AP Biology Review
Unit 1
Water properties: adhesion, cohesion, capillary action, surface tension, high specific heat
Acidic - lots of H+, Basic - lots of -OH
Carbohydrates: CHO, 1 C : 2 H : 1O
Proteins: CHONS, made of amino acids (amine group, carboxyl group, R group)
Lipids: CHOP, also includes phospholipids & steroids, not true polymers, double bonds in fatty acids means unsaturated
Nucleic acids: CHOPN, made up of three parts: a phosphate group, a 5-carbon sugar, and a nitrogenous base. The nitrogenous base can be either a purine (adenine or guanine) or a pyrimidine (cytosine, thymine or uracil).
Unit 2
- Surface Area to Volume Ratio - high ratio means more efficient exchange with outside environment.
- Need to know eukaryotes vs prokaryotes.
- Plasma membrane: selectively permeable, need to know about integral proteins, aquaporins, glycoproteins, and glycolipids.
- Hypertonic - cell shrivels due to high solutes outside, hypotonic - cell bloats due to low solutes outside
Unit 3
- First Law of Thermodynamics: Cells cannot take energy out of thin air. It must harvest it from somewhere.
- Second Law of Thermodynamics: It states that energy transfer leads to less organization. That means the universe tends toward disorder (entropy). In order to power cellular processes, energy input must exceed energy loss to maintain order. Cellular processes that release energy can be coupled with cellular processes that require an input of energy.
- Endergonic reactions - require energy input, exergonic reactions - release energy
- Factors that affect reaction rates - temperature, pH, concentration of product/substrate/inhibitors
- Light-Dependent Reactions: PSII loses electrons which are passed down the ETC and are used to create a proton gradient, which allows for the creation of NADPH and ATP. To replenish the lost electrons water is split into protons, electrons, and oxygen.
- Light-Independent Reactions: carbon fixation (rubisco slams a CO2 onto a ribulose bisphosphate to create two 3-PGA), reduction (ATP and NADPH are used to create G3P), regeneration (ATP is used to create RuBP)
- Glycolysis: glucose is broken down into two pyruvate (ucose + 2 ATP + 2NAD+ → 2 Pyruvic acid + 4 ATP + 2NADH)
- Acetyl CoA Formation: 2 Pyruvic acid + 2 Coenzyme A + 2NAD+ → 2 Acetyl-CoA + 2CO2 + 2NADH
- Krebs Cycle: It begins with acetyl-CoA joining with oxaloacetate to make citric acid and ends with oxaloacetate, 1 ATP, 3 NADH, and 1 FADH2;
- Electron Transport Chain: electrons from the 10 NADH and 2 FADH2 are used to synthesize ATP, final electron acceptor is oxygen
- Anaerobic Respiration: Glycolysis also gives two pyruvates and two NADH. The pyruvate and NADH make a deal with each other, and pyruvate helps NADH get recycled back into NAD+ and takes its electrons. The pyruvate turns into either lactic acid (in muscles) or ethanol (in yeast).
Unit 4
- Signal transduction: reception, transduction, response
- Ligand-gated ion channels in the plasma membrane open or close an ion channel upon binding a particular ligand. This channel opens in response to acetylcholine, and a massive influx of sodium depolarises the muscle cell and causes it to contract.
- Catalytic (enzyme-linked) receptors have an enzymatic active site on the cytoplasmic side of the membrane. Enzyme activity is initiated by ligand binding at the extracellular surface.
- A G-protein-linked receptor does not act as an enzyme, but instead will bind a different version of a G-protein (often GTP or GDP) on the intracellular side when a ligand is bound extracellularly. This causes activation of secondary messengers within the cell. One important second messenger is cyclic AMP (cAMP).
- Interphase: growth phase of the cell (cell grows like regular and chromosomes are duplicated)
- In eukaryotes, checkpoint pathways function mainly at phase boundaries (such as the G1/S transition and the G2/M transition). When damaged DNA is found, checkpoints are activated and cell cycle progression stops. The cell uses the extra time to repair damage in DNA. If the DNA damage is so extensive that it cannot be repaired, the cell can undergo apoptosis, or programmed cell death.
- Mitosis: prophase (nuclear envelope dissolves and chromosomes condense), metaphase (chromosomes line up at the metaphase plate), anaphase (chromosomes are pulled away from the center), cytokinesis & telophase (cytoplasm splits and two cells are formed)
Unit 5
- Law of Segregation: Each gamete gets only one of the copies of each gene.
- Law of Independent Assortment: Each pair of homologous chromosomes splits independently, so the alleles of different genes can mix and match.
- Most sex- linked traits are found on the X chromosome and are more properly referred to as “X-linked.”
- A Barr body is an X chromosome that is condensed and visible. In every female cell, one X chromosome is activated and the other X chromosome is deactivated during embryonic development.
- Incomplete dominance (**blending inheritance): In some cases, the traits will blend. For example, if you cross a white snapdragon plant (genotype WW) with a red snapdragon plant (RR), the resulting progeny will be pink (RW). In other words, neither color is dominant over the other.
- Codominance: Sometimes you’ll see an equal expression of both alleles. For example, an individual can have an AB blood type. In this case, each allele is equally expressed.
- Polygenic inheritance: In some cases, a trait results from the interaction of many genes. Each gene will have a small effect on a particular trait.
- Non-nuclear inheritance: Apart from the genetic material held in the nucleus, there is also genetic material in the mitochondria. The mitochondria are always provided by the egg during sexual reproduction, so mitochondrial inheritance is always through the maternal line, not the male line. In plants the mitochondria are provided by the ovule and are maternally inherited.
- Prophase I
- As in mitosis, the nuclear membrane disappears, the chromosomes become visible, and the centrioles move to opposite poles of the nucleus.
- The major difference involves the movement of the chromosomes. In meiosis, the chromosomes line up side-by-side with their counterparts (homologs). This event is known as synapsis.
- Synapsis involves two sets of chromosomes that come together to form a tetrad (a bivalent). A tetrad consists of four chromatids. Synapsis is followed by crossing-over, the exchange of segments between homologous chromosomes.
- What’s unique in prophase I is that pieces of chromosomes are exchanged between homologous partners. This is one of the ways organisms produce genetic variation.
- Metaphase I
- As in mitosis, the chromosome pairs—now called tetrads—line up at the metaphase plate.
- By contrast, you’ll recall that in regular metaphase, the chromosomes line up individually.
- One important concept to note is that the alignment during metaphase is random, so the copy of each chromosome that ends up in a daughter cell is random.
- Anaphase I
- During anaphase I, each pair of chromatids within a tetrad moves to opposite poles. The homologs will separate with their centromeres intact.
- The chromosomes now move to their respective poles.
- Telophase I
- During telophase I, the nuclear membrane forms around each set of chromosomes.
- Finally, the cells undergo cytokinesis, leaving us with two daughter cells.
- During prophase II, chromosomes once again condense and become visible.
- In metaphase II, chromosomes move toward the metaphase plate. This time they line up single file, not as pairs.
- During anaphase II, chromatids of each chromosome split at the centromere, and each chromatid is pulled to opposite ends of the cell.
- At telophase II, a nuclear membrane forms around each set of chromosomes and a total of four haploid cells are produced.
Unit 6
DNA is made up of repeated subunits of nucleotides. Each nucleotide has a five-carbon sugar, a phosphate, and a nitrogenous base.
The name of the pentagon-shaped sugar in DNA is deoxyribose.
adenine—a purine (double-ringed), guanine—a purine (double-ringed), cytosine—a pyrimidine (single-ringed), thymine—a pyrimidine (single-ringed), uracil - a pyrimidine (single-ringed)
Adenine pairs up with thymine (A–T) by forming two hydrogen bonds.
Cytosine pairs up with guanine (G–C) by forming three hydrogen bonds.
The 5′ end has a phosphate group, and the 3′ end has an OH, or “hydroxyl,” group.
The 5′ end of one strand is always opposite to the 3′ end of the other strand. The strands are therefore said to be antiparallel.
DNA helix twists and rotates during DNA replication, another class of enzymes, called DNA topoisomerases, cuts, and rejoins the helix to prevent tangling.
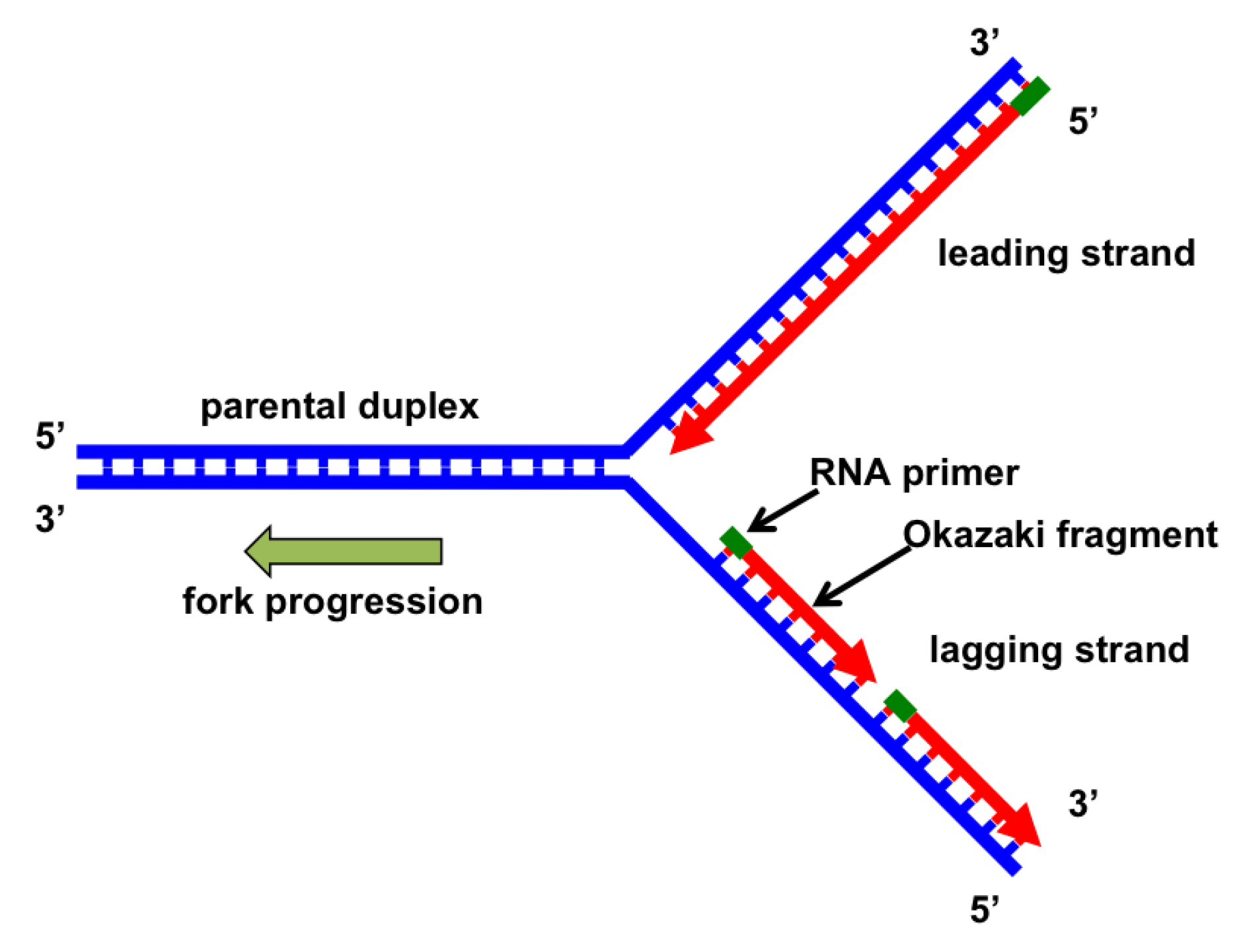
To start off replication, an enzyme called RNA primase adds a short strand of RNA nucleotides called an RNA primer.
DNA polymerase works in the 3’ to 5’ direction while new nucleotides are added in the 5’ to 3’ direction
However, when the double-helix is “unzipped,” one of the two strands is oriented in the opposite direction—3′ to 5′.
Because DNA polymerase doesn’t work in this direction, the strand needs to be built in pieces.
The lagging strand is built in the opposite direction of the way the helix is opening, so it can build only until it hits a previously built stretch. Once the helix unwinds a bit more, it can build another Okazaki fragment.
Messenger RNA (mRNA) is a temporary RNA version of a DNA recipe that gets sent to the ribosome.
Ribosomal RNA (rRNA) makes up part of the ribosomes.
Transfer RNA (tRNA) brings amino acids to the ribosomes. It brings the brings a specific amino acid into place at the appropriate time by matching anticodons to codons. It does by reading the message carried by the mRNA.
The strand that serves as the template is known as the antisense strand.
The other strand that lies dormant is the sense strand, or the coding strand.
RNA polymerase builds RNA by adding nucleotides only to the 3′ side, therefore building a new molecule from 5′ to 3′
In eukaryotes, the introns must be removed before the mRNA leaves the nucleus. This process, called splicing, is accomplished by an RNA-protein complex called a spliceosome. In addition, a poly(A) tail is added to the 3′ end and, a 5′ GTP cap is added to the 5′ end.
Initiation: The start codon is A–U–G, which codes for the amino acid methionine. The tRNA with the complementary anticodon, U– A–C, is methionine’s personal shuttle; when the AUG is read on the mRNA, methionine is delivered to the ribosome.
Elongation: Addition of amino acids is called elongation and when many amino acids link up, a polypeptide is formed.
Termination: The synthesis of a polypeptide is ended by stop codons. There are three that serve as a stop codon. Termination occurs when the ribosome runs into one of these three stop codons.
Structural genes code for enzymes needed in a chemical reaction. These genes will be transcribed at the same time to produce particular enzymes.
The promoter gene is the region where the RNA polymerase binds to begin transcription.
The operator is a region that controls whether transcription will occur; this is where the repressor binds.
The regulatory gene codes for a specific regulatory protein called the repressor. The repressor is capable of attaching to the operator and blocking transcription.
Post-transcriptional regulation occurs when the cell creates an RNA, but then decides that it should not be translated into a protein. This is where RNAi comes into play.
RNAi molecules can bind to an RNA via complementary base pairing. This creates a double-stranded RNA
Post-translational regulation can also occur if a cell has already made a protein, but doesn’t yet need to use it.
Base substitution (point) mutations result when a single nucleotide base is substituted for another. There are three different types of point mutations:
Nonsense mutations cause the original codon to become a stop codon, which results in early termination of protein synthesis.
Missense mutations cause the original codon to be altered and produce a different amino acid.
Silent mutations happen when a codon that codes for the same amino acid is created and therefore does not change the corresponding protein sequence.
Retroviruses like HIV are RNA viruses that use an enzyme called reverse transcriptase to convert their RNA genomes into DNA so that they can be inserted into a host genome.
Unit 7
- Natural Selection: Natural selection is a process by which species of animals and plants that are best adapted to their environment survive and reproduce, while those that are less well adapted die out.
- Evolution: Evolution is the process by which different kinds of living organisms are thought to have developed and diversified from earlier forms during the history of the earth
- Evidence for Evolution: Fossil Record, Biogeography, and Comparative Morphology
- Natural selection requires genetic variation and an environmental pressure.
- Biotic and abiotic factors can affect the direction of evolution.
- As long as a mutation does not kill an organism before it reproduces it may be passed on to the next generation.
- Survival of the fittest is the name of the game, and any trait that causes an individual to reproduce better gives that individual evolutionary fitness
- Another type of selection to be aware of is artificial selection. This is a type of selection where humans directly affect variation in other species.
- With different variation and different environmental pressures, they could each change in different ways and no longer be able to mate. This is called divergent evolution.
- Divergent evolution that occurs quickly after a period of stasis is called punctuated equilibrium.
- Pre-zygotic barriers prevent fertilization.
- A post-zygotic barrier is related to the inability of a hybrid organism to produce offspring.
- Convergent evolution is the process by which two unrelated and dissimilar species come to have similar traits, often because they have been exposed to similar selective pressures.
- A population is in Hardy-Weinberg equilibrium if: it has a large population size, no gene flow, random mating, no mutations, and no natural selection.
- Stanley Miller and Harold Urey simulated the conditions of primitive Earth in a laboratory. They put the gases theorized to be abundant in the early atmosphere into a flask, struck them with electrical charges in order to mimic lightning, and organic compounds similar to amino acids appeared.
- The original life-forms were simply molecules of RNA. This is called the RNA-world hypothesis.