MICB 211 Chapter 11 Notes
Human Microbiome
Microbiota
-ski, mucous membranes constantly in contact with microorganisms present in environment
ex: digestive tube from mouth through intestines and out the other end is constantly exposed to environmental microbes
-non-sterile areas of body are microenvironments colonized by certain microbial species
-in general, same bacterial species colonize the same anatomical sites in all people
microbiota
-some organisms establish permanent residence within host while some are more transient and present for limited periods time
-microbiota of human body is very complex and consists of more than several hundred species of microorganisms
-bacteria are most numerous and obvious microbial components of microbiota
-archaea colonize the lower intestinal tract
Host and Microbiota interactions
-association between host and microbiota is dynamic
relationship that benefits both host and bacteria
Bacteria Benefits
-bacteria in microbiome utilize host in number of ways
-host environment is a continuous source of nutrients — stable environment
-host plays a protective role against damaging conditions (desiccation, temperature extremes)
-utilize host as mode of transportation
Host Benefit
-minimizing hosts susceptibility to pathogens via microbial antagonism
-occurs when colonization of native microbiota inhibits colonization by pathogens
occurs through occupying adherence sites, utilizing nutrients, chemical inhibition and modulating pH and oxygen availability
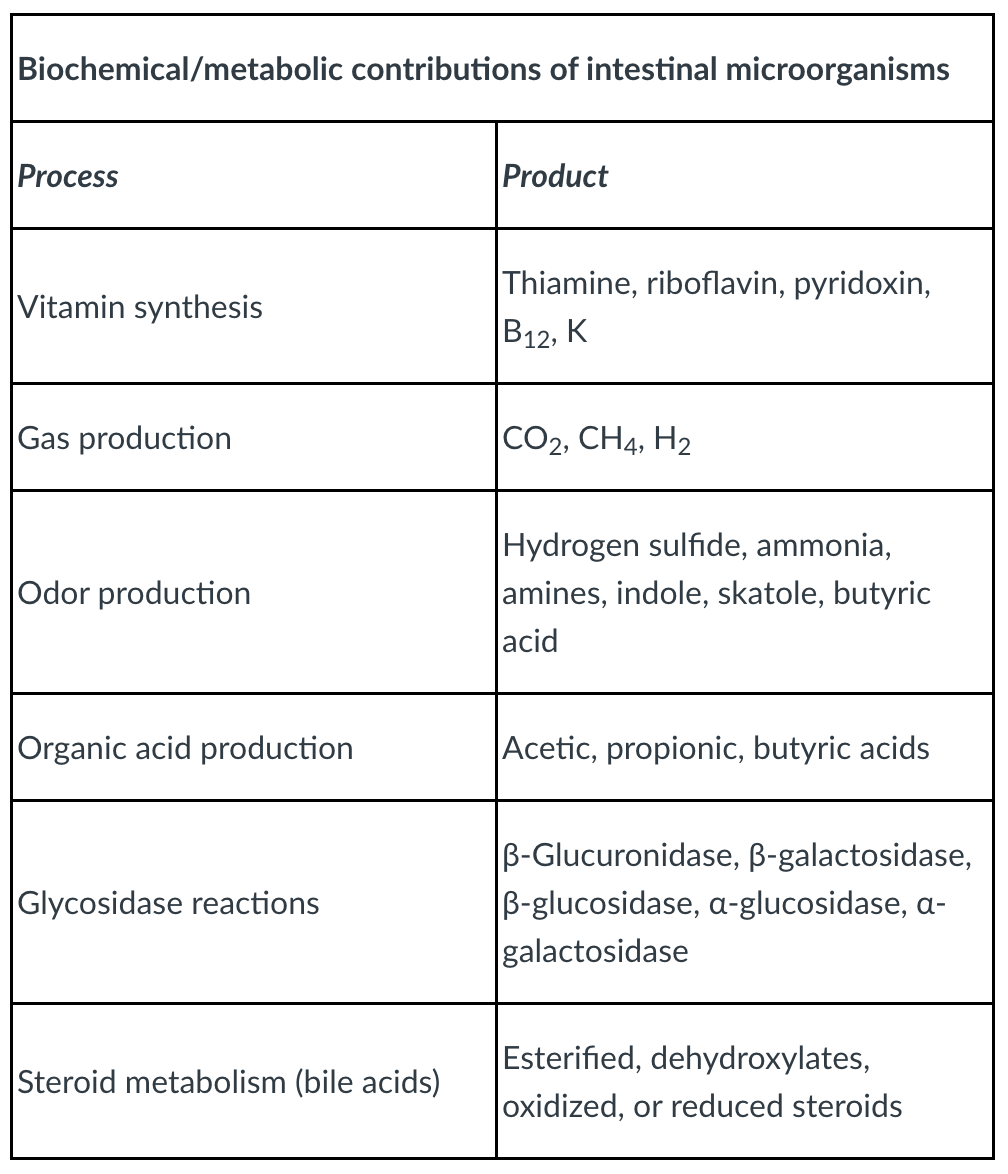
-nutritional synergism
bacteria synthesize and excrete vitamins (vitamin K, B12)
can be absorbed as nutrients by host cells
bacteria capable of converting bile acids to steroids
can be utilized by host
-microbiota stimulates immune system via induction of antibody response
low levels of antibodies can cross react with certain related pathogens → prevent infection or invasion
Microorganisms of Microbiota are Adapted to their Host
-bacteria of microbiota have adapted to survive specifically within their host
-physical association between microbiota and host likely involves biochemical interactions between bacterial surface components (ligands/adhesins) and host cell molecular receptors
-bacteria of microbiota resist innate human defences
don’t elicit strong adaptive immune responses that leads to their death
Harmful Aspects of Host-Microbiota Relationship
-not all host-microbiota interactions are beneficial
-translocation of bacteria from one niche to another can be harmful
ex: E.coli in GI tract is fine but can cause UTIs when in urinary tract
-certain normal residents of microenvironments in body may not be harmful when host is healthy but may become harmful or pathogenic under conditions when host is immunocompromised or suffers from external ailment
opportunistic pathogens
take advantage of host attenuated conditions to establish infections
Host-Microbiota Interactions that may Help or Harm Host
-intestinal microbiota can affect the brain (gut brain axis)
bidirectional
-brain acts on GI and immune functions that help to shape gut’s microbial makeup
-gut microbes affect inflammatory responses and make neuroactive compounds
including neurotransmitters and metabolites → both have effects on brain
-results in positive or negative effects on brain functions and disorders
ex: prevention or development of anxiety and MS
Chronology and Source of Microbiota for Humans
-fetal environment is sterile and prior to birth → humans are free of microorganisms
-microbiota obtained from environment immediately after birth as a result of
passage through birth canal
exposure to other humans (and pets)
ingestion of food and fluids
inhalation of air-borne microorganisms
Factors that Affect Microbiota Composition
-diet is a major contributor to microbial makeup
ex: infant’s diet of milk encourages establish of lactic acid bacteria as part of microbiota
-infection contributes to microbial diversity and composition
infection results in temporary increase in pathogen numbers but antibiotic treatment is another major factor
-antibiotics result in decrease of microbiota
in GI tract → leaves host susceptible to colonization by opportunistic bacteria
microbiota eventually re-establishes itself after antibiotic therapy
Physiological Niches of Human Microbiome
Location of Microbiota
-microbiota bacteria are located at a particular anatomical site
certain species of bacteria are invariably in one locale and never in another
exhibits a tissue preference for colonization
ex: production of stomach acids, bile salts, lysozyme, certain bacteria are unable to colonize regions of GI tract
at colonization sites, bacteria attaches to host receptors using specific bacterial ligands
these host receptors that interact with bacterial ligands are expressed only at certain locations in the host’s body
some members of microbiota are capable of constructing bacterial biofilms on surface of tissue
bacteria are also capable of growth on preexisting biofilms
biofilms are a mixture of microorganism
Microbiota of Gastrointestinal Tract
-majority of microbiota in human body resides in GI tract
stomach, small intestine, large intestine
-intestinal microbiota is a complex ecosystem containing several hundred bacterial species
majority of bacteria are anaerobes
ex: E.coli, bacteroides, lactobacillus
-stomach is very acidic
barrier to microbial growth
bacterial count of stomach is very low → devoid of any significant microbiota
-only a few acid-tolerant bacteria can be cultured from stomach
ex: H.pylori is one of the few bacteria that can colonize stomach wall
causative agent of gastric ulcers and gastric cancers
-bacteria occupy lumen, overlie the epithelial cells, adhere to mucosa of intestines
-in normal hosts, duodenal microbiota is sparse
most organisms are derived from oral cavity and pass through gut with each meal
-last part of small intestine (ileum) contains a moderately mixed microbiota
-microbiota of large intestine is dense and contains a diverse population of bacteria
viewed as anaerobic bacterial fermentation chamber
any oxygen that may be present is quickly consumed by facultative anaerobes
-at birth, entire intestinal tract is sterile but bacteria enters with first feeding on infant
initial colonizing bacteria varies with food source of infants
Essentiality of Microbiota
Gnotobiotic Organisms
-germ-free animals and birds
-colonies of gnotobiotic animals and birds established by isolating first-generation offspring using sterile techniques
-offspring raised in germ-free environments
-mammals
delivering offspring via c-section
-birds
exterior surface of egg is sterilized
-characteristics of gnotobiotic animals
abnormal anatomical and physiological features
underdeveloped lymphatic tissue and poorly developed immune systems
thin intestinal walls
low antibody titer
higher susceptibility to pathogens
reduced susceptibility to certain disease processes which are dependent on activity of microbes
Studying Microbiota
-measuring microbial diversity
Darwinian Tree of Life (TOL)
-displays evolutionary relationships between all forms of cellular life
-history of life is like a tree with multiple branches arising from a common trunk all the way to the tips of the youngest twigs, symbolic of present diversity of living organisms
-branch points present points of divergence or points at which diversity is generated from a common ancestor
-each fork of an evolutionary/phylogenetic tree is an ancestor common to all lines of decent branching from that fork
-2 organisms closely-related to each other share a common ancestor that represents a relatively recent branch point on the tree of life
-last common ancestor shared by all current life referred to as LUCA
Building a Tree of Life
-two closely-related organisms have to be similar with respect to structural characteristic while two distantly-related organisms have to be very different with respect to the same characteristic
-some essential attributes of an evolutionary indicator/chronometer:
structure should be found in all forms of life
so all organisms can be compared to each other using the same structure
structure should be able to accommodate variation over time without loss or change in function
variations should accumulate slowly so structure retains traces of ancestral patterns over billions of years of evolution
changes in structural feature should be measurable
-a good characteristic for tracing vertical evolutionary relationships might be sequence of nucleotides in RNA component of small ribosomal subunit (SSU rRNA)
all organisms must synthesize proteins, ribosomes are found in all forms of cellular life
structure of SSU rRNA already known to exhibit differences/variations between organisms without loss of ribosomal function
SSU rRNA only on component of complex translational machinery of a cell → has to interact with many other molecules involved in protein synthesis
SSU rRNA nucleotide sequences expected to vary slowly over evolutionary time
differences between nucleotide sequences of different SSU rRNA molecules could be measured in order to construct a phylogenetic tree
Woese Tree of Life
-using SSU rRNA as an evolutionary indicator/chronometer → data best represented as a TOL not with two major branches but with 3 branches
prokaryotes → domains, bacteria, archaea
Determining Composition of Microbiota
-researchers use 16S rRNA gene sequence to measure microbial diversity
-different ways to define microbial diversity and dependent on parameters measured
-diversity can be a measure of
richness
absolute number of different species in community
abundance and evenness
relative prevalence of that species in community compared to other species
more proportional the distribution, the more more even the community is as well
phylogenetic distance
how closely related the species are to each other on a phylogenetic tree
-higher the richness, evenness, phylogenetic distance → more diverse the community