AP Bio Unit 6 Gene expression and regulation ultimate notes
DNA and RNA structure
discovery of DNA structure
in the 1950’s Rosalind Franklin performed X-ray Crystallography of DNA
her work revleaed a pattern that was regular and repetitive
during the same time, Edwin Chargaff analyzed DNA samples from different species
he found the following rule held true for all species
the amount of adenin equals the amount of theyme (2 H bonds)
the amount of cytosine equals the amounts of guanine (3 H bonds)
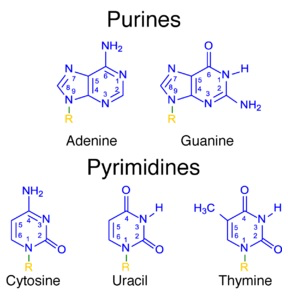
Nucleotide structure
purines
double ring structure (A,G)
Pyrimidines
single ring structure (C,U,T)
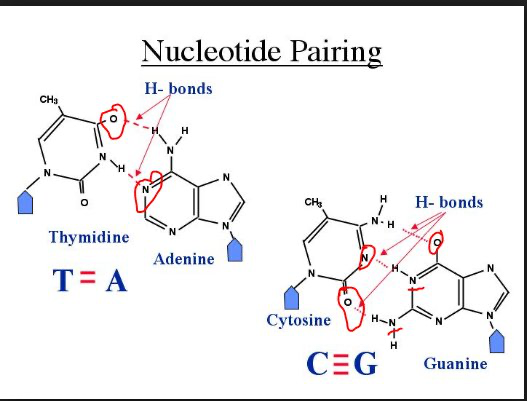
nucleotide pairing
the base pairs are held together by hydrogen bonds
adenine and thymine have two hydrogen bonds
cytosine and guanine have three hyrdrogen bonds
discovery of DNA structure
watson and crick combined the findings of franklin (helix shape) and chargaff (base pairing) to create the first 3D, double helix model of DNA
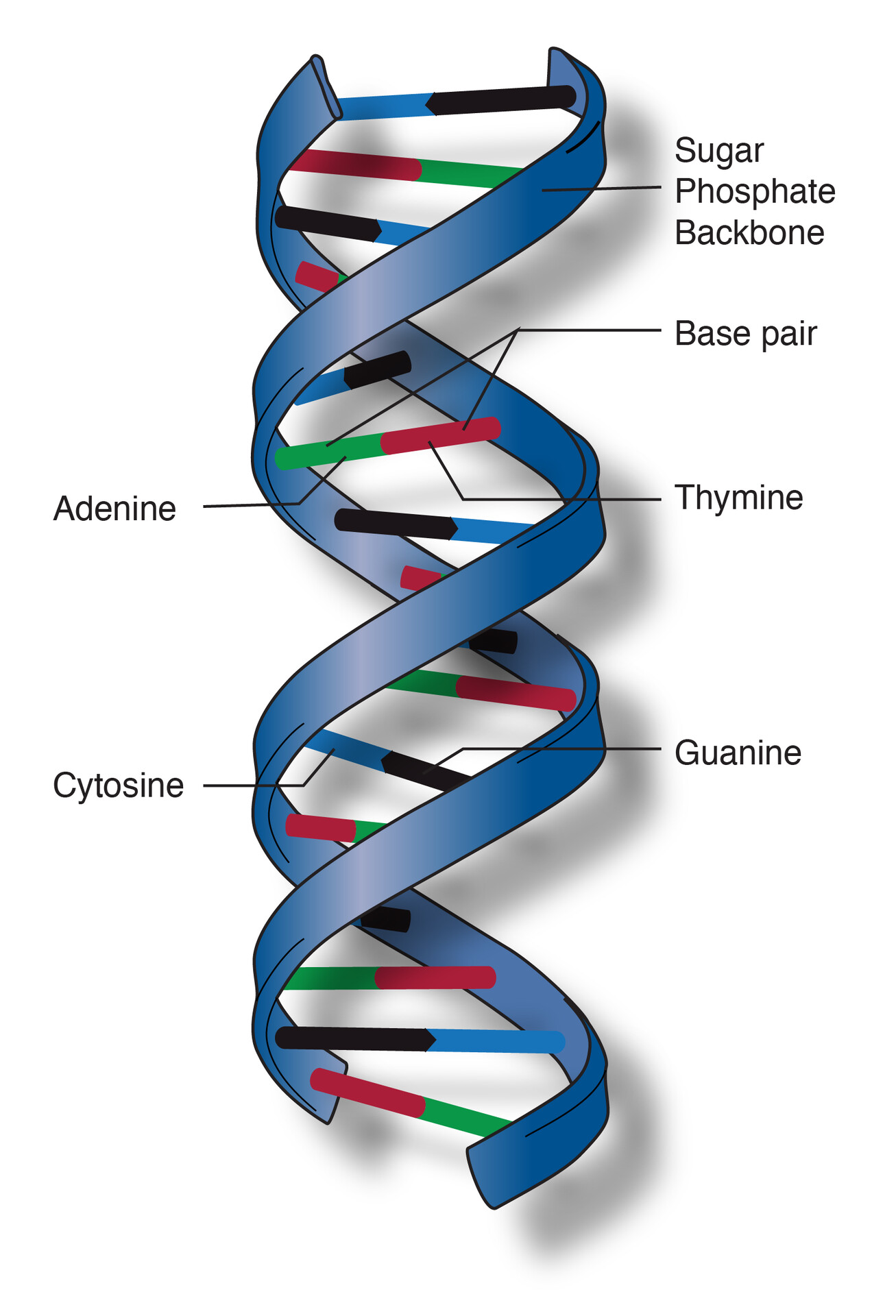
key features of DNA structure
DNA is a double stranded helix
a. backbone:
sugar-phosphate
b. center:
nucleotides pairing
DNA strands are antiparallel
a. one strand runs 5’ to 3’, other strand runs in opposite, upside-down direction 3’ ti 5’
b. 5’ end: free phosphate group
c. 3’ end: free hydroxyl group
key function of DNA
DNA is the primary source of heritable information
genetic information is stored in and passed from one generation to the next through DNA
exception: RNA is the primary source of heritable information in some viruses
prokaryotice vs eukaryotic DNA
Eukaryotic cells | prokaryotic cells |
DNA found in nucleus | DNA is in nucleoid region |
linear chromosomes | Chromosomes are circular |
prokaryotes (and some eukaryotes) also contain plasmids. they are small, circular DNA molecules that are seperate from the chromosomes |
plasmids
plasmids replicate independently from the chromosomal DNA
primarily found in prokaryotes
contain genes that may be useful to the prokaryote when it is in a particular environment, but may not be required for survivial
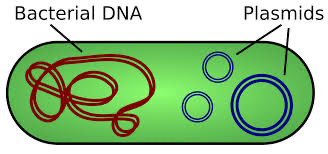
plasmids can be manipulated in laboratories
plasmids can be removed from bacteria, then a gene of interest can be inserted into the plasmid to form recombinant plasmid DNA
when the recombinant plasmid is inserted back into the bacteria the gene will be expressed
bacteria can exchange genes found on plasmids with neighboring bacteria
once DNA is echanged the bacteria can express the genes acquired
helps with survival of prokaryotes
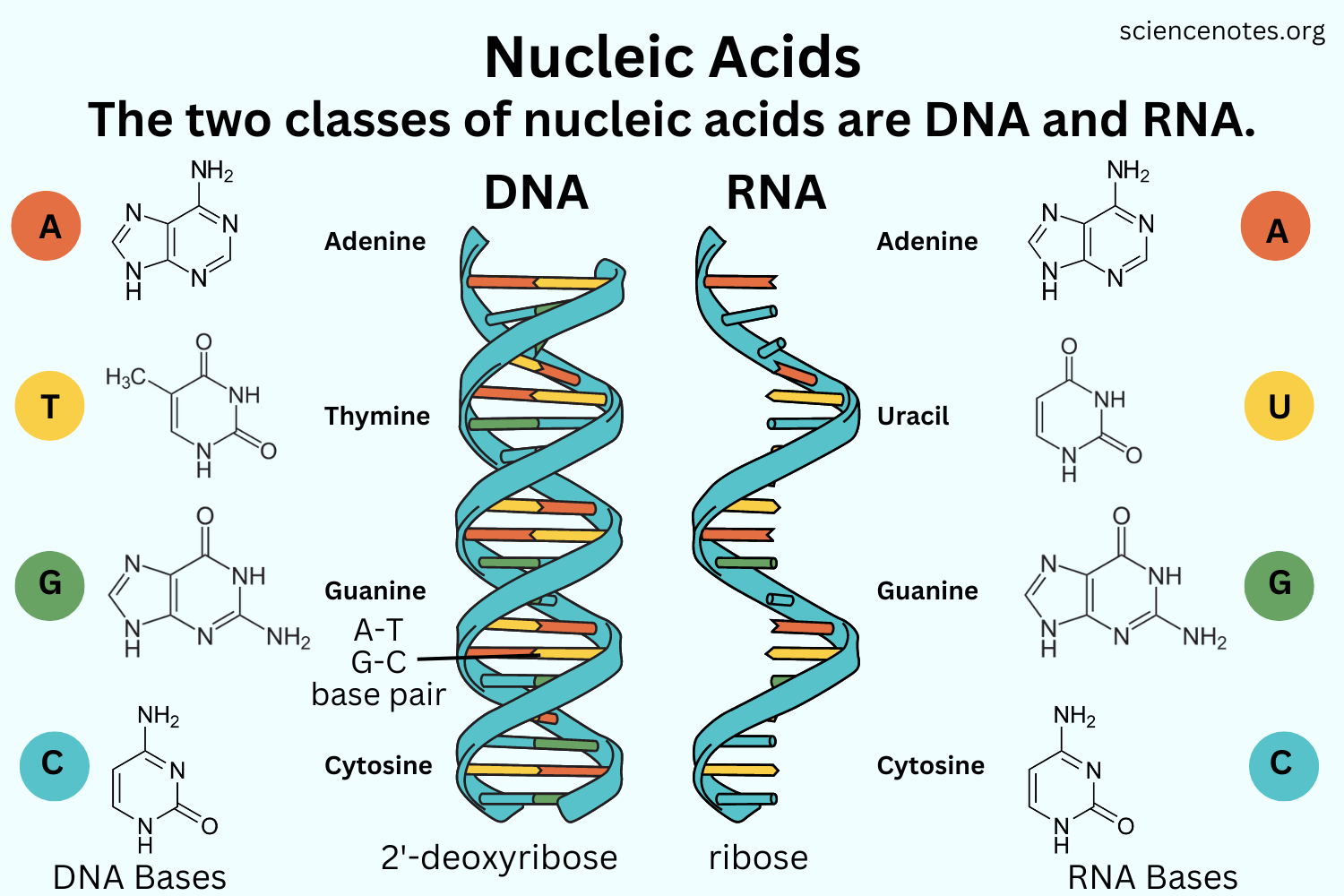
RNA | DNA |
ribonucleic acid | deoxyribonucleic acid |
single stranded | double stranded |
A=U C=G | A=T C=G |
DNA Replication
models of DNA Replication
there were 3 alternative models for DNA replication
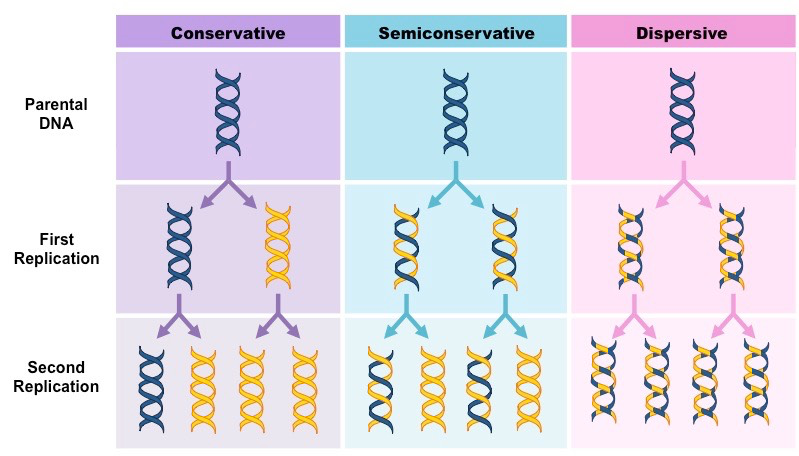
conservative model
the parental strands direct synthesis of an entirely new double stranded molecule
the parental strands are fully “conserved”
semi conservative model
the two parental strands each make a copy of itself
after one round of replication the two daughter molecules each have one parental and one new strand
dispersive model
the material in the two parental sterands is dispersed randomly between the two duaghter molecules
after one round of replication the daughter molecules contain a random mix of parental and new DNA
which model is correct?
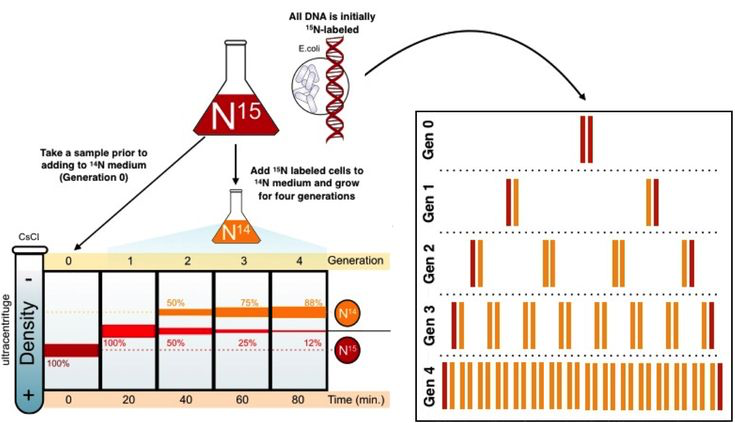
in 1954 meselson and stahl performed an expirment using bacteria
process:
bacteria was cultured with a heavy isotope, 15^N
bacteria was transferred to a medium with 14^N, a light isoptope
DNA was centrifuged and analyzed after each replication
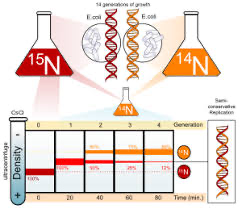
semi conservative model
by analyzing samples of DNA after each generation, it was found that the parental strands were following the semi-conservative model
steps in DNA Replication
DNA replication begins at sites called origins of replication
various proteins attach to the origin of replication and open the DNA to form a replication fork
helicase will unwind the DNA strands at each replication fork
to keep the DNA from re-bonding with itself, proteins called single strand binding proteins (SSBPs) bind to the DNA to keep it open
topoisomerase will help prevent strain ahead of the replication fork by relaxing supercoiling
the enzume primase initiates replication by adding short segments of RNA, called primers, to the parental DNA strand.
the enzyme that synthesize DNA can only attach new DNA nucleotides to an existing strand of nucleotides
primers serve as the foundation for DNA synthesis
antiparallel elongation
DNA polymerase III (DNAP III) attaches to each primer on the parental strand and moves in the 3’ to 5’ direction
as it moves, it adds nucleotides to the new stran in the 5’ to 3’ direction
the DNAP III that follows helicase is known as the leading strand and it only requires one primer
the DNAP III on the other parental strand that moves away from helicase is known as the lagging strand and requires many primers
the leading stranf is synthesized in one continous segment, but since the llagging strand moves away from the replication fork it is synthesized in chunks
okazaki fragments: segments of the lagging strand
after DNAP III forms an okazaki fragment, DNAP I replaces RNA nucleotides with DNA nucleotides
DNA ligase: joins the okazaki fragments forming a continous DNA strand
putting it all together video:
problems at the 5’ end
since DNAP III can only add nucleotides to a 3’ end, there is no way to finish replication on the 5’ end of a lagging strand
over many replication, this would mean that the DNA would become shorter and shorter
how arer the genes on DNA protected from this?
telomeres: repeating units of short nucleotide sequences that do not code for genes
form a cap at the end of DNA to help postpone erosion
the enzyme telomerase adds telomeres to DNA
proofreading and repair
as DNA polymerase adds nucleotides to the new DNA strand, it proofreads the bases add
if errors still occur, mismatch repair will take place
enzymes remove and replace the incorrectly paired nucleotide
if segments of DNA are damaged, nuclease can renove segments of nucleotides and DNA polymerase and ligase can replace the segments
Transcription and RNA Processing
proteins
proteins are polypeptides made up of amino acids
amino acids are linked by peptide bonds
Gene expression: the process by which DNA directs the synthesis of proteins
includes two steag: transciption and translation
DNA → RNA → protein
(transciption) (translation)
Transciption and Translation
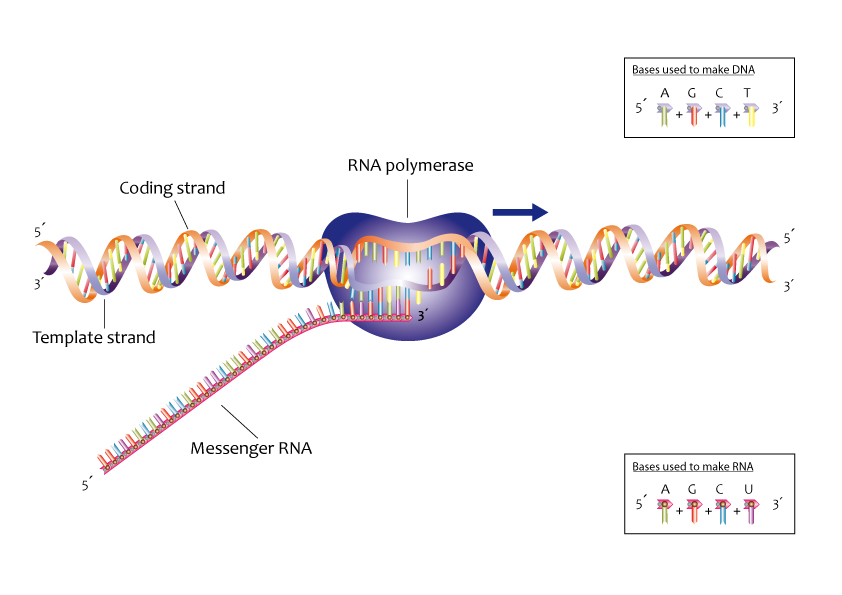
transciption: the synthesis of RNA using information from DNA
allows for the “message” of the DNA to be transcribed
occurs in the nucleus
translation: the synthesis of a polypeptide using information from RNA
occurs at the ribosome
a nucleotide sequence becomes an amino acid sequence
types of RNA
Messenger RNA (mRNA)
Ribosomal RNA (rRNA)
Transfer RNA (tRNA)
Messenger RNA (mRNA)
Messenger RNA is synthesized during transcription using a DNA template
mRNA carries information from the DNA (at the nucleus) to the ribosomes in the cytoplasm
Transfer RNA (tRNA)
transfer RNA molecules are important in the process of transltion
each tRNA can carry a specific amino acid
can attach to mRNA via their anticodon
a complementary codon to mRNA
allow information to be translated into a peptide sequence
Ribosomal RNA (rRNA)
rRNA helps form ribosomes
helps link amino acids together
The Genetic Code
DNA contains the sequence of nucleotides that codes for proteins
the sequince is read in groups of three called the triplet code
during transcription, only one DNA strand is being transcribed
known as the template strand (also known as the noncoding strand, minus strand, or antisense strand)
mRNA molecules formed antiparallel and complementary to the DNA nucleotides
Base pairing: A = U and C = G
the mRNA nucleotide triplets are called codons
codons code for amino acids
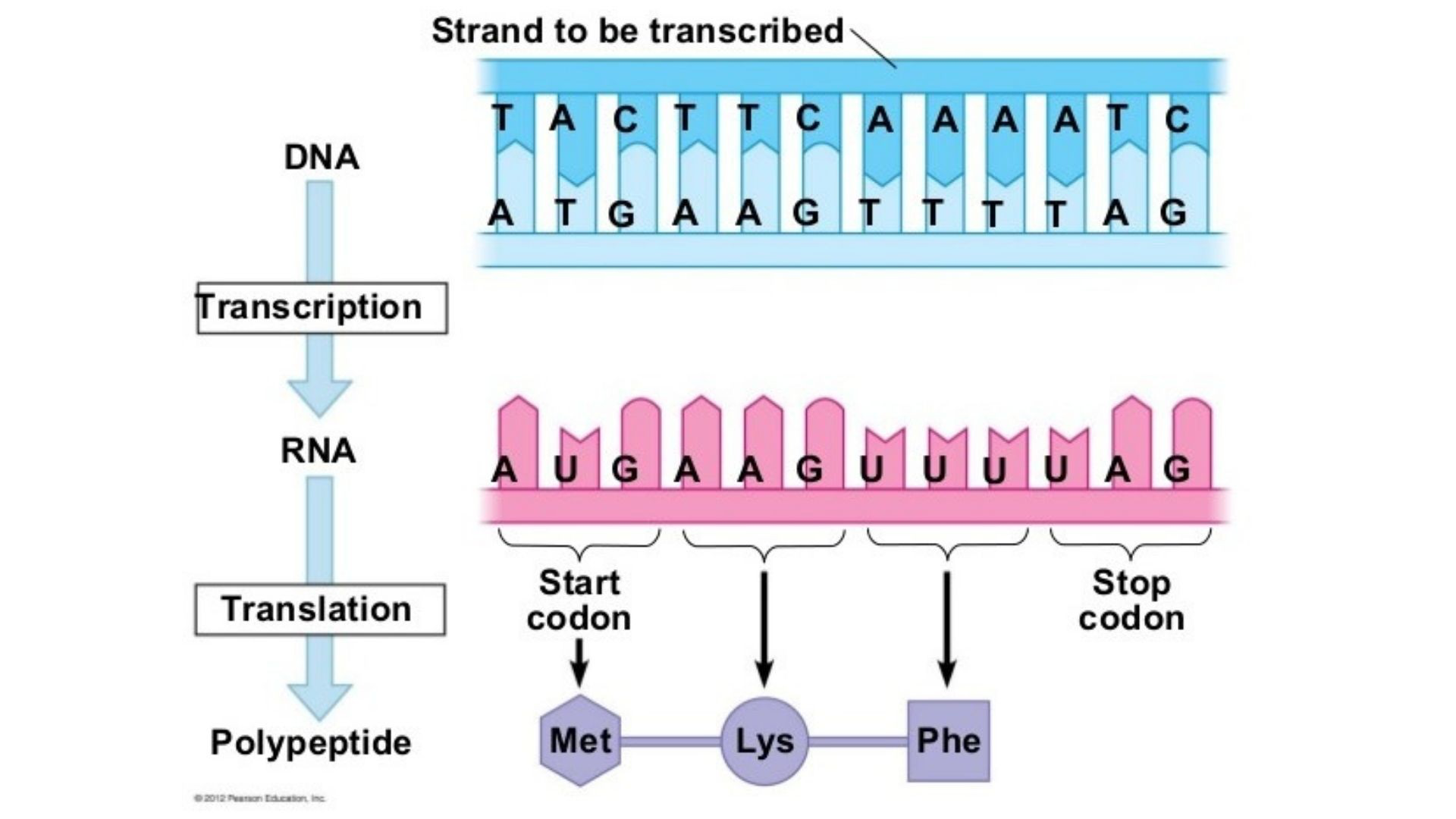
64 different codon combinations
61 code for amino acids
3 are stop codons
universal to all life
redundancy: more than one codon code for each amino acid
reading frame: the codons on the mRNA must be read in the correct groupings during translation to synthesize the correct proteins
example: the fat cat ate the rat
if the reading fram shifts even one letter, it will produce a completely different outcome
Hef atc ata tet her at
Steps of Transcription
there are three steps
initation
elongation
termination
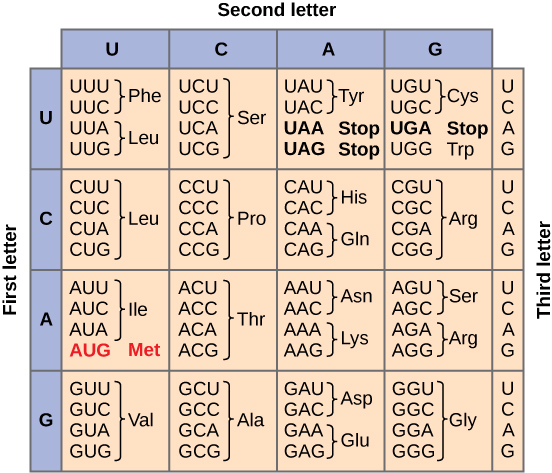
Step 1: Initiation
transcription begins when RNA polymerase molecules attach to a promoter region of DNA
do not need a primer to attach
promoter regions are upstream of the desired gene to transcribe
Eukaryotes:
promoter region is called TATA box
transcription factors help RNA polymerase bind
Prokaryotes:
RNA polymerase can bind directly to promoter
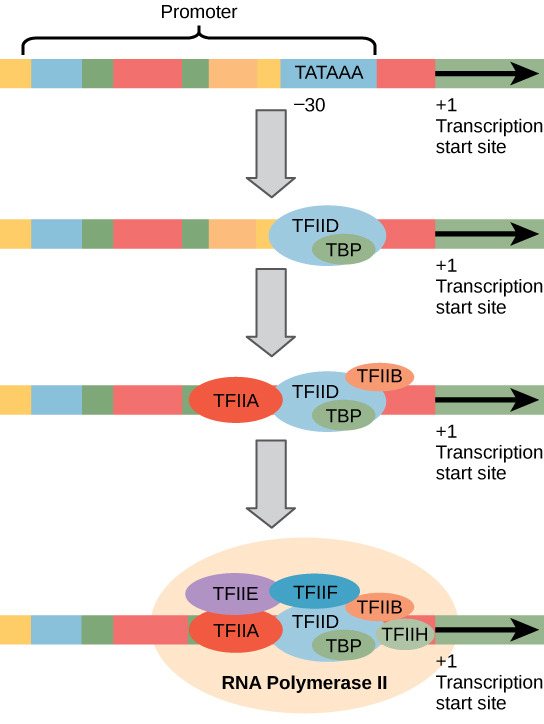
Step 2: Elongation
RNA polymerase opens the DNA and reads the triplet code of the template strand
moves in the 3’ to 5’ direction
the mRNA transcript elongates 5’ to 3’
RNA polymerase moves downstream
only opens small sections of DNA at a time
pairs complementary RNa nucleotides
the growing mRNA strand peels away from the DNA template strand
DNA double helix then reforms
a single gene can be transcribed simultaneously by several RNA polymerase molecules
helps increase the amount of mRNA synthesized
increases protein production
Step 3: Termination
Prokaryotes:
transcription proceeds through a termination sequence
causes a termination signal
RNA polymerase detaches
mRNA transciption is released and proceeds to translation
mRNA does NOT need modifications
Eukaryotes:
RNA polymerase transcribes a sequence of DNA called the polyadenylation signal sequence
codes for a polyadenylation signal (AAUAAA)
releases the pre-mRNA from the DNA
must undergo modification before translation
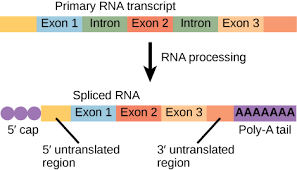
Pre-mRNA modifications
there arre three modification that must occur to eurkaryotic pre-mRNA before it is ready for translation
5’ cap
Poly-A tail
RNA splicing
5’ cap (GTP): the 5’ end of the pre-mRNA recieves a midified guanone nucleotide “cap”
Poly-A tail: the 3’ end of the pre-mRNA revieces 50-250 adenin nucleotides
both the 5’ cap and the poly-A tail function to:
help the mature mRNA leave the nucleus
help protect the mRNA from degradation
help ribosomes attach to the 5’ end of the mRNA when it reaches the cytoplasm
RNA Splicing: sections of the pre-mRNA, called introns, are removed and then expns are joined together
a. introns: intervening sequence, do not code for amino acids
b. exons: expressed sections, code for amino acids
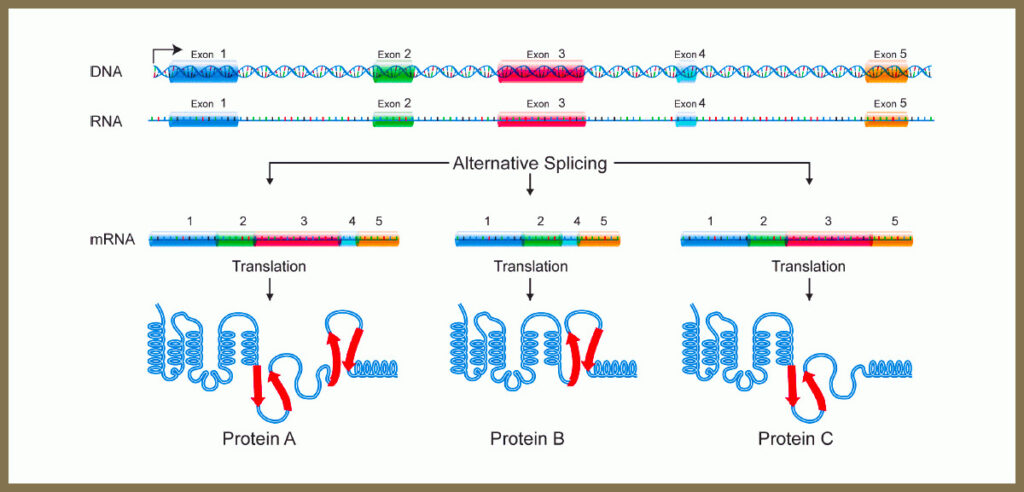
why does splicing occur?
a single gene can code for more than one kind of polypeptide
known as alternative splicing
snRNPs: small nuclear RNA
Spliceosome: several snRNPs that recognize splice site sequence and cut the gene
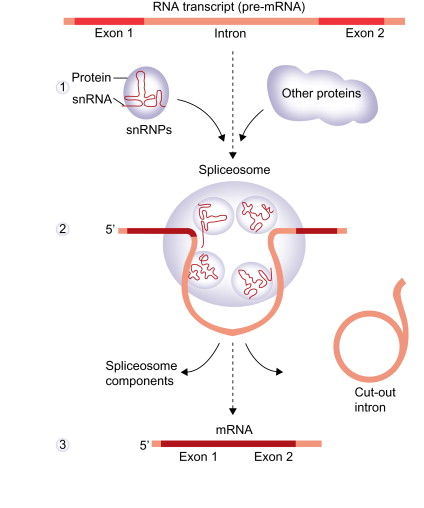
once all modifications have occured, the pre-mRNA is now considered mature mRNA and can leave the nucleus and procees to the cytoplasm for translation at the ribosomes
Translation
Translation: the synthesis of a polypeptide using information from the mRNA
occurs at the ribosome
a nucleotide sequence becomes an amino acid sequence
tRNA is a key player in translating mRNA to an amino acid sequence
Transfer RNA
tRNA has an anticodon region which is complementary and antiparallel to mRNA
tRNA carries the amino acid that the mRNA codon codes for
ACU codes for Thr
the enzyme aminoacyl-tRNA synthetase is responsible for attaching amino acids to tRNA
when tRNA carries an amino acid it is “charged”
Ribosomes
translation occurs at the ribosome
ribosomes have two subunits: small and large
prokaryotic and eukaryotic ribosomal subunits differ in size
prokaryotes: small subunits (30s) large subunit (40s)
eukaryotes: small subunit (40s) large subunits (60s)
the large subunit has three sites: A, P, and E
A site: amino acid site
holds the next tRNA carrying an amino acid
P site: polypeptide site
holds the tRNA carrying the growing polypeptide cahin
E site: exit site
Translation
translation occurs in three stages:
initiaition
elongation
termination
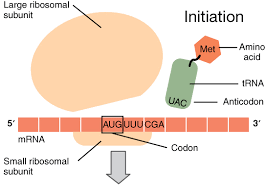
Step 1: Initation
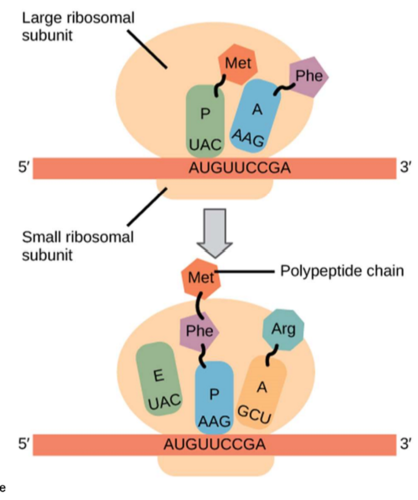
translation begins when the small ribosomal subunit binds to the mRNA and a charged tRNA binds tot he start codon, AUG, on the mRNA
the tRNA carries methionine
next, the large subunit binds
note: the first tRNA carrying Met will go to the P site, every other tRNA will go to the A site first
Step 2: Elongation
elongation starts when the next tRNA comes into the A site
mRNA is moved through the ribosome and its codons are read
each mRNA codon codes for a specific amino acid
codon charts are used to determine the amino acid
since all organisms use the same genetic code, it supportd the idea of common ancestry

Elongation occurs in steps:
codon recognition: the appropriate anticodon of the next tRNA goes to the A-site
peptide bond formation: peptide bonds are formed that transfer the ploypeptide to the A site tRNA
translocation: the tRNA in the A site moves to the P site, the tRNA in the P site goes to the E site. The A site is open for the tRNA
Step 3: Termination
termination occurs when a stop coodn in the mRNa reaches the A site of the ribosome
stop codons do not code for amino acids
the stop codon signals for a release factor
hydrolyzes the bond that holds the polypeptide to the P site
polypeptide releases
all translational units disassemble
Protein Structures
primary: chain of amino acids
secondary: coils and folds due to hydrogen bonds forming
tertiary: side chain interaction
quaternary: 2+ polypeptide chains interacting
Protein folding
as translation takes place, the growing polypeptide chain begins to coil and fold
genes determine the primary structure
primary structure determines the final shape
some polypeptide require chaperone proteins to fold correctly and some require modification before it can be functional in the cell
Prokaryotes
no introns
circular, naked DNA in the cytoplasm
transcription and translation are simultaneous
Eukaryotes
introns and exons
linear, wound DNA in the nucleus
- 1 hour from DNA to protein
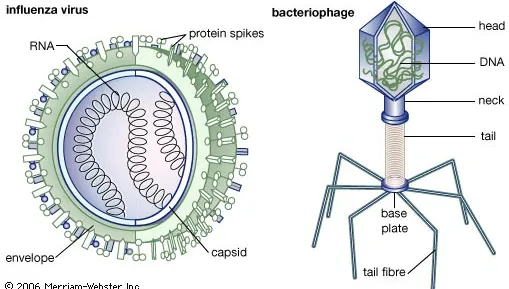
Virus Vocabulary
parasites: need host “machinery”
capsids: crystal-like protein shell
assimilation: virus takes over host and reprograms host cells to produce the viral proteins
bacteriophages: viruses that infect bacteria
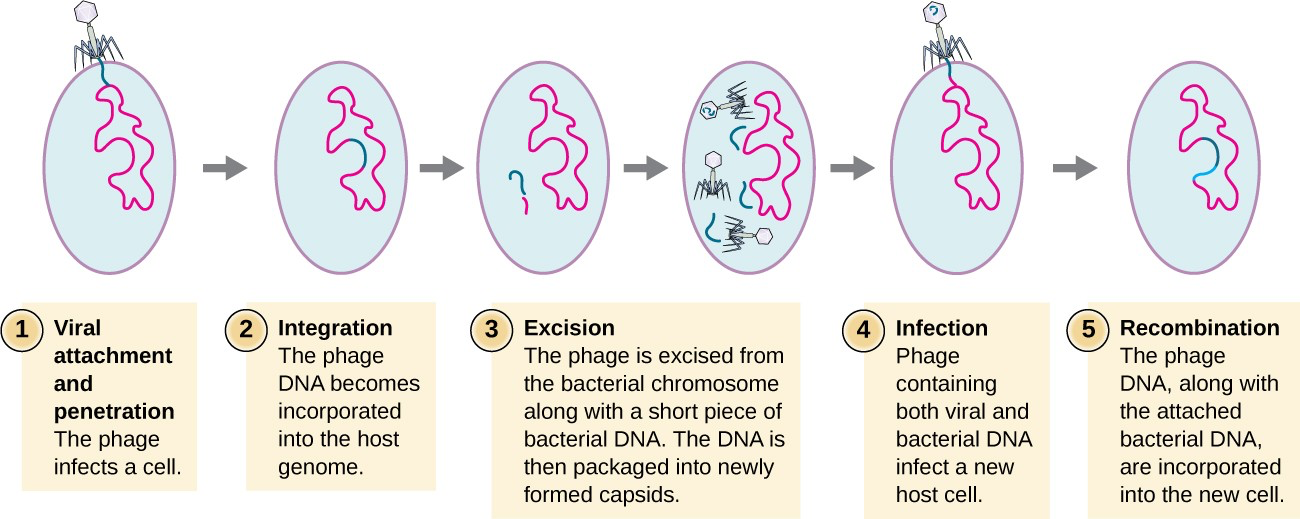
lytic: actively reproduce virus in bacteria and release virus by rupturing host
lysogenic: integrate viral DNA into bacterial DNA
prions: misfolded proteins transmitted
central dogma violation
Lytic cycle

Lysogenic cycle
Retroviruses
retroviruses, like HIV, are an exception to the standard glow of genetic information
information flows from RNA to DNA
uses an enzyme known as reverse transcriptase
couples viral RNA to DNA
DNA then becomes part of the RNA
Prions
infectious proteins that caus other proteins to misfold, leading to severe neurodegenerative diseases. Unlinke bacteria, viruses, or fungi, prions lack DNA or RNA and are purely protein based.
Regulation of Gene expression & Cell Specialization
Gene Expression
prokaryotes and eukaryotes must be able to regulate which genes are expressed at any given time
genes can be turned “on” or “off” based on environmental and internal cues
on/off refers to whether or not transcriptioin will take place
allows for cell specialization
Bacterial Gene Expression
Operons: a group of genes that can be turned on or off
operons have 3 parts:
promoter: where RNA polymerase can attach
operator: the on/off switch
genes: code for related enzymes in pathway
operons can be repressible or inducible
repressible (on to off): transcription is usually on, but can be repressed (stopped)
inducible (off to on): transcription is usually off, but can be induced (started)
Regulatory gene: produces a repressor protein that binds to the operator to block RNA polymerase from transcribing the gene
always expressed, but at low levels
binding of a repressor to an operator is reversible
Allosteric Regulation
allosteric activator: substrate binds to allosteric site and stabilizes the shape of the enzyme so that the active sites remain open
allosteric inhibitor: substrate binds to allosteric site and stabilizes the enzyme shape so that the active sites are closed (inactive form)
repressible operons
example: the Trp operon
the trp operon in bacteria controls the synthesis of tryptophan
since it is repressible, transcription is active
it can be switched off by a trp repressor
allosteric enzyme that is only active when tryptophan binds to it
when too much tryptophan builds up in bacteria, tryptophan is more likely to bind to the repressor turning it active, which will then temporarily shut off transcription for tryptophan.

Inducible operon
example: the lac operon
the lac operon controls synthesis of lactase, an enzyme that digests lactose (milk sugar)
since it is inducible, transcription is off
a lac repressor is bound to the operatore (allosterically active)
the inducer for the lac repressor is allolactose
when present it will bind to the lac repressor and turn the lac repressor off (allosterically inactive)
the genes can now be transcribed
Eukaryotic Gene Expression
the phenotype of a cell or organism is determined by a combination of genes that are expressed and the levels that they are expressed
differences between cell types is known as differential gene expression
eukaryotic gene expression is regulated at different stages
chromatin structure:
if DNA is tightly wound it is less accessible for transciption
how can it be modified?
histone acetylation adds acetyl groups to histones, which loosens the DNA
DNA methylation adds methyl groups to DNA, which causes the chromatin to condense
epigenetic inheritance:
chromatin modifications do not alter the nucleotide sequence of the DNA, but they can be heritable to future generations
modifications can be reversed, unlinke mutations
explains why one identical twins may inherit a disease while the other does not
transcription initiation:
once chromatin modifications allow the DNA to be more accessible, specific transcription factors bind to control elements
sections of non coding DNA that serve as binding sites
gene expression can be increased or decreased by binding of activators or repressors to control elements
RNA processing:
alternative splicing of pre-mRNA
translation initiation
translation can be activated or repressed by initiation factors
MicroRNA’s and small interfering RNA’s (siRNA) can bind to mRNA and degrade it or block translation
Eukaryotic Development
during embryonic development, cell division and cell differentiation occurs
cells become specialized in their structure and function
morphogenesis: the physical process that gives an organism its shape
how do cells differentiate during early development?
cytoplasmic determinants: substances in the maternal egg that influence cells
inductioni: cell to cell signals that can cause a change in gene expression
both cytoplasmic determinants and induction influence pattern formation
a “body plan” for the organism
homeotic genes map out the body structures
as cells differentiate, apoptosis plays a critical role
apoptosis: programmed cell death
allows structures to take their form
example: if apoptosis did not occur during the development of human hands and feet we would be born with webbed fingers and toes
ubiquitin: “death tag”
marks unwanted proteins with a lab which are broken does in proteasomes (waste disposers)
Mutations and Biotechnology
Mutations
Mutations: changes in the genetic material of a cell, which can alter phenotypes
primary source of genetic variation
normal function and production of cellular products is essentialn
any disruption can cause new phenotypes
changes can be large scale or small scale
large scale: chromosomal changes
small scale: nucleotide substitutions, insertions, or deletions
Small Scale Mutations
point mutations: change a single nucleotide pair of a gene
substitution: the replacement of one nucleotide and its partner with another pair of nucleotides
silent: change still codes for the same amino acid (remember: redundancy in the genetic code)
missense: change results in a different amino acid
nonsense: change results in a stop codon
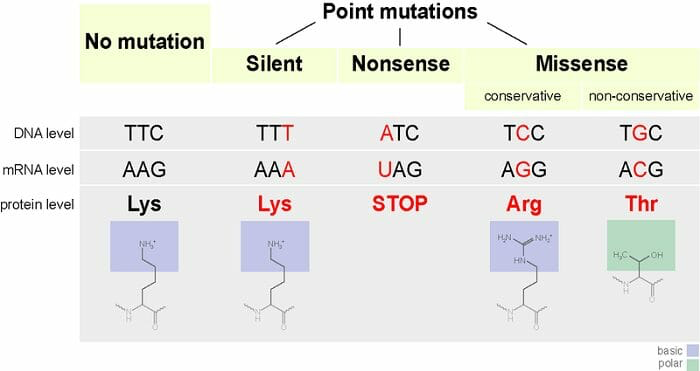
Small Scale Mutations
frameshift mutation: when the reading frame of the genetic information is altered
disastrous effects to resulting proteins
insertion: a nucleotide is inserted
deletion: a nucleotide is removed
Large Scale Mutations
mutations that affect chromosomes
nondisjunction: when chromosomes do not deperate properly in meiosis
results in the incorrect number of chromosomes
example: down syndrome- trisomy 21
translocation: a segment of one chromosome moves to another
inversions: a segment is reversed
duplications: a segment is repeated
deletions: a segment is lost
Natural selection
any time mutations occur, they are subject to natrual selection
genetic changes can sometimes enhance the survival and reproduction of an organism
Increasing Genetic Variation
prokaryotes can exchange genetic material through horizontal gene transfer
if there is a mutation that is beneficial to the survival and reproduction of that prokaryote then it can also be transferred
transformation: uptaking of DNA from a nearby cell
transduction: viral transmission of genetic material
conjugation: cell to cell transfer of DNA
transposition: movement of DNA segments within and between DNA molecules
Transformations
will DNA or proteins transform bacteria?
DNA
first evidence that DNA is the genetic material
Hershey & Chase
grew two viruses with radioactively labeled macromolecules in either proteins or DNA
Biotechnology
both DNA and RNA can be manipulated through genetic engineering
Gel Electrophoresis: a technique used to separate DNA fragments by size
DNA is loaded into wells on one end of a gel and an electric current is applied
DNA fragments are negatively charged so they move towards the positive electrode
PCR (polymerase chain reaction): a method used in molecular biology to make several copies of a specific DNA segment
segments of DNA are amplified
results can be analyzed using gel electrophoresis
DNA Sequencing: the process of determining the order of nucleotides in DNA
Sanger Sequencing: determines DNA sequences by selectifvely incorperating chain-terminating dideoxynucleotides (ddNTPs), generating NDA fragments of varying lengths that are analyzed using capillary electrophoresis.
Modern Sequencing: uses massively parallel sequencing to generate millions of short reads simultaneously, making it faster, cheaper, and more scalable for whole-genome and high-throughput sequencing applications
Biotechnology
Biotechnology Today
genetic engineering
maniupulation of DNA
if you are going to engineer DNA & genes & organisms, then you need a set of tools to work with\
Bacteria
Bacteria review
one-celled prokaryotes
reproduce by mitosis
binary fission
rapid growth
generation every 20 minutes
10^8 (100 million) colony overnight
dominant form of life on earth
incredibly diverse
Bacterial genome:
single circular chromosme
haploid
naked DNA
no histone proteins
4 million base pairs
4300 genes
1/1000 DNA in eukaryote
transformation:
bacteria are opportunists
pick up naked foreign DNA wherever it may be hanging out
have surface transport proteins that are specialized for the uptake of naked DNA
imports bits of chromosomes from other bacteria
incorperate the DNA bits into their own chromosome
express genes
transformation
form of recombination
Plasmids
small supplemental circles of DNA
5000-20000 base pairs
self-replicating
carry extra genes
2-30 genes
genes for antibiotic resistance
can be exchanged between bacteria
bacterial sex
rapid evolution
can be imported from environment
how can plasimids help us?
a way to get genes into bacteria easily
insert new gene into plasmid
insert plasmid into bacteria = vector
bacteria now expresses new gene
bacteria makes new proteins
biotechnology
plasmids used to insert new genes into bacteria
How do we cut DNA?
restriction enzymes
resitriction endonucleases
discovered in 1960’s
evolved in bacteria to cup up foreign DNA
“restrict” the action of the attacking organism
protection against viruses & other bacteri a
bacteria protect their own DNA by methylation & by not using the base sequences recognized by the enzymes in their own DNA
Restriction enzymes
action of enzyme
cut DNA at specific sequences
restriction site
symmetrical “palindrome”
produces protrudinf ends
sticky ends
will bind to any complementary DNA
many different enzymes
named after organism they are found in
ex: EcoRI, HindIII, BamHI, SmaI
discovery of restriction enzymes
restriction enzymes are named for the organism they come from: EcoRI = 1st restriction enzyme found in E.Coli
restriction enzymes:
cut DNA at specific sites
leave “sticky ends”
Sticky ends
cut other DNA with same enzymes
leave “sticky ends” on both
can glue DNA (genes) together at “sticky ends”
why mix genes together? gene produces protein in different organism or different individual
the code is universal
since all living organisms use the same DNA, use the same code book, and read their genes the same way
Copy (& Read) DNA
tranformation
insert recombinant plasmid into bacteria
grow recombinant bacteria in agar cultures
bactiera make lots of copies of plasmid
“cloning” the plasmid
production of many copies of inserted gene
production of “new” protein
transformed phenotype
Uses of genetic engineering
genetically modified organisms (GMO)
enabling plants to produce new proteins
protect crop from insects: BT corn
corn produces a bacterial toxin that kills corn borer (caterpillar pest of corn)
extend growing season: fishberries
strawberries with an anti-freezing gene from flounder
improve quality of food: golden rice
ricde producing vitamin A improves nutritional value
Cut, Paste, Copy, Find……
word processing metaphor
cut
restriction enzymes
paste
logase
copy
plasmids
bacterial transformation