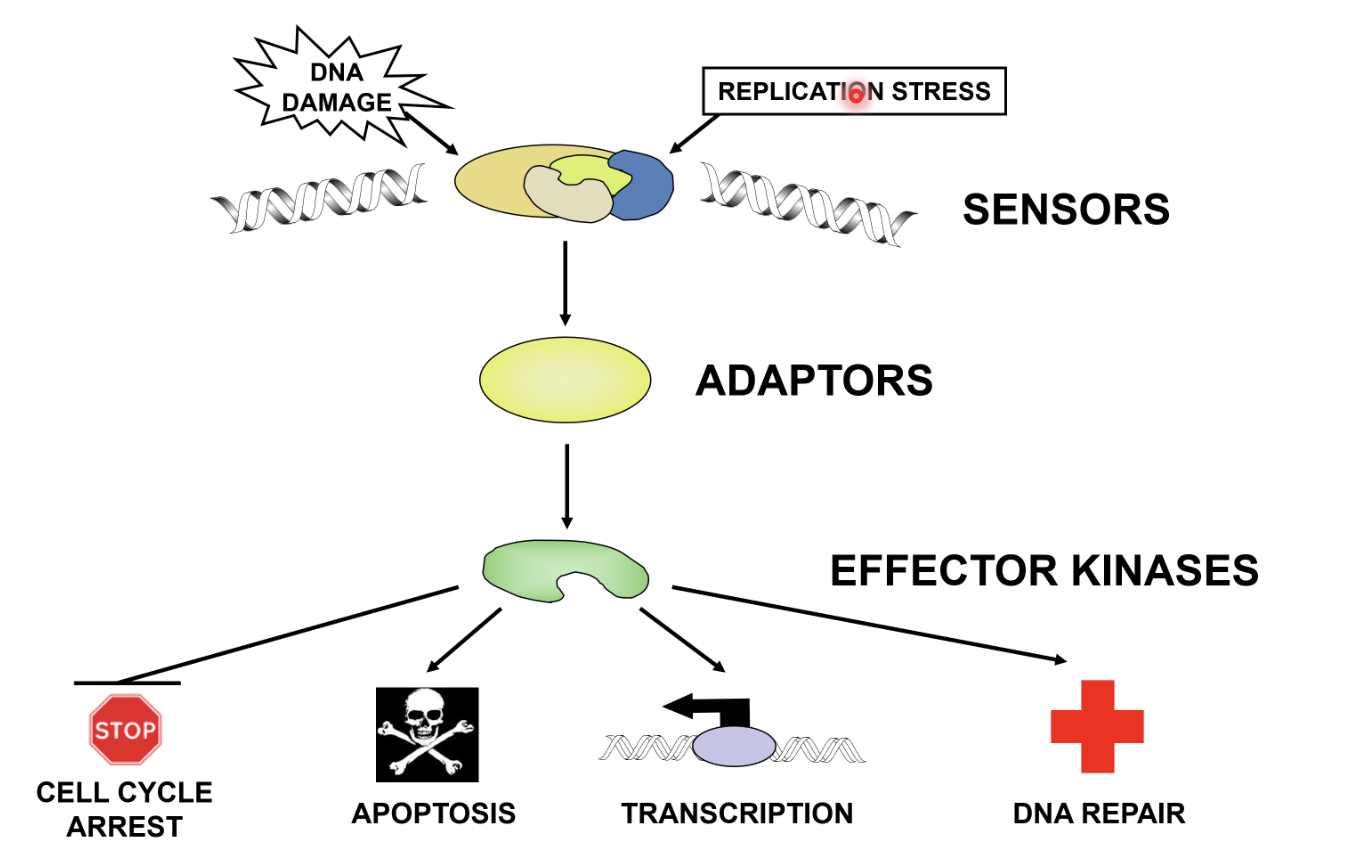
Sensors - detection
Adaptors - signal to effectors
Effector kinases - causes cell cycle arrest, apoptosis, transcription, or DNA repair
Main forms of DNA damage
SSBs by radiation, oxidative species
Mismatches - from replication, homologous recombination, cytosine deamination to uracil, 5 Me-C to thymine, A to hypoxyanthine (codes like G)
Damaged bases due to radiation, oxidative species, alkylation particularly methylations, diamination (C-U)
Missing bases (abasic sites) - particularly purines, occurs spontaneously especially if the base is alkylated; also an intermediate in the action of DNA glycosylases
Altered bases - pyrimidine dimers, alkylated DNA, oxidised bases
DSBs by radiation
Intra-strand crosslinks due to UV light and cancer drugs
Inter-strand crosslink due to cancer drugs and metabolites (eg aldehydes)
Alkylation of phosphate backbone
Main DNA lesions (most to least common)
SSBs
Depurination
O6 methyl guanine
Thymine glycol
Cytosine deamination
DSBs
DNA crosslinks
More studies are looking into endogenous sources of DNA damage as these events are more likely than damage from the environment
Endogenous sources - mainly hydrolytic and oxidative reactions, DNA replication errors, SSBs generated by recombination, DNA repair intermediates
DNA mutagenesis
Deletions, insertions, substitutions
Transitions - pyrimidine substituted for a different pyrimidine or purine substituted for another purine
Transversion - pyrimidine substituted for purine or purine substituted for pyrimidine
Direct reversal of damage
Photolyase - found in bacteria, fungi, plants, and certain animals, but not humans or placental mammals
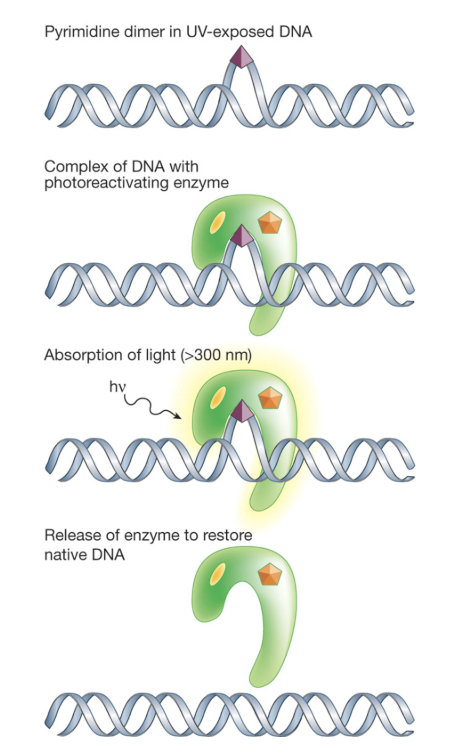
Strong affinity for structure of damaged lesions and can reverse base damage caused by UVB radiation
Pathway has two cryptochromes (FAD and FADH-) which can absorb blue light (300-500nm)
FADH- gets excited, transferring an electron to the UV damaged site
Breaks chemical bonds causing DNA damage and returns pyrimidines to original configuration
Removal of O6 alkyl groups by suicide protein O6-ATase
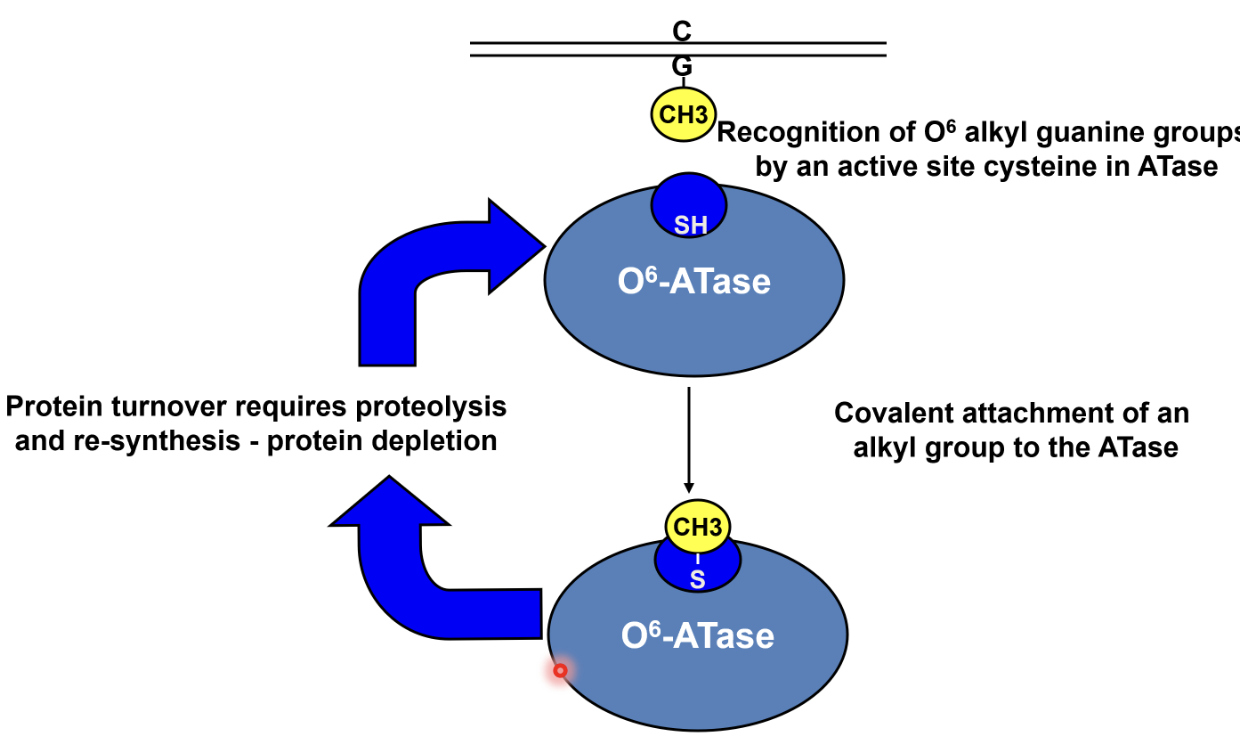
O6 alkyl guanine is recognised by the ATase which transfers the alkyl group to a cysteine residue in its active site, causing covalent attachment of alkyl group to ATase
This corrects the damage but inactivates the ATase, hence protein turnover requires proteolysis and resynthesis
Temozolomide induces DNA methylation in the treatment of brain cancers, however cancer cells tend to upregulate this ATase as a resistance mechanism
Drug development investigating inhibitors of these ATases to prevent this, but there are issues with bypassing the blood brain barrier and tumour evolution
Base excision repair
There is redundancy in the pathway, eg. O6-alkyl guanine can also be repaired via BER (which may also be a reason for resistance)
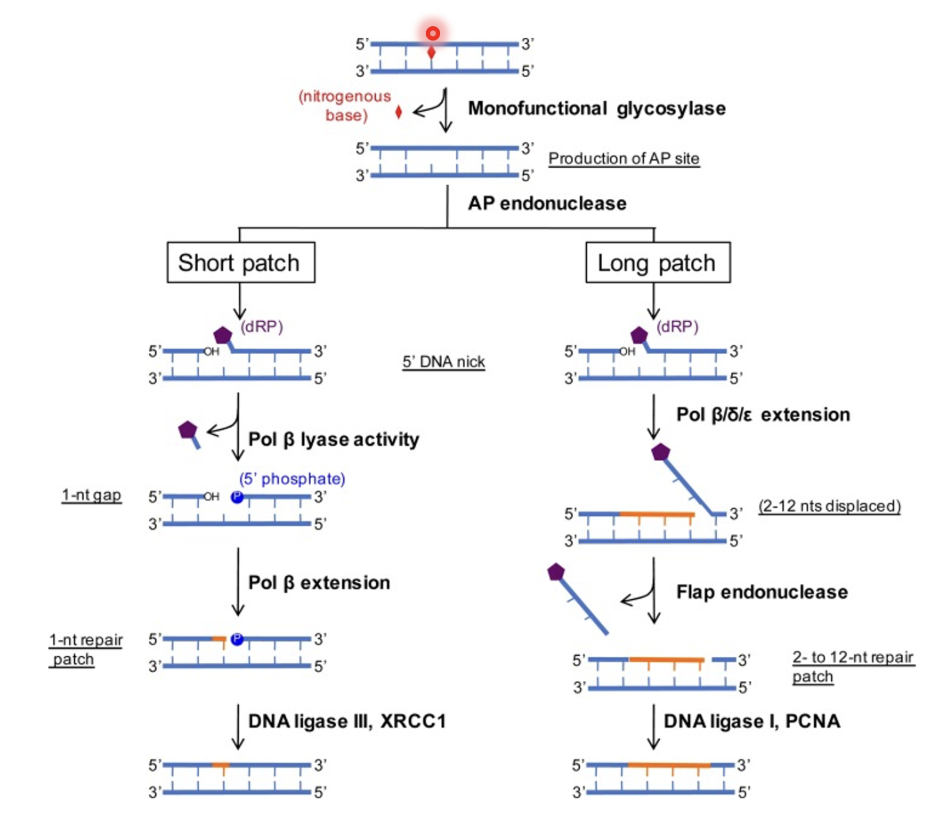
Removal of damage nitrogenous base
Initiated by monofunctional glycosylases which have high specificity for small chemical base modifications (eg deaminated cytosine, oxidised guanine…)
Nitrogenous base gets removed by glycosylase, producing an AP site (abasic site)
AP endonuclease cleaves the AP site, exposing a 3’-OH on one side of the cleavage site and deoxyribose phosphate (dRP) on the other side
This triggers either short or long patch BER (preference for either is not well understood)
Short patch
DNA pol β (has dRP lyase activity) removes the phosphorylated ribose to restore normal DNA biochemistry by exposing 5’-phosphate but leaves a 1-nt gap
Pol β extension fills the gap and DNA ligase III/XRCC1 seals the nick
Long patch
DNA pol β/δ/ε extends from the 3’-OH for 3-12 nt which generates a displaced flap
Flap endonuclease cleaves the flap and the dRP is thus removed
Nick is sealed by DNA ligase I/PCNA
Lesion recognition
The enzymes have a motor protein that slides rapidly up and down the double helix
Damaged base can be recognised by shape, hydrogen bonding potential and electric charge distribution
When sliding along the phosphate backbone they can flip out bases from the interior of the helix
Once flipped into the enzyme binding pocket, damaged bases but not normal bases get cleaved by glycosylase
Normal bases will have a short residency time in the active site but damaged bases have a long residency which permits the enzyme to attack the N-glycosidic bond to cleave the base
Damage can be recognised from seconds to minutes
Nucleotide excision repair
UVB crosslinks adjacent pyrimidines in DNA which must be repaired by NER
Xeroderma pigmentosum is a condition associated with defects in NER causing severe photosensitivity, skin and non-skin cancers, and neurological abnormalities
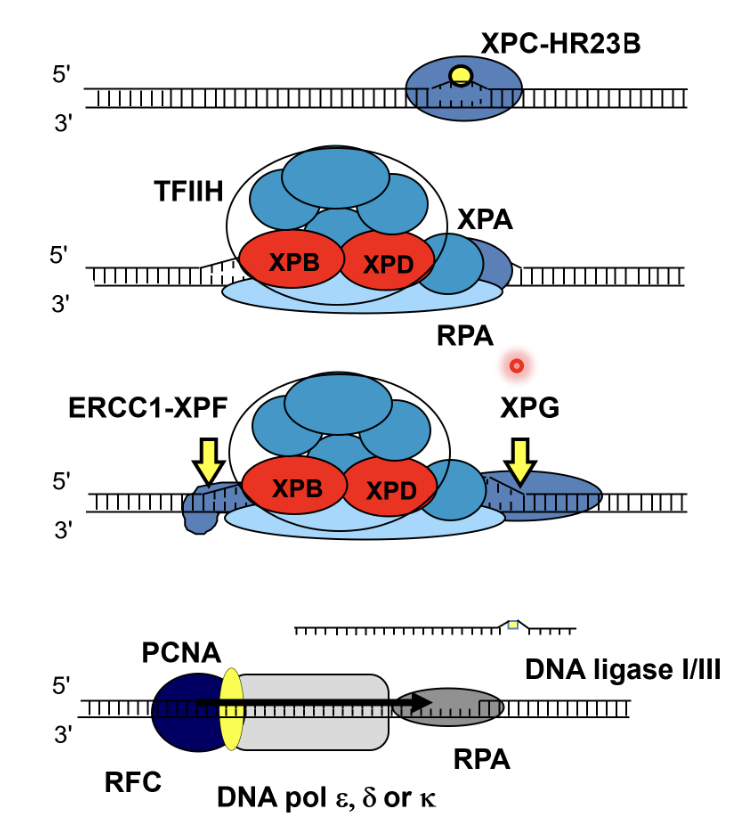
DNA damage is recognised by XPC-HR23B and the DNA duplex is opened up
TFIIH (related to another form of TFIIH involved in initial RNA pol II transcription), XPA (structural role), XPB and XPD helicases (create a bubble structure of DNA) and RPA (single stand binding protein) are recruited to the damaged strand
Bubble structure creates landmarks whereby endonucleases ERCC1-XPF (cuts 5’ to the damage) and XPG (cuts 3’ to the damage) can excise the strand
ERCC1-XPF cuts first, followed by DNA synthesis, and then XPG cuts the flap (rather than a flap endonuclease)
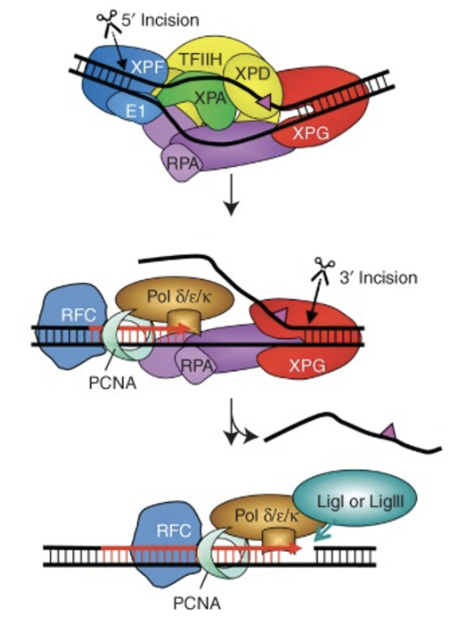
20-30nt stretch of DNA strand containing the damage is removed, leaving a gap which gets filled by DNA pol ε/δ/κ and PCNA (which is supported by RFC acting as a clamp loader) using the undamaged strand as a template
The nick is sealed by DNA ligase I/III
Damage detection
Detects bulky, large chemical damage
Damage is detected by structural distortion and defamation of the DNA structure as bulky chemical damage alters base pairing and DNA geometry, causing local unwinding/denaturation and lack of base pairing at the damaged site
XPC recognises the lack of base pairing
TFIIH is recruited by direct interaction with XPC
XPB (moving 3’ to 5’) and XPD (5’ to 3’) helicases unwind DNA around the lesion
XPD particularly acts as a damage verification helicase and moves towards the damage and its unwinding activity is arrested by DNA damage, generating a static bubble which can be cut by the endonucleases
If there is no damage XPD would continue to translocate along the DNA and continue unwinding which collapses the NER apparatus, avoiding incision
NER can be transcription coupled
A subpathway of NER operates independently of XPC-HR23B as it is triggered by RNA pol II arrest at a lesion
Cockayne syndrome A and Cockayne syndrome B proteins (CS factors) are needed to couple the arrest of RNA pol II to the NER factory
Cockayne syndrome A protein is a motor protein which hydrolyses ATP and can move things around on DNA, could possibly push RNA pol II back away from the lesion to allow space for the NER apparatus to work
They may be involved in recruited and positioning other NER proteins
NER on UV damage induced regions are is more efficient in rapidly transcribed reghions of the genome due to RNA pol II
Cells degrade RNA pol II as a last resort if transcription coupled repair fails which may enable global genome repair function
Cockayne syndrome - characterised by growth failure, impaired develoopment, photosensitivity, premature aging, hearing loss/eye abnormalities, sometimes associated with Xeroderma pigmentosum
Due to defective transcription coupled repair (80% due to defective CSA, some due to defect in core NER factors)
Tend to be protected from skin tumours and reach maturity before phenotypic consequences
One theory is that these individuals do not deal with RNA pol II stalling well and its breach of DNA damage causes permanent stalling and a gradual breakdown of the gene expression profile
DNA damage tolerance
Nearly all DNA lesions block DNA pol synthesis and replication in S phase
Achieved by translesion synthesis (TLS) - approx 16 types of TLS pols in humans
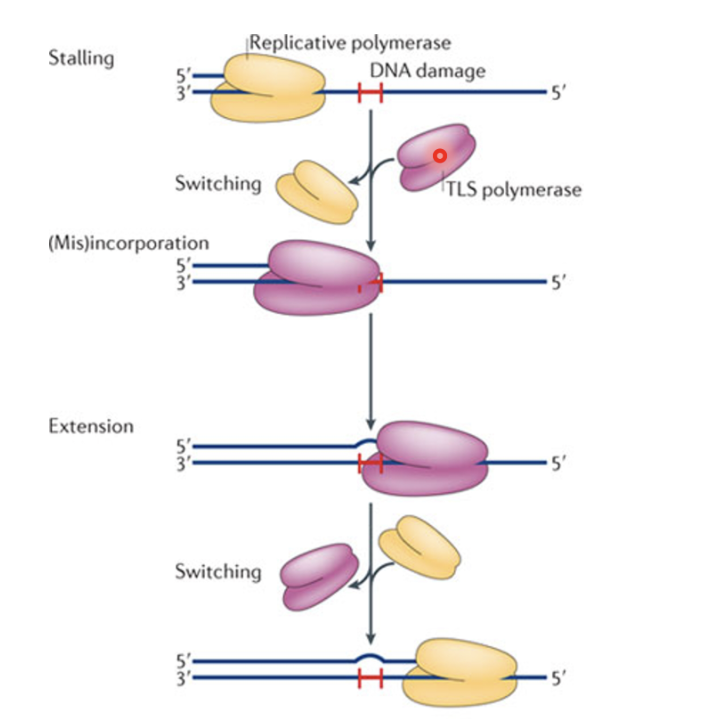
Replicative pol stalls and switches with TLS pol which can accomodate damage due to low processivity and continue extension, leading to mutation, they later switch back with the replicative pol after passing over the damaged region
Switch from replicative to TLS pol controlled by PCNA modification in eukaryotes
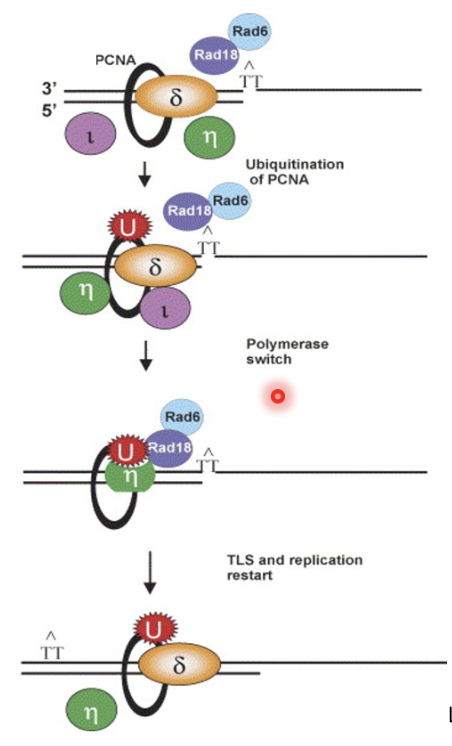
Upon arrest, replicative pol is removed/remodelled at PCNA, causing monoubiquitination of K164 on PCNA by Rad18 and Rad6
Causes recruitment of TLS pols (Pol ι and η) which continues extension and later switches back with replicative pol
Deubiquitination of PCNA is time consuming if it at all occurs, so not strictly necessary for re-exchange of TLS pol for replicative pol
Pol η has evolved to insert AA opposite TT adduct so does not necessarily cause mutation
When TLS by Pol η goes wrong it causes Xeroderma pigmentosum variant form, characterised by defect in conversion of newly synthesised DNA from low to high MW after UV irradiation
DNA interstrand crosslink repair and Fanconi anaemia
Fanconi anaemia
Recessive chromosomal instability syndrome
Bone marrow failure and childhood AML
Cancer predisposition up to x10,000
Developmental abnormalities
Genetically heterogeneous - 22 genes identified
FANCA and FANCC mutations (most common) - less severe phenotypes
Carrier freq ~1:200
Exclusive hypersensitivity to replication blocking DNA damage exemplified by DNA interstrand cross linking (ICL-inducing) agents
Defects in ICL repair leads to Fanconi anaemia
Sources - endogenous aldehydes, oxidised lipids… etc
Broad clinical implications
FANCD1 (BRCA2), Rad51C - high penetrance breast cancer susceptibility alleles
FANCJ, FANCN - low penetrance breast cancer susceptibility alleles
FANCQ (XPF) - multiple inherited cancer prone/developmental disorders
FA pathway
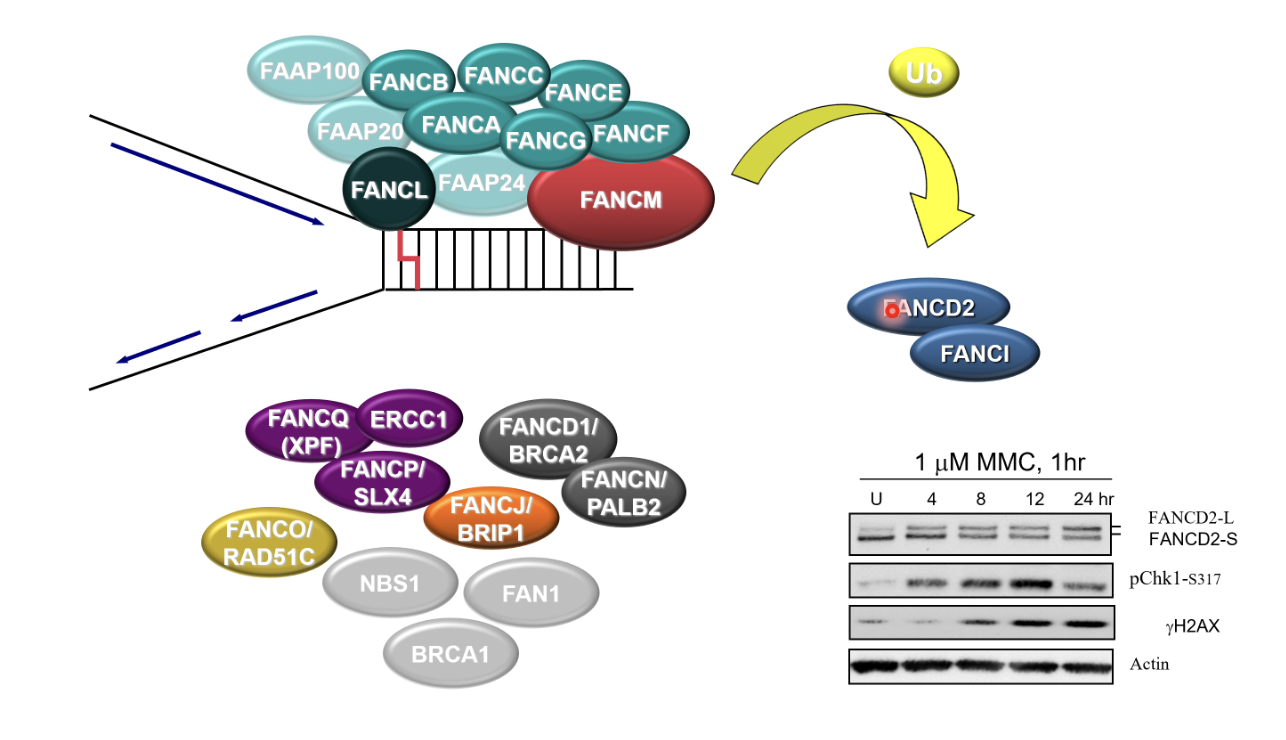
Activation (likely via phosphorylation) of FA core complex
FANKL monoubiquitinates FA ID complex (FANCD2/FANCI)
FANCD2/FANCI bind to dsDNA around the damaged replication fork and bind to it to recruit other proteins
ICL incision, TL synthesis, HR
But only FANCM (translocase), FANCL (E3 ubiqutin ligase) and XPF (nuclease) have enzyme activity
Convergence of replication forks is a requirement to initiate rep-couple ICL repair
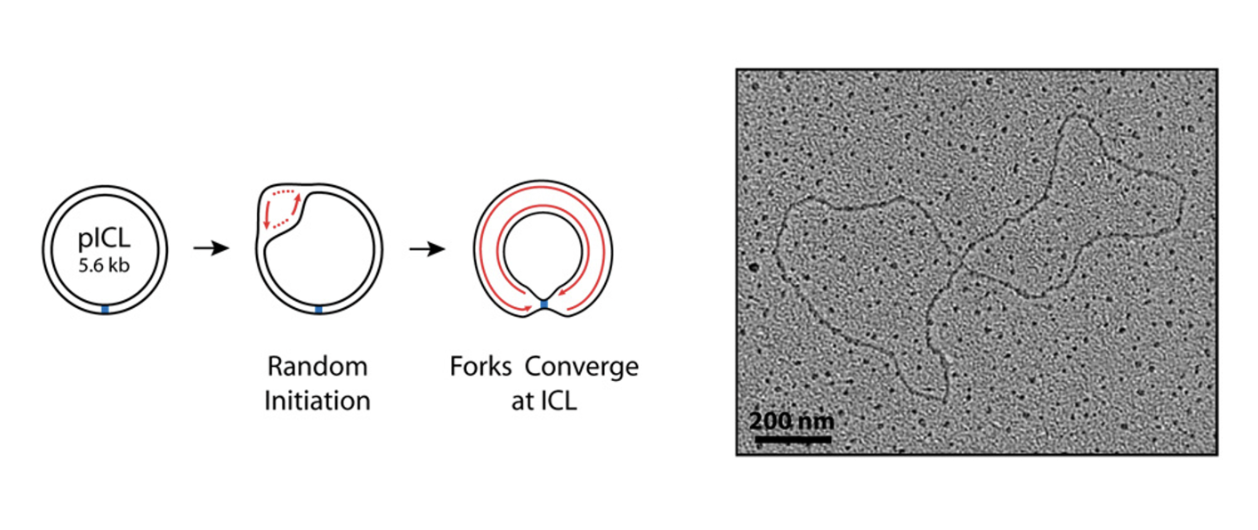
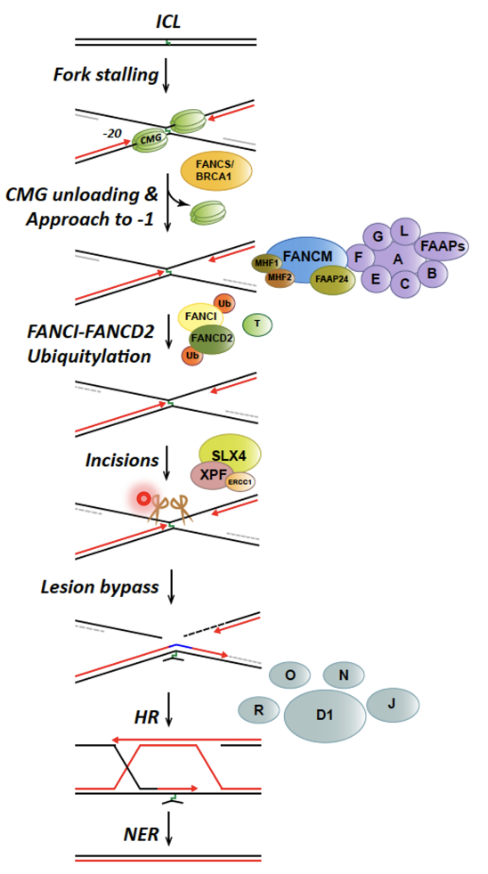
Convergence of replication forks signals the presence of the lesion and triggers FA pathway
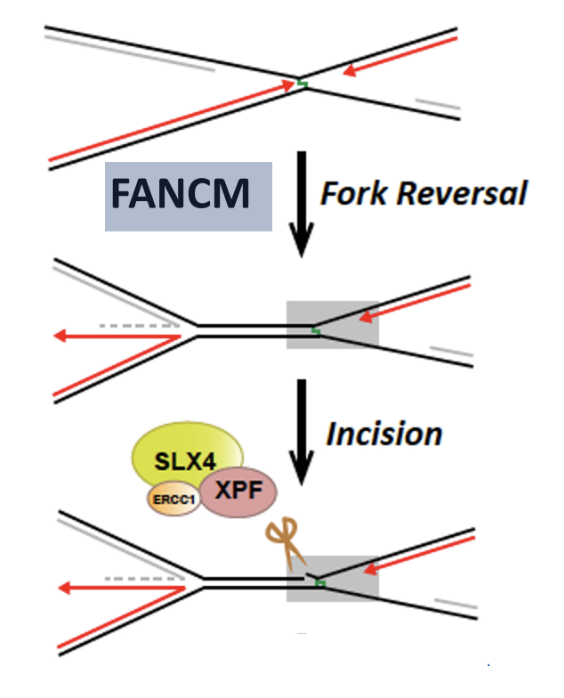
FANCM may be function to rewinds the DNA behind the stalled fork (fork reversal) which generates a region of dsDNA adjacent to the crosslink
Disassembly of the replisome (CMG unloading)
Activation of FA core complex
FANCI-FANCD2 ubiquitylation
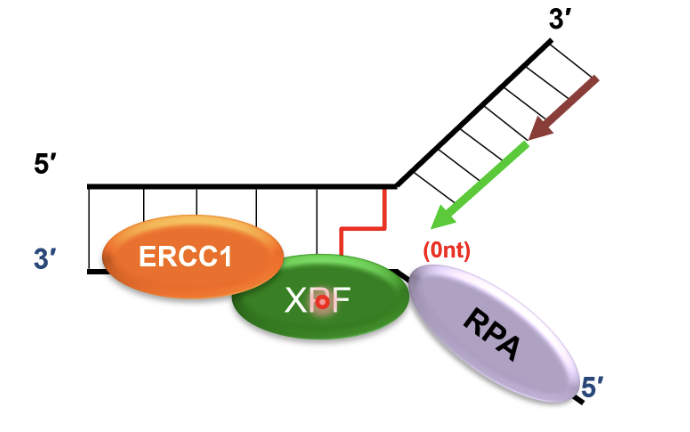
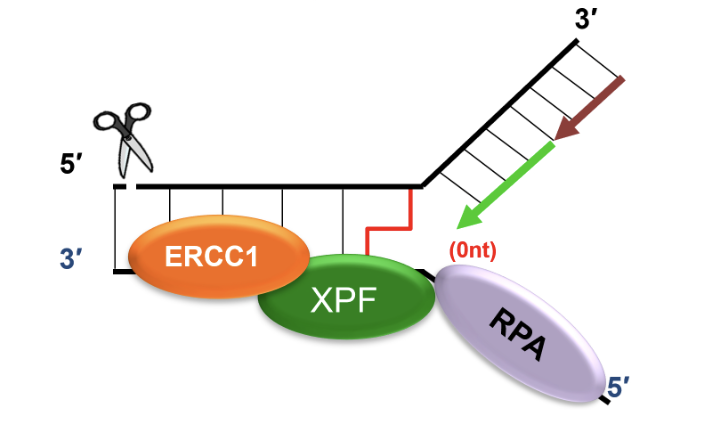
Recruits SLX4 and ERCC1-XPF to cleave DNA crosslink, releasing one of the damaged chromatids
dsDNA adjacent to crosslink produced by FANCM is an excellent substrate for SLX4 and ERCC1-XPF mediated cleavage
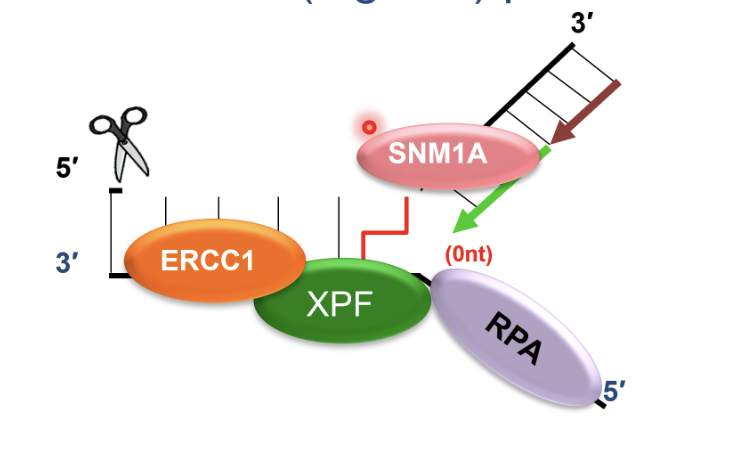
SNM1A recruited to ERCC1-XPF incised intermediate by interacting with PCNA using its PIP motif
This is a damage tolerant 5’-to-3’ exonuclease that digests through and past the incised intermediate, leaving a single nucleotide and providing a downstream template for subsequent TLS complete repair of crosslink chromatid
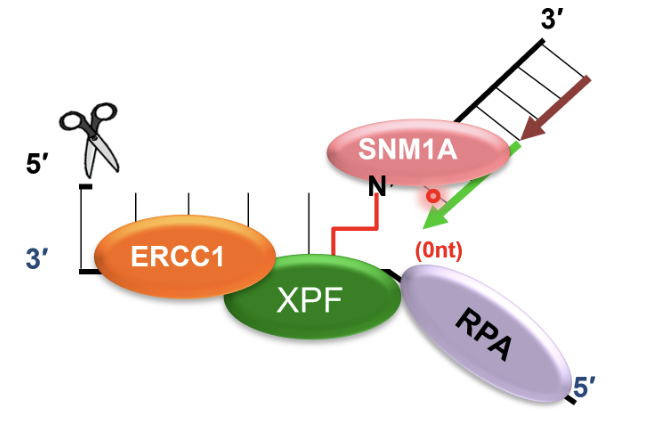
TLS polymerases take the stalled fork and extend it past the cross link in an error prone manner
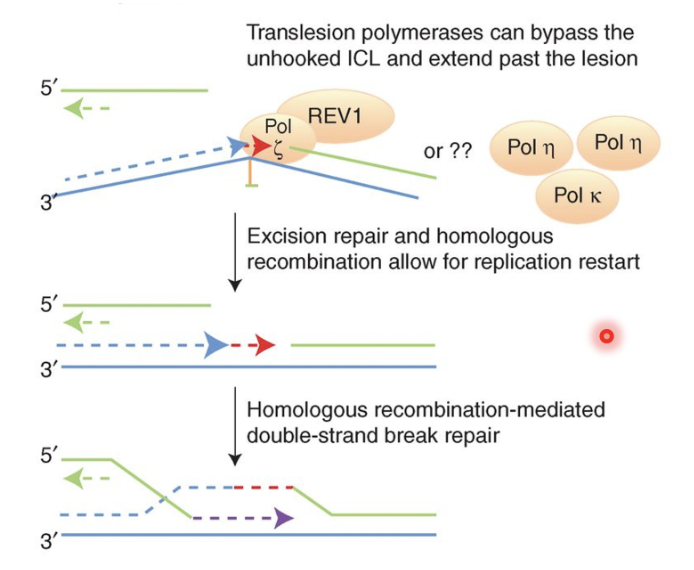
TLS pol ζ recruited to ERCC-1 incised and SNM1A-resented ICL and synthesises DNA past ICL, repairing first chromatid
This produces an intact chromatid and a broken chromatid (DNA DSB) which is repaired by BRCA2 and other HR factors using the intact chromatid as a template
Remnant of the initial crosslink may be removed by NER