M1L3 Post translational modifications in DNA damage response
Main PTMs - protein methylation, acetylation, phosphorylation, ubiquitination/ubiquitylation (and ubiquitin-like proteins), poly(ADP-ribosyl)ation/PARylation
Covalent, reversible
Histones are the platform for PTMs and chromatin remodeling
Methylation
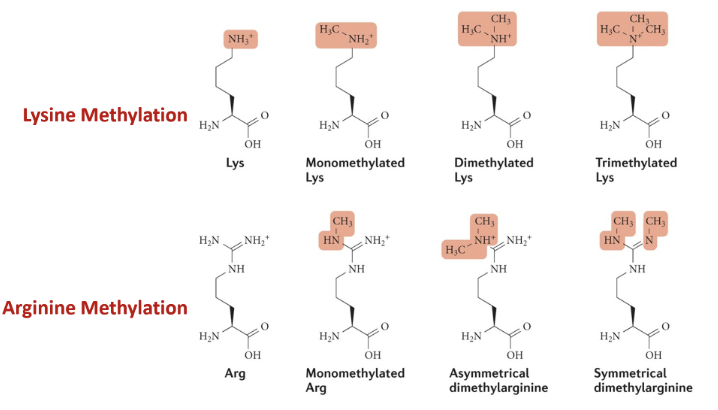
Lysine methylation - monomethylated, dimethylatied, trimethylated
Adding methylation is increasing hydrophobicity - compacts chromatin by disfavouring exposure to the aqueous environment, thus burying hydrophobic regions and allowing histone tails to interact strongly via hydrophobic contacts)
Hydrophobic patches can also act as docking sites for chromatin-modifying proteins and deterring the binding of factors that cause euchromatinisation
Does not negate the positive charge from protonated amine group
Arginine methylation - monomethylated, asymmetrical or symmetrical dimethylation
Writer (methyltransferase) —> reader (methyl binding protein) —> eraser (demethylase)
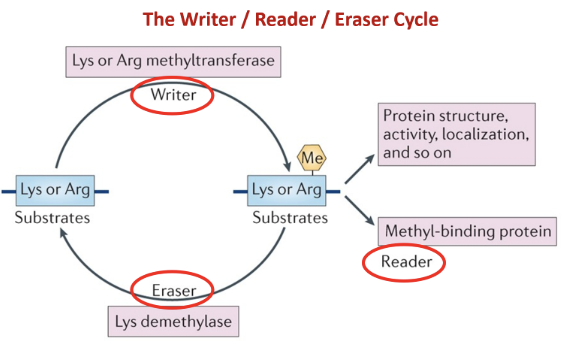
Methylation determines chromatin state (DNA condensation for gene silencing) and generates binding surface for DDR proteins (to remove methylation for damage repair)
Methylation does not just work on histones but also on proteins involved in DDR pathways to activate them
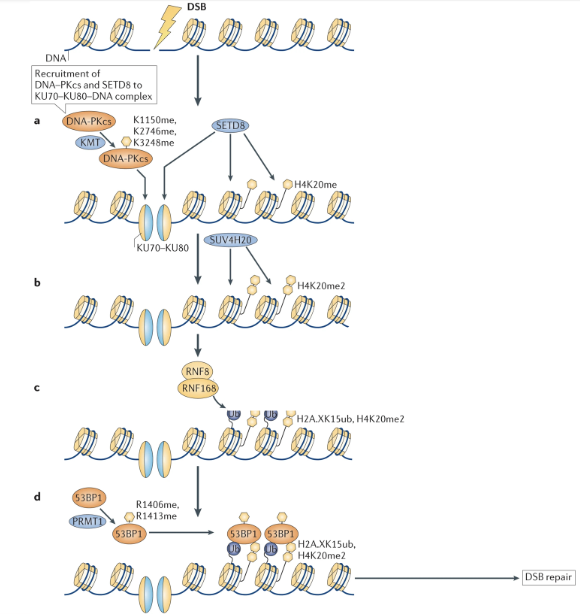
When there is a DSB, DNA damage is detected by Ku70-Ku80 complex which binds to the broken ends of the DNA
DNA-PKcs are recruited to the complex and are methylated at K1150, K2746, and K3248 by lysine methyltransferase (KMT)
This stabilises the DNA-protein complex and creates binding sites for downstream enzymes
SET8 (histone methyltransferase) can bind to the site and add a monomethyl group to K20 of H4 (H4K20me)
SUV4H20 adds another group to produce H420me2
Methyl groups act as landing pads for DDR proteins
E3 Ub ligases RNF8/RNF168 ubiquitylate H2A (H2AK15ub)
H4K20me2 + H2AK15ub define a specialized chromatin environment at DNA breaks which recruits 54BP1
53BP1 is methylated (R1406me, R1413me) by the PRMT1, augmenting its histone binding ability and stabilising its binding to the lesion
53BP1 initiates DSB repair pathways
Acetylation
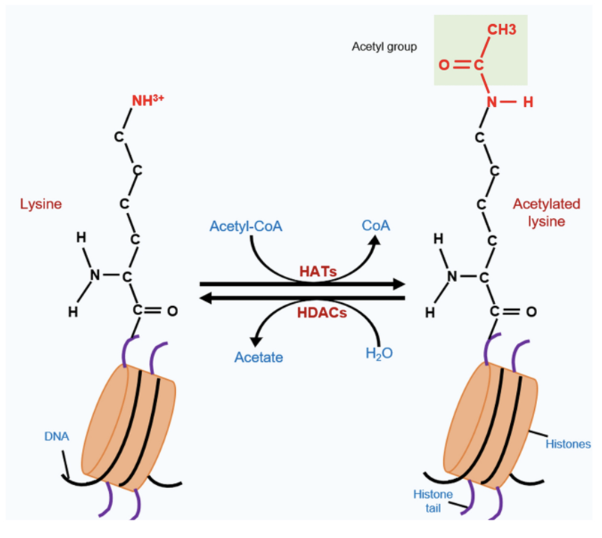
Operates on lysine
Donor molecule is acetyl-CoA - transfers acetyl group to histone acetylatransferase (HAT) which acetylates lysine (eg. Tip60, CBP/p300, GCNS)
Neutralised positive charge of lysine, thus much more disruptive than methylation and opens up DNA structure
Chromatin relaxation and generates binding sites for DDR proteins
Erasers - histone deacetylases (HDACs) eg. zinc-dependent HDAC1-11 and NAD+-dependent sirtuins
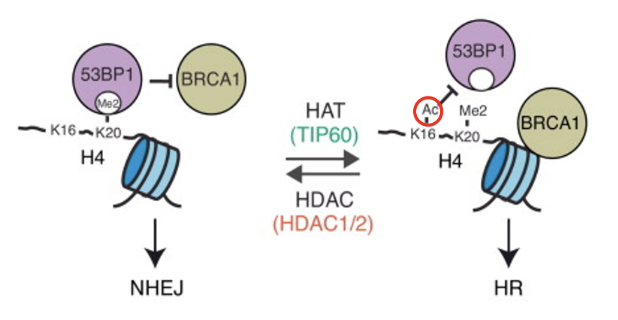
Acetylation/methylation status affects the DDR pathway that is initiated
When K20 is dimethylated in histone H4, it recruits 53BP1 to bind to the methyl patch which blocks the activity of BRCA1, triggering NHEJ
When K16 is of H4 is acetylated by Tip60, 53BP1 can not bind and BRCA1 carries out HR
Deacetylation of K16 by HDAC1/2 will favour NHEJ once again
Phosphorylation
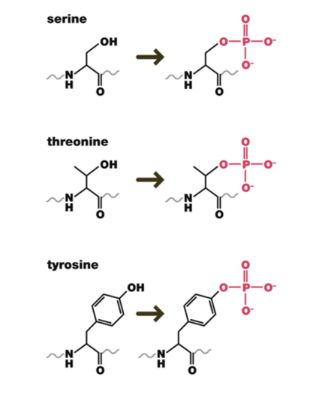
Phosphate groups can be added to the hydroxyl group of serine, threonine, or tyrosine (these amino acids are very common on the outer surface of proteins, providing many sides of the protein for kinases to act on)
Cascade of phosphorylation events in DDR (preceding kinase licenses the activity of the next downstream kinase)
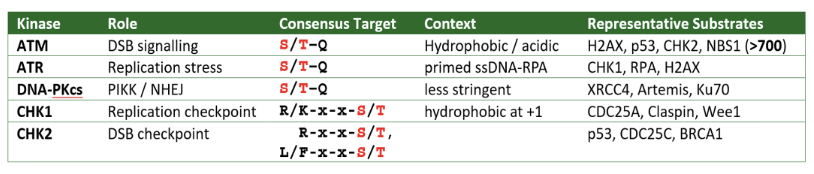
Kinases phosphorylate very specific sites
Rapid response and reversibility (DDR phosphatases, eg. PP2A, PP4C, PP6, WIP1)
Core operating system of DDR
Immediate activation of repair/checkpoint proteins
Signal amplification across chromatin
Temporal control due to balance from phosphatases
Substrate abundance and diversity
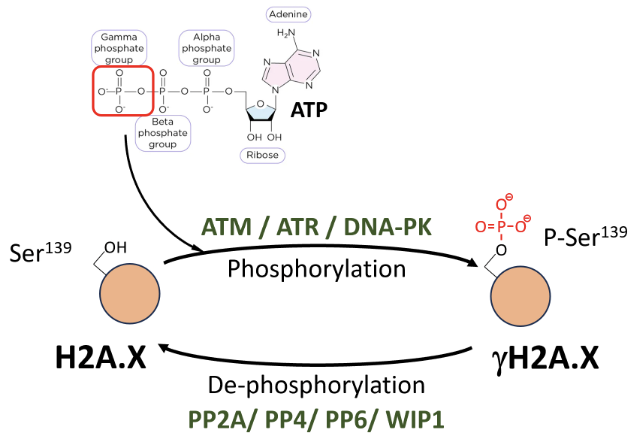
γH2A.X is formed by P-Ser139 modification (by ATM/ATR/DNA-PK) on H2A.X - most used DNA DSB marker
Phosphate is deposited by transferring the gamma phosphate group from ATP to the hydroxyl group of the amino acid substrate
ATM repairs DSBs, ATR function is related to replication stress and generation of ssDNA, DNA-PK is involved in NHEJ
Dephosphorylated by PP2A, PP4, PP6, WIP1
Active ATM diffusion-driven spread
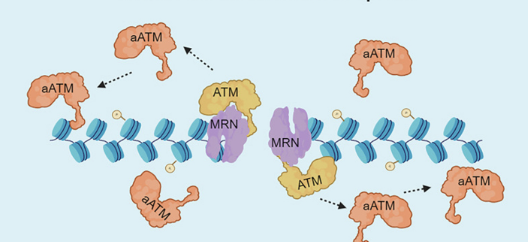
MRN complex (MRE11–RAD50–NBS1) recognises the break and recruits ATM kinase
Upon binding to the MRN-DNA complex, ATM autophosphorylates and gets activated (aATM)
aATM dissociates from MRN and diffuses along nearby chromatin, phosphorylating H2AX across a domain surrounding the break
Phosphorylation mediated activation of CHK kinases for DDR
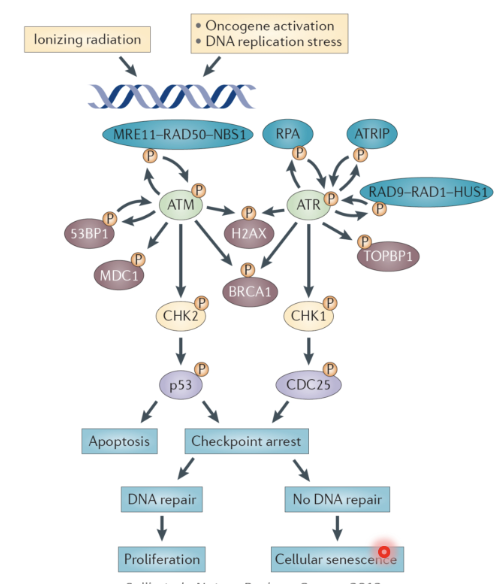
DSBs activate the ATM pathway
MRN complex (MRE11–RAD50–NBS1) detects and binds to DSB
MRN recruits ATM and activates it by promoting its monomerisation and autophosphorylation activity
Active ATM phosphorylates:
H2AX - marker for DSB
53BP1, MDC1, BRCA1 → mediate DSB repair and checkpoint signalling
CHK2 - checkpoint kinase
CHK2 phosphorylates p53 which can trigger apoptosis or checkpoint arrest
If the cell cycle is arrested DNA may be repaired or the cell may senesce if repair does not happen
Replication stress activates the ATR pathway
Stalled replication forks are characterised by ssDNA bound by RPA
ATR binds to RPA-ssDNA via its partner ATRIP
RAD9-RAD-1-HUS1 is loaded on to the DNA and ATR is activated
ATR phosphorylates
H2AX to mark the DSB
BRCA1 and TOBP1 to coordinate repair
CHK1 - checkpoint ikinase
CHK1 phosphorylates CDC25 which arrests the cell cycle, allowing for repair
If the lesion is unrepaired the cell may senesce
Phosphorylation mediated activation of CHK2
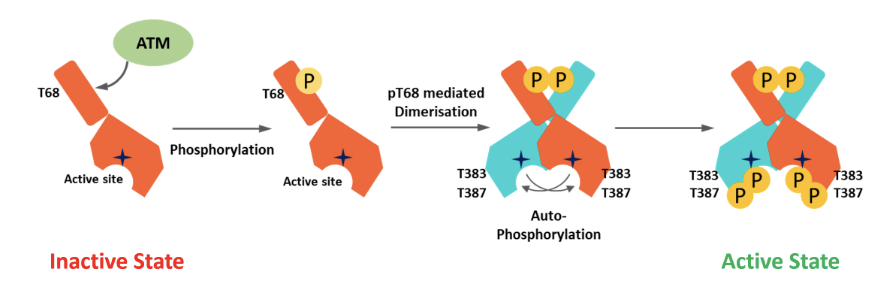
In the inactive state CHK2 is monomeric and the kinase domain in the C terminus is blocked by the N lobe
Active ATM phosphorylates at T68, causing pT68 mediated dimerisation and subsequent autophosphorylation
Active kinase is fully phosphorylated at T383 and T387
Ubiquitination
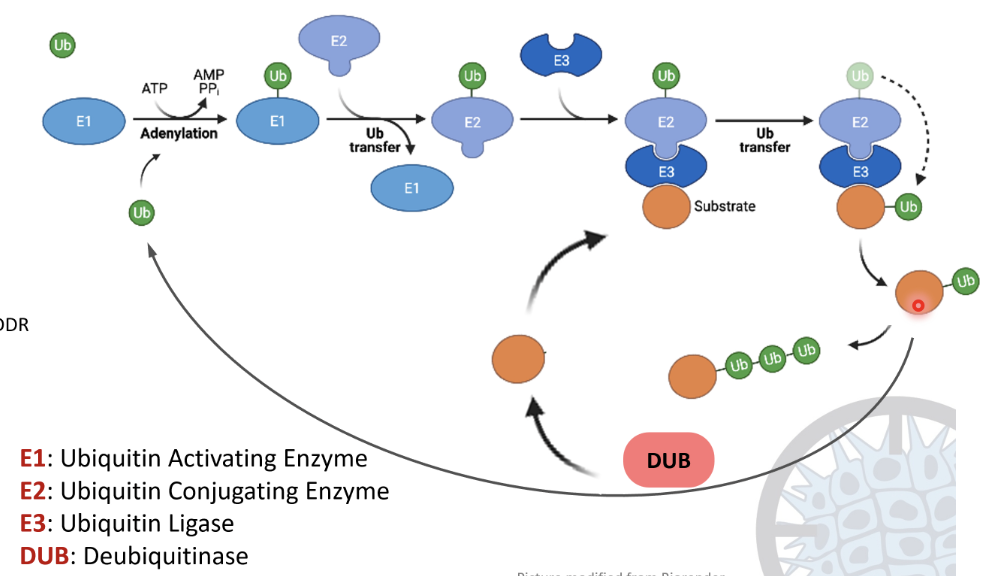
Ubiquitination cascade
E1 ubiquitin activating enzyme activates ubiquitin using ATP, forming a Ub-AMP intermediate and releasing PPi
Ub forms a strong thioester bond with a cysteine residue on E1
Ub is transferred from E1 to the E2 ubiquitin conjugating enzyme by forming a thioester bond
E3 ubiquitin ligase recognises a substrate protein and binds to both the substrate and E2-Ub
A covalent isopeptide bond forms between the C-terminus of ubiquitin and the lysine side chain of the substrate
The process can repeat to form a polyubiquitin chain
Deubiquitinases (DUBs) can hydrolyse the Ub linkage to recycle the substrate and Ub
Ub has two glycine residues at the C terminus which can bind to a lysine on the substrate, forming a covalent isopeptide bond
A Ub that is already bound to a substrate can bind to another Ub to form a polyubiquitin chain by binding to a lysine on the other Ub
Due to 7 functional lysine residues on Ub, there is a diversity of possible polyubiquitinated substrates
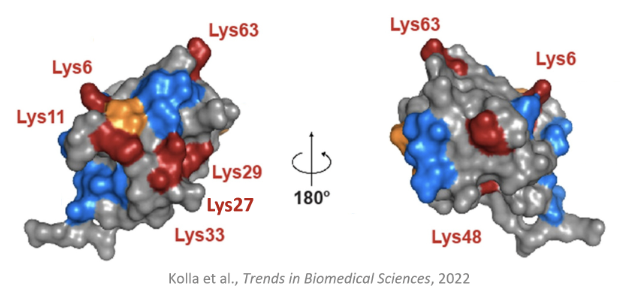
Substrates can be monoubiquitylated, multi-monoubiquitylated, form homotypic chains (chains of the same Ub), mixed or branched heterotypic chains (of different Ubs)
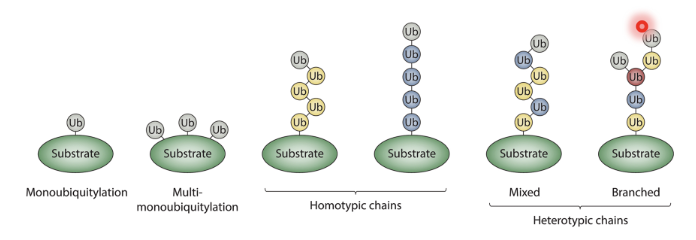
Depending on the site of ubiquitylation it can signal for degradation by proteasome, DDR… etc
Ub-K48 chains almost always associated with proteasomal degradation
Monoubiquitination of specific proteins is a mark of DNA damage
A single monoubiquitination event is usually not enough to trigger DDR, several nucleosomes should be monoubiquitinated to trigger a response
Need many ubiquitination events, not necessarily formation of chains to mount DDR
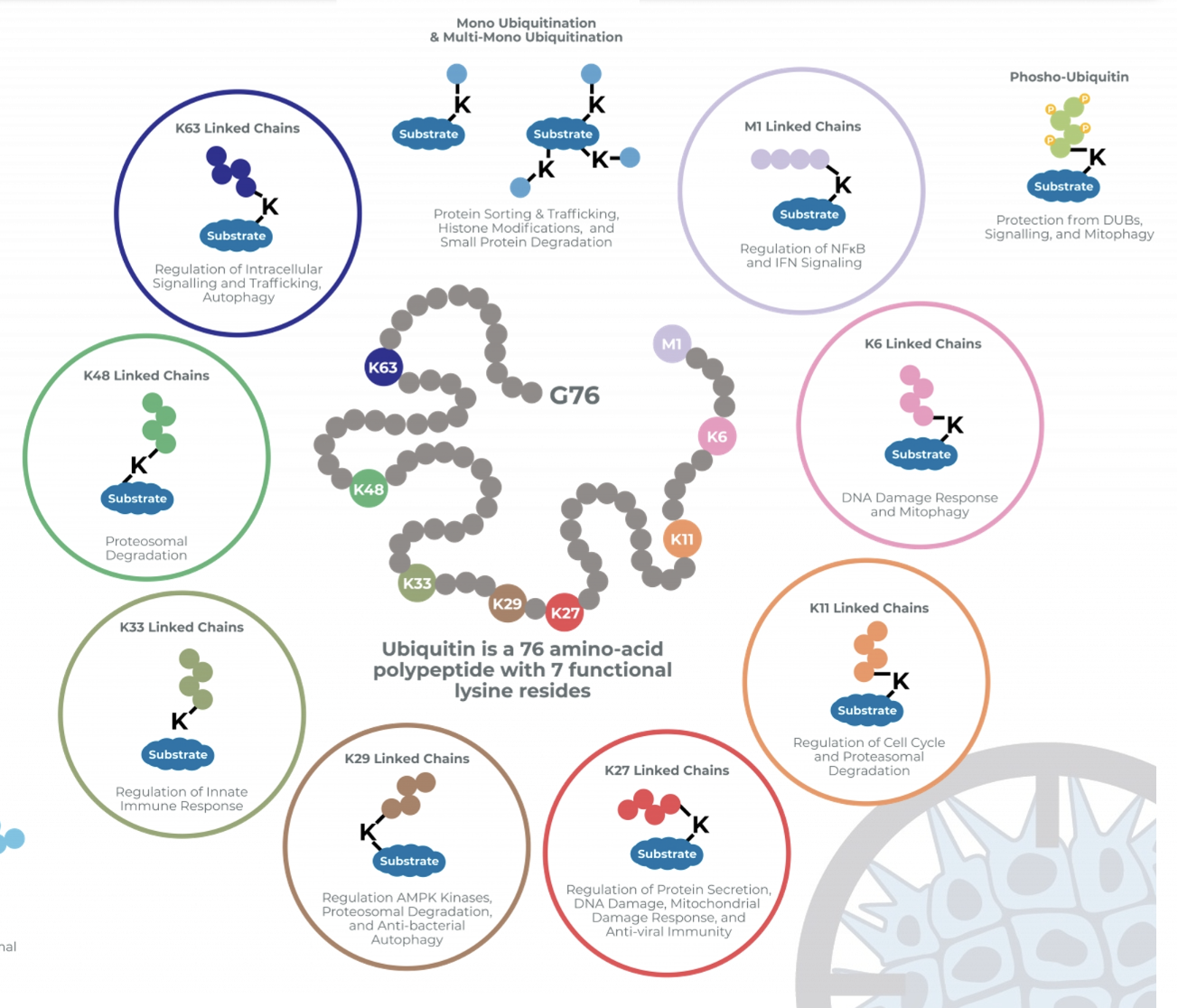
Protein ubiquitination upon DNA damage
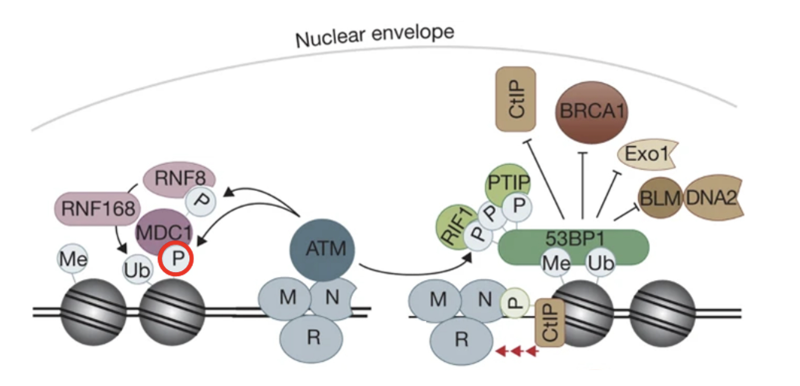
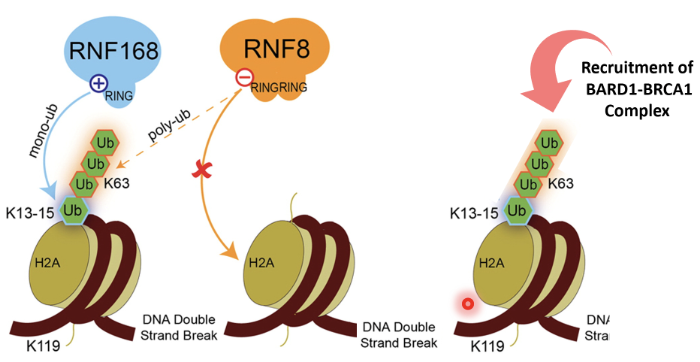
RNF8 - bridge between phosphorylation and ubiquitination
It is an E3 Ub ligase with a RING domain to identify E2 enzymes
It can also recognise phosphorylation sites
MRN identifies DSB and recruits ATM
ATM phosphorylates H2AX which recruits MDC1
MDC1 phosphorylated by ATM
RNF8 binds to phosphorylated MDC1 as a reader and gets activated
RNF8 ubiquitinates histones (mainly H2A and H2AX) at lysine residues K13–K15
Monoubiquitination marks serve as docking sites for RNF168 (another RING-type E3 ubiquitin ligase)
RNF168 is recruited to RNF8-mediated Ub marks and extends the chains by adding K63-linked polyubiquitin chains on the same histones
This serves as recognition platforms for repair proteins
53BP1 recognizes H2A ubiquitination + methylation marks (H4K20me2) and promotes NHEJ
BRCA1–BARD1 complex recognizes ubiquitinated chromatin and triggers HR
PARylation
Addition of ADP ribose to substrate
Acceptors are DNA, RNA, Glu, Asp, Ser, Thr, Arg, Cys
ADP-ribosylation cycle
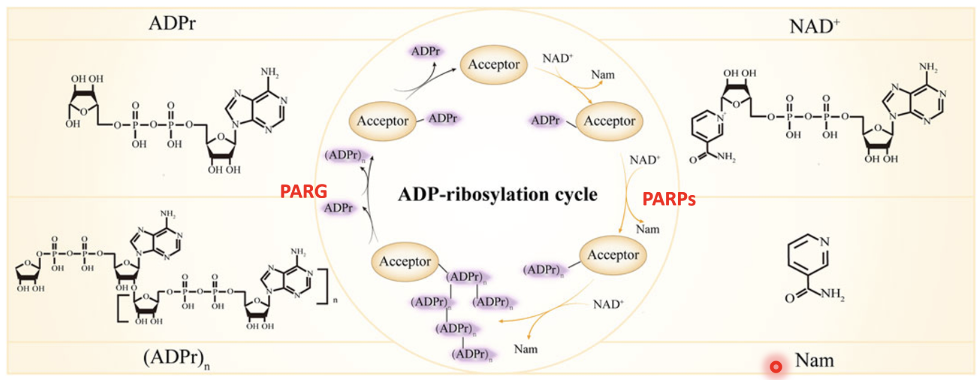
PARPs use NAD⁺ as a cofactor and transfer an ADP-ribose group from NAD⁺ to specific amino acids on target proteins (acceptors), leaving Nam as a byproduct
The first ADP-ribose added to a protein forms mono-ADP-ribosylation (ADPr)
PARP enzymes can extend this modification by linking multiple ADP-ribose units together via glycosidic bonds, forming long branched polymers (PAR chains) - (ADPr)n.
Once DNA repair is complete, the modification is reversed by PARG (poly[ADP-ribose] glycohydrolase) which hydrolyses the ribose–ribose bonds within the PAR chain, removing ADP-ribose units and returning the protein to its unmodified state
Especially important in SSBs
Protein PARylation upon DNA damage
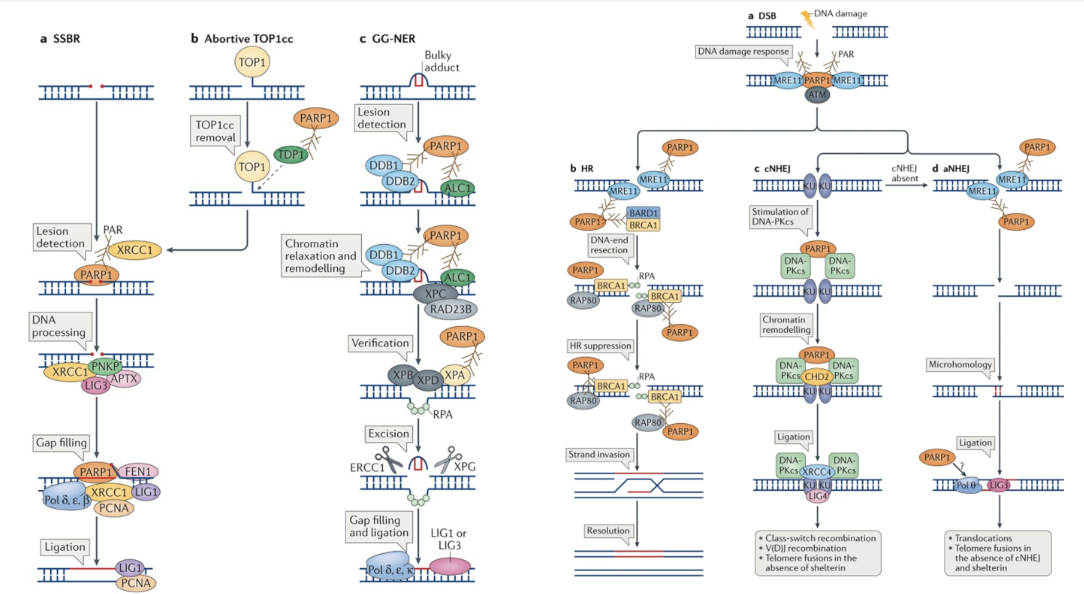
SSB repair
PARP1 detects single-strand breaks (SSBs) and becomes auto-PARylated
This creates a negatively charged platform to recruit XRCC1, a scaffold protein that coordinates DNA repair
XRCC1 recruits PNKP (for end processing), LIG3 (ligation), APTX (to remove damaged ends), and FEN1, Pol δ/ε/β, PCNA for gap filling
LIG1 seals DNA backbone
Abortive TOP1cc — Trapped Topoisomerase 1 complex
Topoisomerase I (TOP1) can get covalently trapped on DNA (TOP1cc lesions).
PARP1 detects these complexes and helps TDP1 remove the trapped TOP1 protein.
PARP1-driven PARylation also relaxes chromatin, allowing other repair enzymes to access the DNA.
GG-NER — Global Genome Nucleotide Excision Repair
In nucleotide excision repair, bulky DNA adducts (like UV-induced thymine dimers) are recognised by DDB1–DDB2 and XPC–RAD23B complexes.
PARP1 PARylates itself and these complexes, promoting chromatin remodelling to allow lesion verification and excision.
XPB/XPD (helicases) and XPA/RPA (verification) act next, followed by ERCC1–XPF/XPG (nucleases) to remove the damaged strand.
Finally, DNA polymerases δ, ε, κ, and ligases (LIG1/LIG3) fill and seal the gap
DSB detection
PARP1 acts alongside ATM and the MRN complex (MRE11–RAD50–NBS1) to detect DSBs.
PARP1 adds PAR chains (PARylation) to recruit and organize downstream repair factors.
This modification facilitates chromatin relaxation and ATM activation.
HR
During HR, PARP1 interacts with MRE11, BRCA1, BARD1, and RPA.
PARylation promotes DNA end resection, which is necessary for strand invasion.
However, excessive PARP1 activity can also suppress HR by competing with BRCA1 complexes.
NHEJ
PARP1 facilitates KU70/80 and DNA-PKcs binding to DNA ends.
This activates chromatin remodeling (via CHD2) and end ligation by LIG4–XRCC4.
PARylation helps stabilize this complex and coordinate repair of blunt or incompatible DNA ends.