Host Basics_ Mycobacterium smegmatis

An Introduction to the Host Bacteria, Mycobacterium smegmatis
As a network of researchers, SEA-PHAGES explores the genetic diversity and relationships of populations of bacteriophages that infect Actinobacteria, incluidng the bacterium Mycobacterium smegmatis (M. smegmatis, or colloquially known as “smeg”). There are many different strains of M. smegmatis (isolated from different environmental or clinical samples). The M. smegmatis
strain widely used in SEA-PHAGES is called M. smegmatis mc2155. If you intend to isolate phages using this bacterium, then this will be your ‘host’ for phage isolation.
M. smegmatis is one of more than 100 species that make up the genus Mycobacterium, classified within the phylum Actinobacteria. Mycobacteria were named this because they sometimes have a fungus-like appearance under the microscope—although they are definitely not fungi! The Mycobacterium genus contains many species that affect human health, including M. tuberculosis and M. leprae, which are the causative infectious agents of tuberculosis and leprosy, respectively. M. smegmatis, however, is a saprophyte and is found in environmental samples, including soil and compost. Although they share the same genus, M. smegmatis and M. tuberculosis have many other differences, including vastly different doubling times: approximately 3 hours for M. smegmatis and 24 hours for M. tuberculosis. Some bacteriophages are capable of infecting both species, whereas others infect only one or the other.
M. smegmatis will grow and divide on many nutrient-rich media. In your research, you will grow M. smegmatis on 7H9 media supplemented with albumin and dextrose (AD) at 37 ˚C. Under these conditions, M. smegmatis grows at its fastest, dividing approximately once every 3 hours. This means that it typically takes about 3–4 days for an individual colony growing from a single cell on an agar plate. However, when preparing a bacterial lawn, you can start by adding many bacterial
cells (e. g., 107 or more) so that a lush lawn grows within 20–24 hours. M. smegmatis will also grow at 30 ˚C, but it divides more slowly and therefore requires more time to form either a colony or a lawn. For reproducibility of experiments, it is important that cultures used for a given set of experiments be prepared at the same temperature.
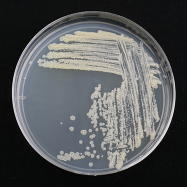
M. smegmatis mc2155 colonies are tan-white in color and dry, rough, and wrinkled in texture (Figure 4.0-4). A liquid culture of M. smegmatis inoculated from a single colony will take 3–5 days to form a saturated liquid culture, exhibiting a tan-white color. The strain you will work with, M.
smegmatis mc2155, is not inhibited by the antibiotic ampicillin or the antifungal cyclohexamide. Therefore, these can be added to the growth media to prevent other microorganisms from growing in your cultures.
On average, 60 % of attempts to isolate phage from soil by enrichment with M. smegmatis yield phage.
 |
|---|
Figure 4.0-4. Mycobacterium smegmatis growing on an agar plate. |
If you are using this bacterium as your host for phage isolation, refer back to the list below for the specific growth and culture requirements for your experiments.
Growth media:
Liquid: 7H9, supplemented with albumin dextrose (AD) and Tween80 (see instructions for culturing bacteria that require Tween80)
Agar: L-agar plates
Top Agar: 7H9
Growth temperature: 37˚C, (or 30˚C)
Antimicrobials: carbenicillin (50 μg/ml), cycloheximide (10 μg/ml)
Phage Buffer: 10 mM Tris (pH 7.5), 10 mM MgSO4, 68 mM NaCl, 1 mM CaCl2, (10% glycerol, optional)
Restriction Enzymes: BamHI, ClaI, EcoRI, HaeIII, HindIII, MseI.
Note: Isoschizomers, which are different restriction enzymes that recognize the same DNA sequence, may be used in place of any of the enzymes listed above.