DNA Running Notes
9/26
Chromatin vs Chromosome (different forms of DNA)
think of Raiz’s moving houses analogy
Chromatin (use form)
contain histone proteins
histone proteins act like spools for yarn (tightly wound, super thin)
exist in groups of 8
Chromosome (moving form)
Most cells in the body are NOT-DIVIDING
when cells are not dividing (making proteins)
karyotype
(the picture of the complete set of chromosomes)
chromosomes are stained → banding pattern (stripes) become visible
homologous chromosomes
same size/shape, same banding pattern (when stained), same centromere position
same genes coding for the same traits @ the same loci
alleles within gene loci MAY or MAY NOT be identical
somatic cells = body cell (skin cell, liver cell, eye cell, etc.)
2-3% of DNA actually codes for protein
TQ: Know how to draw dividing and non-dividing cells (diploid)
9/29
Central Dogma
DNA → RNA → Protein (mRNA), rRNA, tRNA, RNAi, other???
3’ → OH; 5’ → PO4
always a bond btwn 3’ OH and 5’ PO4
starts @ 5’ and ends at 3’
DNA is antiparallel
3 phosphates → exergonic reaction powers an endergonic one (coupled)
removing 2 phosphates will give energy to power DNA poly to put new nucleotides onto the growing DNA strand.
DNA replication
does not go from end to end, too long and time-consuming → instead there are POINTS OF ORIGIN (where the enzymes will start replication)
semiconservative - half old NA, half new daughter strands
helicase will open the DNA from the point of origin from two different directions
point of origin is part of “non-coding DNA”
unwind only a little bit to protect from damage
topoisomerase
in front of helicase
keeps the DNA from getting bunched up
breaking covalent bonds within the DNA
ssbp (single strand binding proteins)
at the replication fork and as you open up the DNA, the nitrogen base pairs will want to come together (due to h-bond attraction)
ssbp’s will grab and bind the outside of the DNA to keep it apart to allow DNA polymerase to come and read to make new strands
DNA polymerase
multiple exist within a replication bubble at a time (not shown b/c would be cluttered)
can only build a new DNA strand in the 5’ to 3’ direction
cannot just PUT in a new nucleotides, must have nucleotides already laid down with a substrate to covalently bond with
NEEDS A PRIMER (short piece of RNA already put down on the DNA to act like a substrate)
analogy: clay pot making analogy
primase enzyme puts down RNA nucleotides FIRST (5-10 nucleotides in length)
stick 5’ end of the new strand to the 3’ end of the primer (still follows the 5’ → 3’)
leading strand → built continuously, rep fork continues to OPEN MORE
two mirror images are present
9/30
RNA primers are not identical (deoxyribose vs ribose sugar, T vs U) to the parent DNA → must be removed
Active site for DNA polymerase did not evolve to build 3’ → 5’ or read 5’ → 3’
If nucleotide is attached to base DNA, weak hydrogen bond interaction will mean after the DNA moves, the nucleotide will just fall off
Understand polarity of DNA and 5’ → 3’ for the Quiz
Polarity of DNA
DNA poly reads 3’ → 5’, BUILDS 5’ → 3’
Primase puts primers on both sides at the same time (@ point of origin)
Leading strand happens first, Lagging strand happens AFTER as more DNA is unzipped by helicase
Can’t open all DNA at the same time b/c nucleotides are at risk to be damaged
After the new nucleotides are added, DNA is double stranded again → needs to be rewinded (histones)
As DNA poly runs into a primer, it’ll knock it off and then continue to attach DNA nucleotides OVER IT
DNA poly can’t make phosphodiester bond → ligase
DNA poly can make bond between 3’ to 5’ but can’t make the bond between an existing 5’
can’t make bond in front, has to make bond behind (you know what i mean)
active site structure doesn’t allow for it
ligase
“seals the nicks” between the Okazaki fragments, between Okazai fragments and leading, and between leading and lagging
like cement, no nucleotides are placed
reducing activation energy, hydrolyzing to create the bond between the nucleotides
know how to recognize nucleotides
enzyme structures and their functions (like WHAT ARE THEY DOING)
10/3
Haploid and diploid has nothing to do with double-arm and single arm
Bikini Diagram
DNA poly knocks out RNA primers, puts in new nucleotides
can only bind 5’ to existing 3’ OH
b/c DNA poly is an enzyme with specific active site, cannot create covalent (phosphodiester bond) between existing 3’ to 5’
primers fall apart and get reused after getting knocked out
DNA replication only happens before cell division
10/6
G-C (3 hydrogen pairs, remember C is the third letter)
A-T(2 hydrogen pairs, T is for two)
C and T are (pyrimidines, ONE ring)
A and G are (purines, TWO rings)
DNA replication only happens when cell receives signal to divide
3 types of RNA diagram
Transcription + Translation Intro Video
10/7
aminoacyl-tRNA synthetase
coupled reaction
functional efficiency of enzyme
review this sheet
Doesn’t matter if DNA is paired with DNA or anticodon with mRNA codons → antiparallel structure (TEST QUESTION)
61 anti-codons (don’t forget the 3 stops because STOP codons exist because tRNA doesn’t exist with the appropriate anticodon
Why are there redundancies in codons vs amino acids (64 vs 20)
Wobble hypothesis
when some of the tRNA comes third position (nucleotide in codon) doesn’t matter as much, can suffice → wobble
first two nucleotides matter the most
Other theory
some triplets occur at higher percentages
possibly to slow the ribosome down, sometimes it works too quickly that it’ll put the wrong amino acid in
Transcription
Transcription Initiation Complex (T.I.C.)
study the diagram
If you need a LOT of mRNA to be transcribed (bc you need a lot of enzyme to be produced) → purpose of T.I.C.
Remember that the start codon and stop codon are not at the ends of the exon
In eukaryotes every gene has a unique promoter that regulates it
will talk about bacteria and operons later
Promoter
starts transcription and sets up TIC
puts RNA poly in right position
Hard part for students to understand!!
rocks and boulder lookin things on transcription diagram are all PROTEINS (Transcription Factors, TFs)
Transcription factors are what give RNA poly access to certain genes
TATA box (located inside of promoter)
thymine-adenine repeated
TATA binding protein (type of TF) will bind to TATA box
Basal TFs
TFs are premade, in existence of the cell in loads (in cytoplasm and vesicles, ready to go)
they’re not activated though
Cell might have something bind → signals transmitted like dominoes → ends up ACTIVATING TFs (i.e. phosphorylation, goes from off to on)
No need for individual Basal TFs for every gene (cells are roughly 6-10 types)
Analogy: BTF is like the structure that rockets launch off of, RNA is the rocket
PROMOTERS ARE NOT TRANSCRIBED, only found in DNA and coded for during transcription
Upstream, u have sequences of DNA that also assist with regulating
enhancers boost frequency of transcription through stabilizing TF complex
the larger the protein complex → the greater the amount of transcription will be (remember its all temporary)
silencers
if repressor protein binds to the silence region, prevents accumulation of proteins → less likely to be transcribed

General Order (in order to ensure that RNA poly lands in the correct location)
TATA binding protein
Basal TF
activate in different combination to activate different gene
set number per cell
Activators
Co-activators
start and end with an exon (transcription ends at the very end of an exon)
Primary Transcript
parts before the AUG and after all the exons/introns are transcribed
Termination sequence:
UTR:
UNTRANSLATED REGION
hairpin loop, mRNA folds in onto itself
UTR + poly A tail, allows mRNA to exist in the cytoplasm longer → translate it for longer (remember multiple ribosomes on mRNA)
Post Transcriptional Modification of mRNA
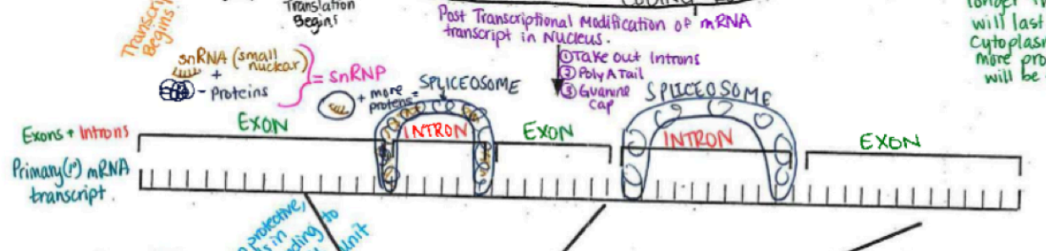
spliceosome is a ribonucleic protein (protein + RNA)
recognize mRNA sequences at the ends of exons and beginnings of introns
binds, then snips introns out → introns are degraded and recycled
exons are joined together
QUESTION: if breaking the bonds between introns and exons, how is this reaction fueled?
Guanine cap (with three phosphates attached) on 5’
think hard hat
may be the key to getting out of the nuclear membrane (remember it is highly selective)
polyA tail (~200 adenine nucleotides long)
protects end of transcript (think steel toed boots)
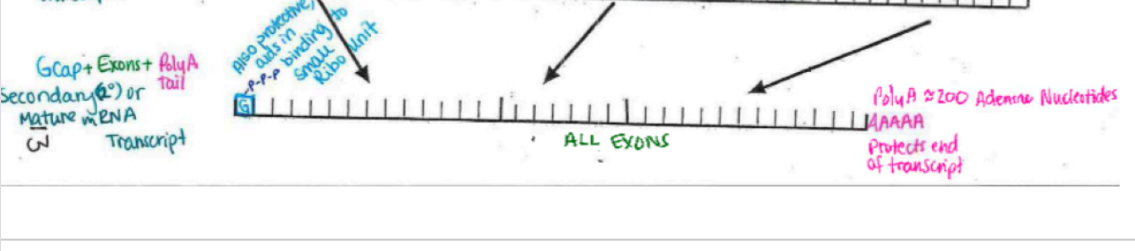
10/9 (i wasn’t here)
Telomeres
10/10
First amino acid is N terminus, last is C terminus (amine and carboxyl group)
Operons
TIC → shows the control of gene expression
Rather than having unique TFs for every gene → combinations of TFs are placed to express genes (combination lock analogy)
combinatorial control
Coordinated Control