Chapter 8: Genome Structure, Chromatin, And The Nucleosome
Function of DNA packing:
readily fits inside the cell
protect DNA from damage
transmission to daughter cells during cell division
organization to each molecule of DNA that regulates DNA accessibility
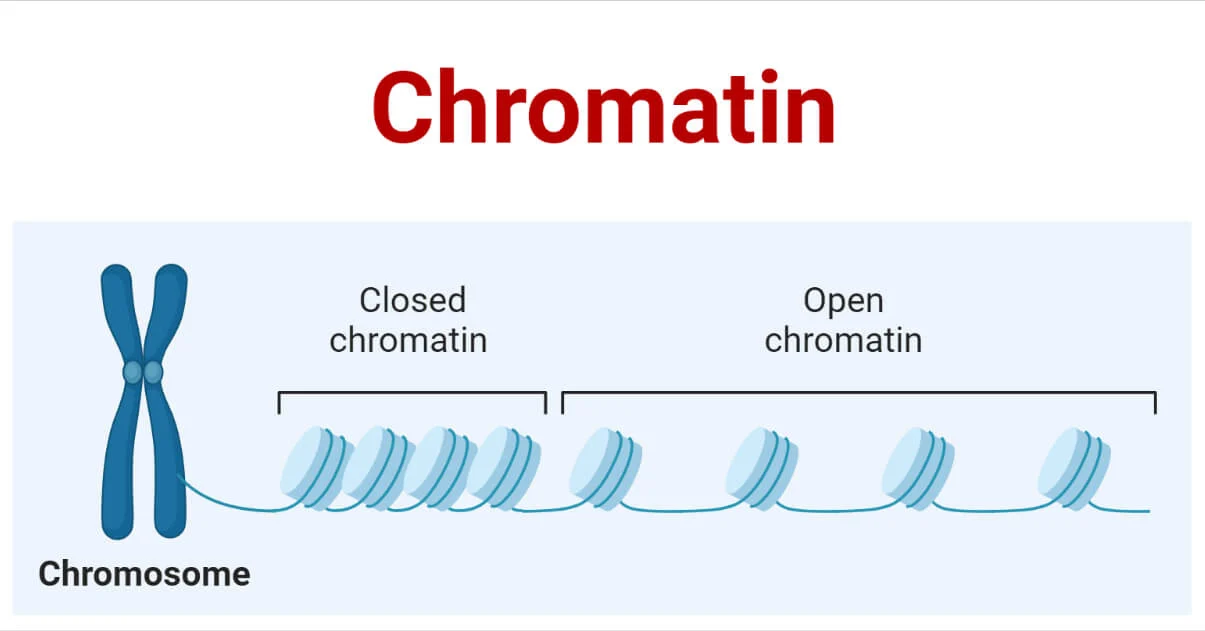
Bacterial DNA packing is the most compact DNA during cell replication
1. Genomic Sequence and Chromosome Diversity
1) Chromosomes can be circular or linear
all eukaryotes have multiple linear chromosomes
most bacteria and archaea have circular chromosomes
2) Number of chromosomes is maintained in all cells
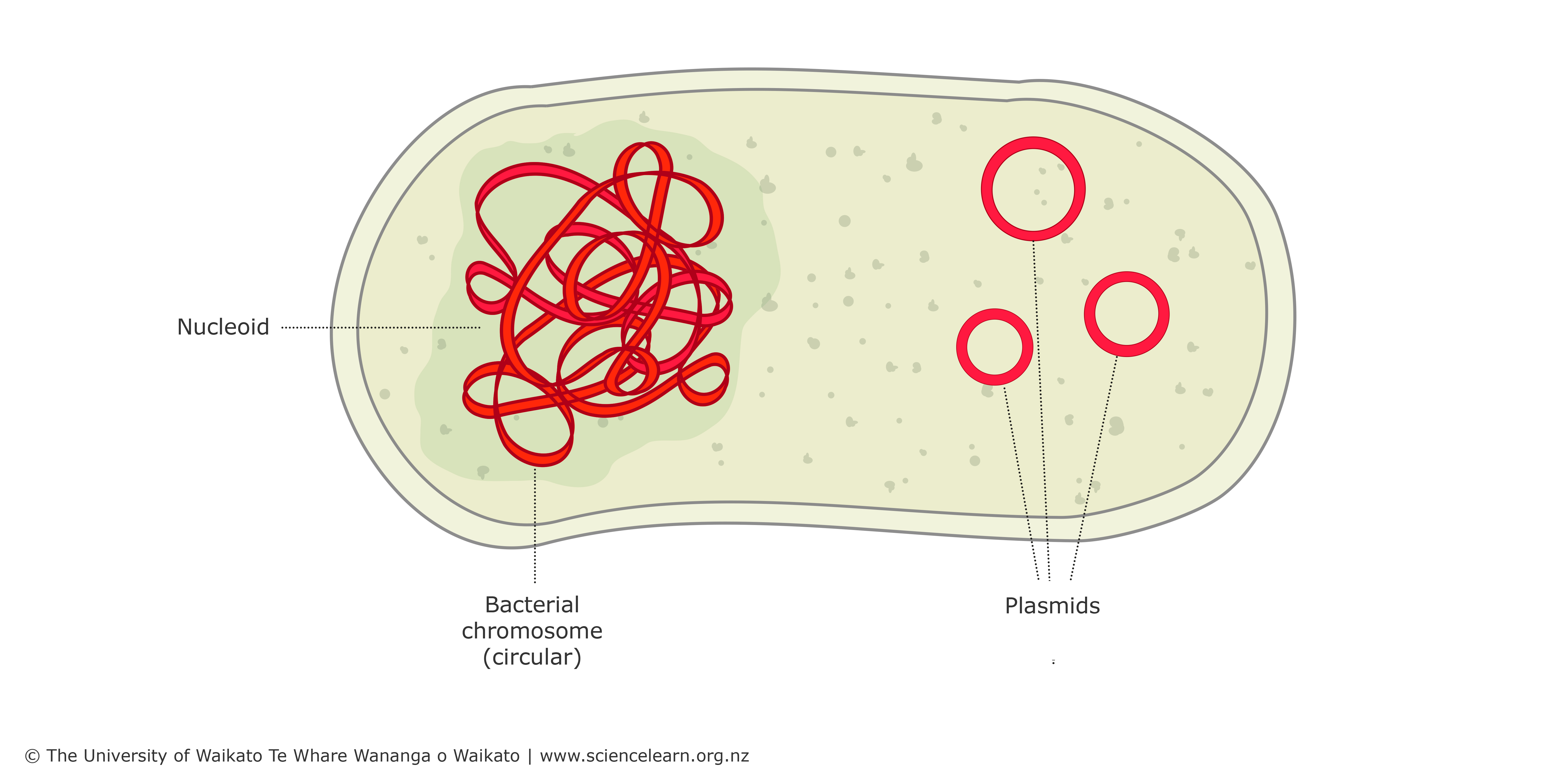
Prokaryotic cells (archaea and bacteria)
typically have one complete copy of chromosomes that is packaged into a nucleoid
often carry one or more plasmids that are not essential for growth but carry genes with desirable traits (ex. antibiotic resistance)
Eukaryotic cells
chromosomes are contained within the nucleus
most are diploids (2n) with each homologous chromosome derived from each parent
gametes are haploids (n)
polyploid cells have more than two copies of each chromosome → high levels of metabolism, difficulties in segregation (plants)
3) Genome size, number of genes or types of genes, and gene density
Genome size
most bacterial genomes: 1-6 Mb
most archaea genome sizes = size range of bacterial genomes
eukaryotes span an extremely wide range of genome sizes
polyploid plants have much larger genomes
C-value is the total amount of DNA per haploid set of chromosomes
minimum genome size does increase with morphological complexity, mostly in lower eukaryotes
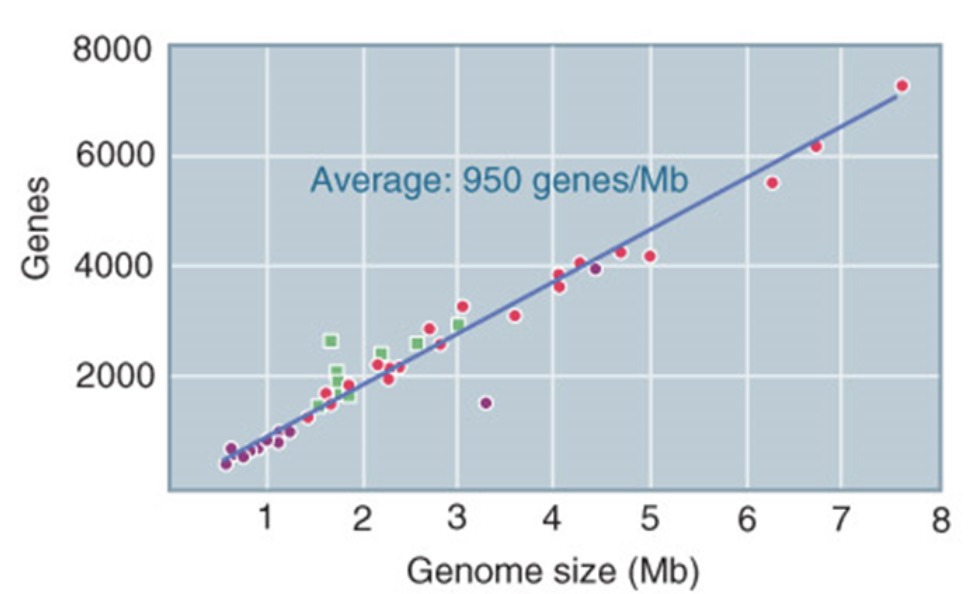
Genome size and gene number
bacteria and archaea have fewer genes than eukaryotes → in prokaryotes gene number strongly correlates with genome size
number of different proteins in human genome >> number of genes
90% of human multi-exon genes go through alternative splicing
in eukaryotes gene number does not correlate with genome size or organism complexity
Gene number vs. types of genes
many genes are duplicated → the number of different gene families (number of unique genes + number of gene families) is much smaller than the total number of genes
proportion of unique genes decreases with increasing genome size → multicellular eukaryotes with larger genome sizes tend to have a larger proportion of genes that are present in multiple copies
Gene size may be correlated to complexity
the amount of DNA devoted to the expression of one gene may be largely different despite the very similar size of the protein encoded
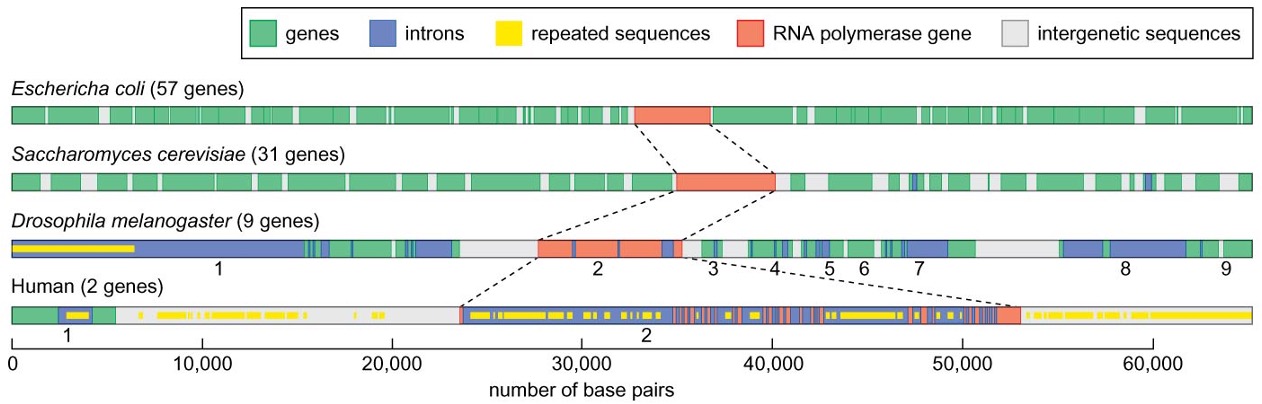
genes from more complex organisms tend to have longer sizes with a larger number of exons and more interrupting DNA in between
exons containing 5’-, 3’-untranslated regions tend to be longer than those encoding proteins
introns vary widely in size among multicellular eukaryotes
4) Non-exon DNA sequences in human genome
Introns
intervening sequences that are removed when primary RNA transcript is processed to give mature RNA product
major reason for increase in gene size in complex organisms
microorganisms (rapidly replicate) may have lost introns due to genomic compaction
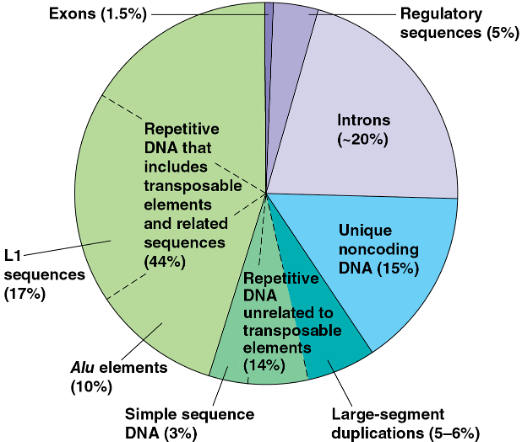
Repetitive DNA
repetitive sequences account for the majority of the human genome
larger genomes within a taxonomic group do not contain more genes but have large amounts of repetitive DNA
prokaryotes contain nonrepetitive DNA almost exclusively
polypeptides are generally encoded by sequences in nonrepetitive DNA
i) Transposons (DNA changing its position)
44% of entire human genome is made up of transposable elements and sequences related to them
may cause genome rearrangements that confer selective advantages, or may be selectively neutral
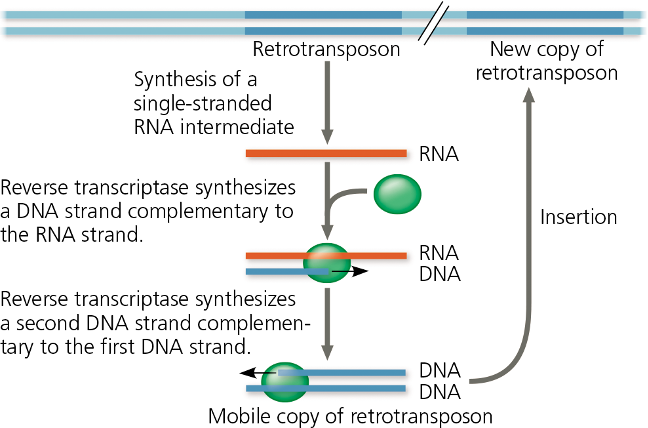
large size of plant genome is also accounted for by extra transposable elements
some currently functional genes originated as transposons and evolved into their present condition after losing the ability to transpose ( >50 genes)
in human and other primates, many transposon related sequences consist of family of Alu elements (10% of human genome) → ~300 NT, do not code for protein, many Alu elements are transcribed into RNA that are thought to help regulation gene expression
17% of human genome is made up of a type of retrotransoposon called LINE-1 (L1) → ~6,500 NT, low rate of transposition, crucial for development of early embryos
some transposons encode proteins that do not carry normal cellular functions
transposons are usually included in “noncoding” DNA category
ii) other repetitive DNA
arisen from mistakes during DNA replication or recombination
~1/3 are duplications of long stretches (10~300 kb)
Simple sequence DNA
contain many copies of tandemly repeated short sequences
short tandem repeat (STR) is a series of repeating units of 2~5 NT → number for STRs can vary among sites
common in centromeres and telomeres, where it probably plays structural roles in the chromosome
Definition of a gene
must include the ability of a gene to the direct production of either functional RNA or protein molecules
no clear-cut 1:1 correspondence between single gene and particular regulatory region
Chromosome Duplication and Segregation
1) DNA elements maintained during cell division in eukaryotes
Origin of replication
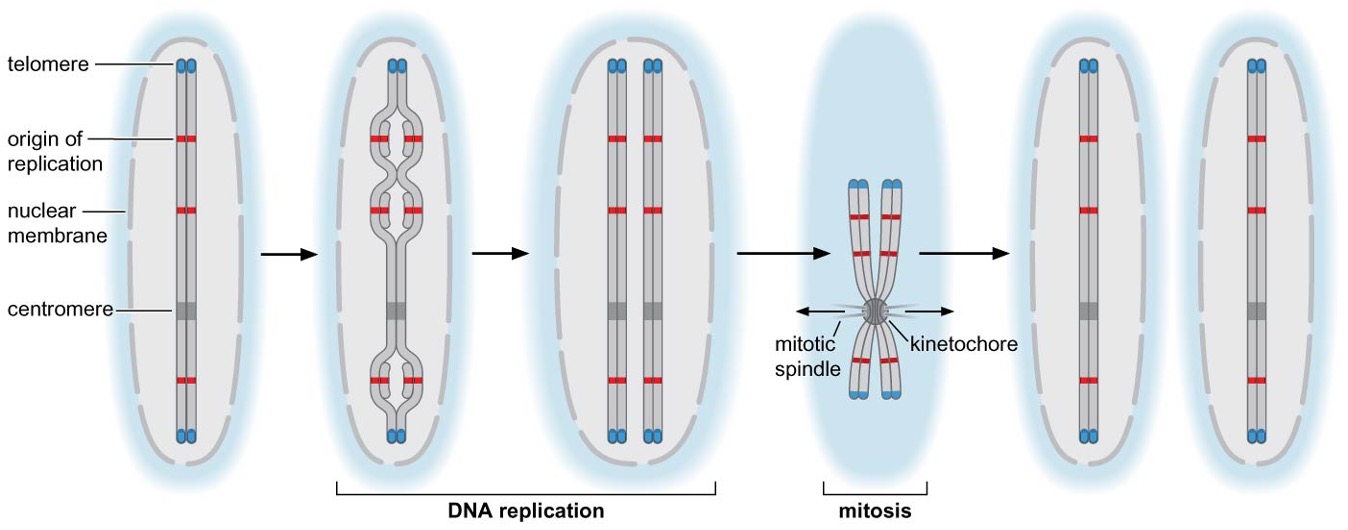
Centromere
required for correct segregation of sister chromosomes after replication
direct formation of kinetochore → binds to spindle microtubules
one chromosome - one centromere
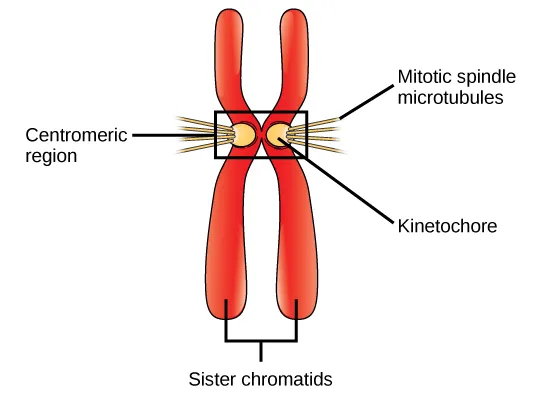
Centromere size and composition vary dramatically among different organisms
Telomere
located at two ends of linear chromosome
bound proteins distinguish natural ends of chromosomes and prevent recombination or degradation
telomerases are recruited to telomeres and allow the cell to replicate end of chromosomes
a portion of the telomere is maintained in a single-stranded form

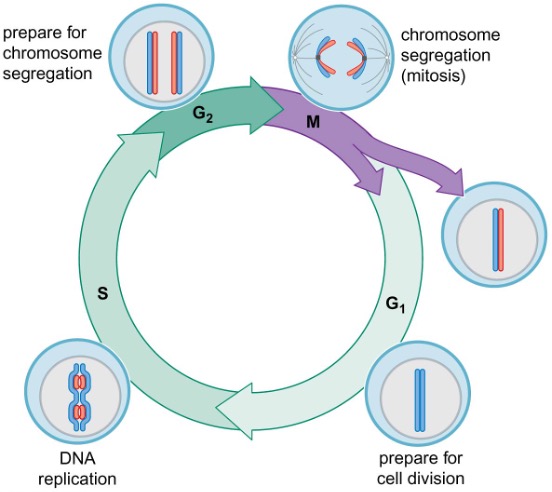
2) Eukaryotic cell cycle
chromosome duplication and segregation occur simultaneously in bacterial cells, and at distinct times in eukaryotic cells
the mitotic cell cycle can be divided into G1, S (: DNA synthesis), G2, and M (: chromosome segregation) phase
gap phases provide time for the cell to prepare for the next phase of the cell cycle and check that the previous phase of the cell cycle has been accomplished appropriately → if not, cell cycle stops at cell cycle checkpoints
 3) Changes in chromatin structure
3) Changes in chromatin structure
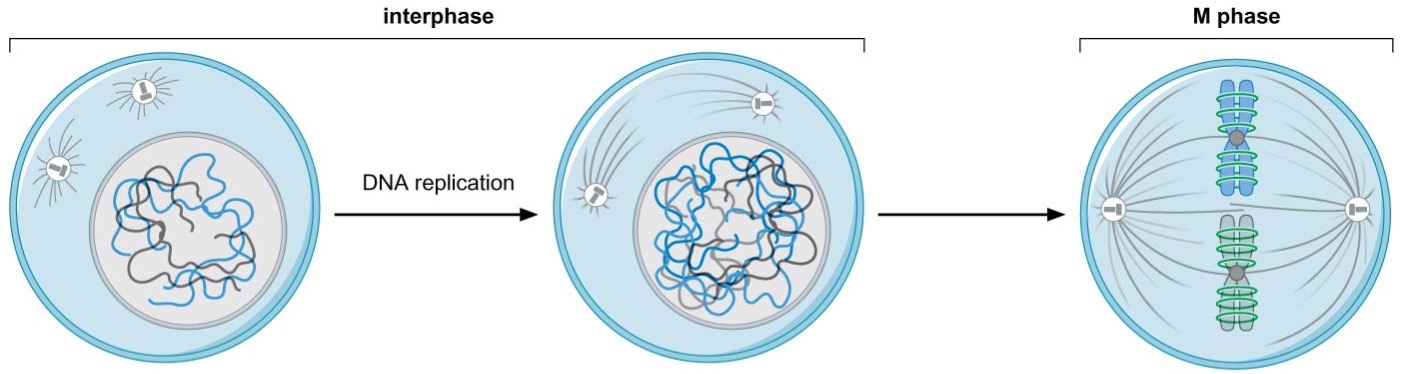
chromosome structure constantly changes throughout the cell cycle
chromosome condensation allows chromosomes to become their most compact form during M phase
chromosomes are decondensed during the rest of the cell cycle (interphase)
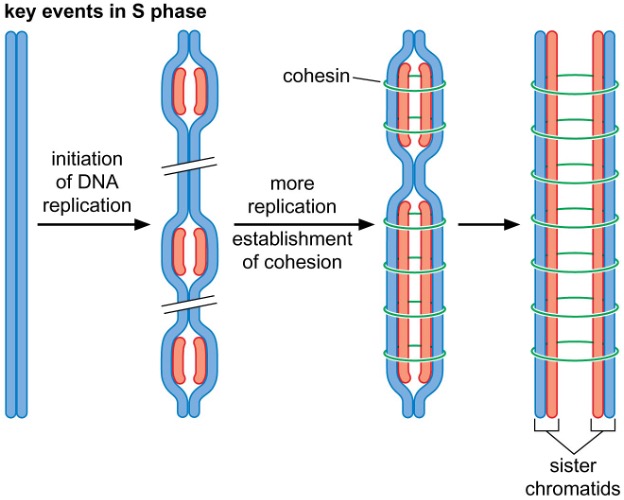
sister-chromatid cohesion is established immediately after replication
transcription is associated with regional changes in chromosome structure
4) Sister chromatid cohesion and chromosome condensation
SMC (structural maintenance of chromosome) proteins and non-SMC proteins form multiprotein complexes that link two DNA helices together
Cohesin
core of SMC1-SMC3 associated with smaller non-SMC proteins
connects sister chromatids during mitosis
separase breaks down cohesin
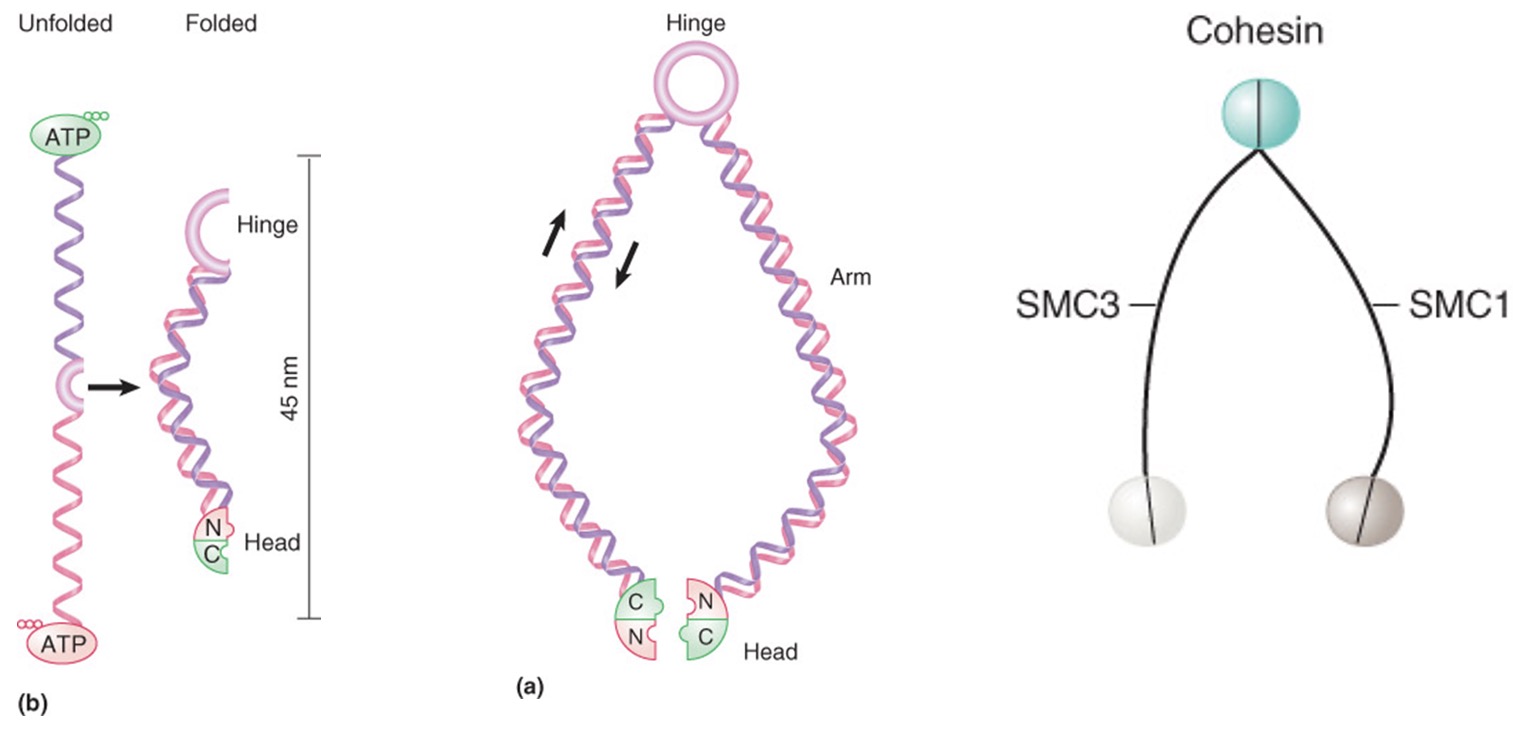
Models for DNA linking by cohesins
Cohesins may form extended structure in which each monomer binds DNA and connects via the hinge region, allowing two different DNA molecules to be linked, and head-head interactions would create tetrameric structure
Cohesion dimers interact at both head and hinge regions to form circular structure that could DNA molecules together by encircling them
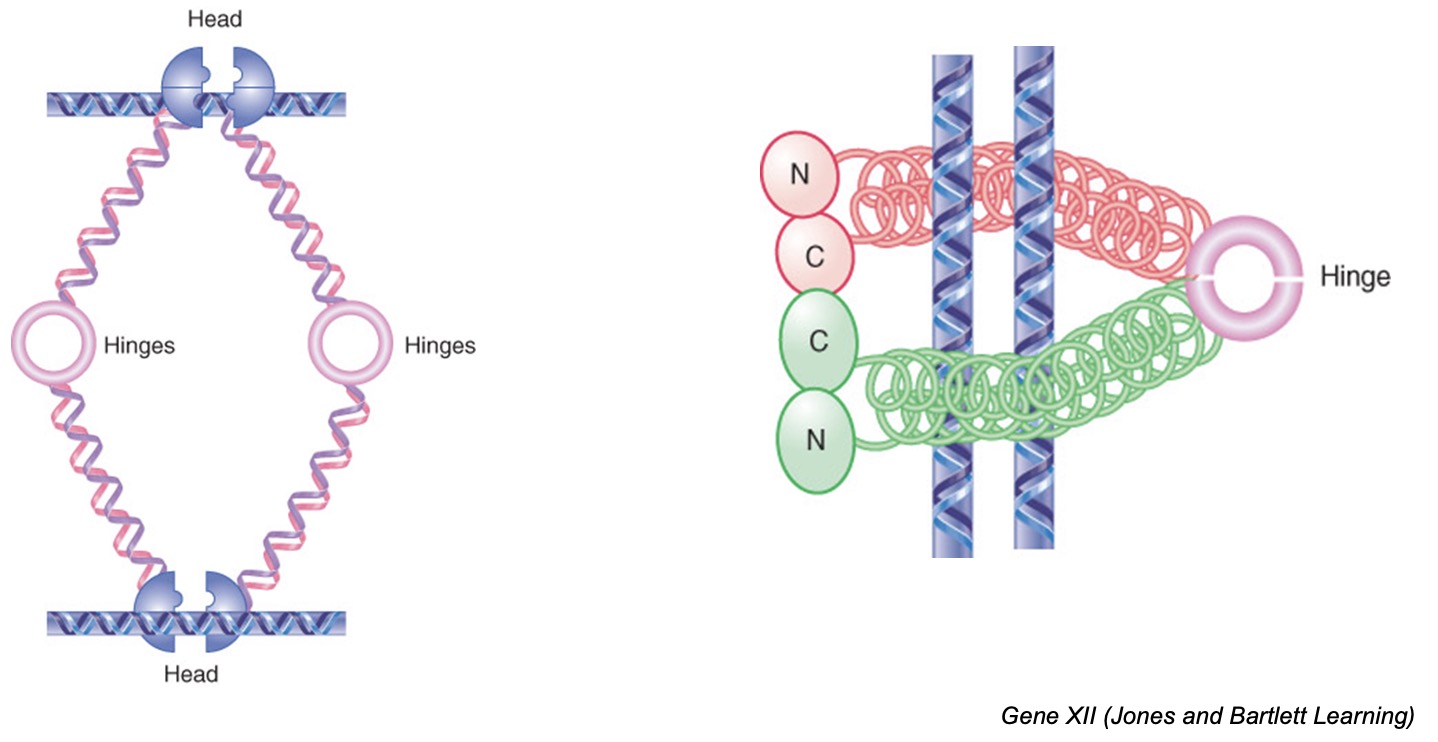
Condensin
core of SMC2-SMC4 associated with non-SMC proteins
condensins are responsible for chromatin condensation at mitosis by introducing positive supercoils into DNA such that chromatin is more tightly coiled
condensins may form a dimer, with interacting hinge domains, that pulls together distant sites on the same DNA molecule
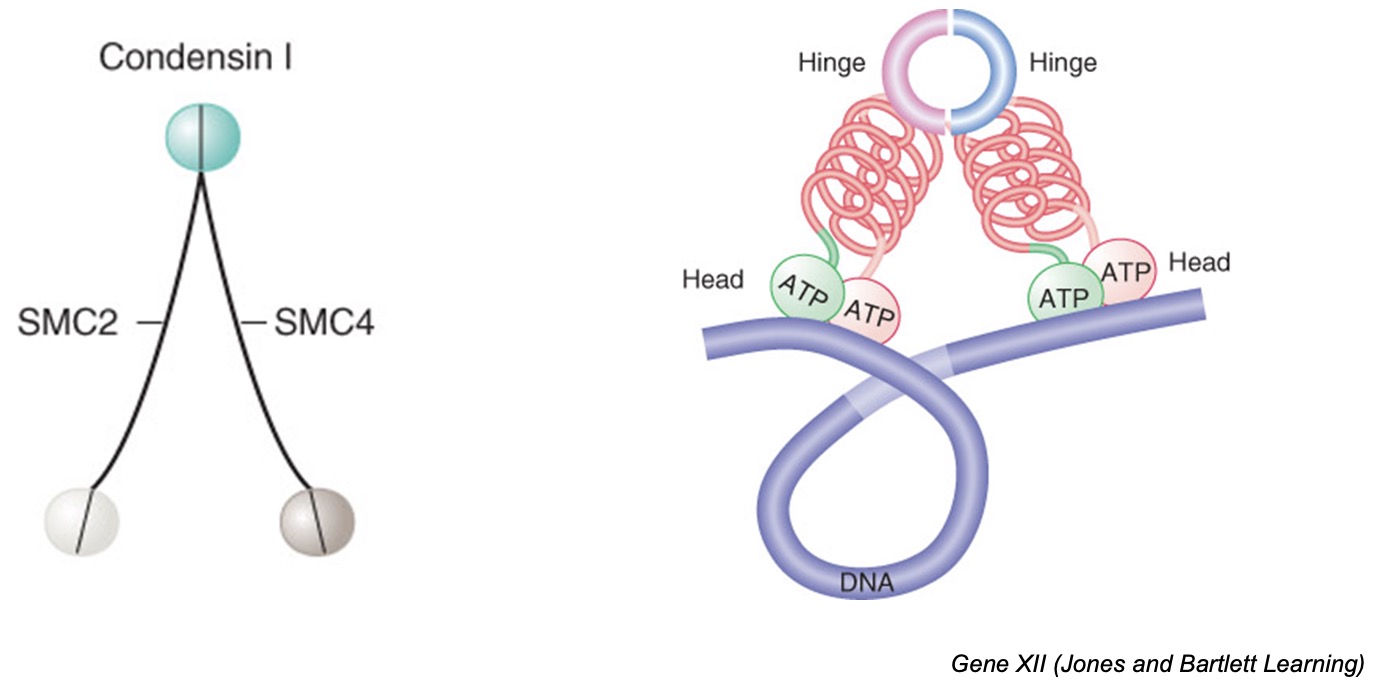
5) Mitosis
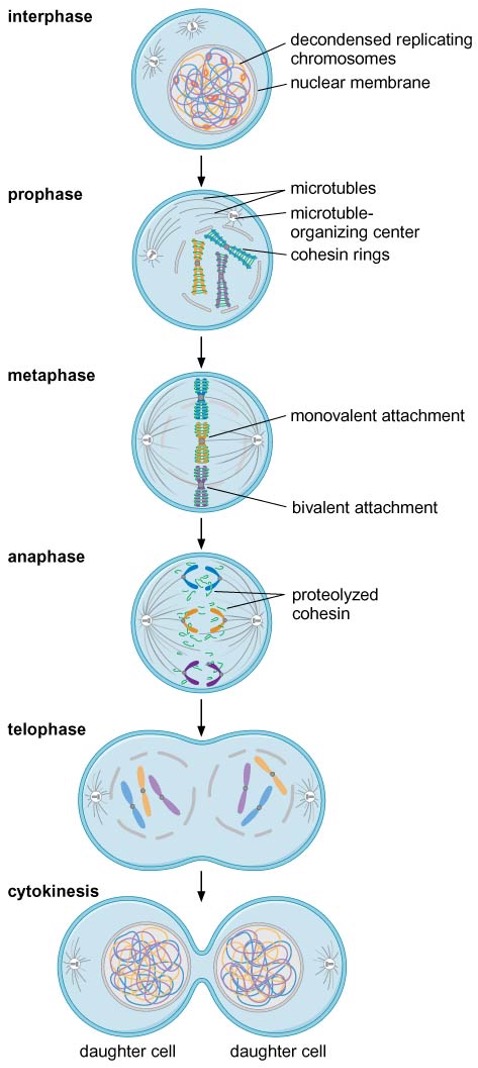
Prophase
condensin and type II topoisomerases condense chromosomes into highly compact form
nuclear envelope breaks down
Metaphase
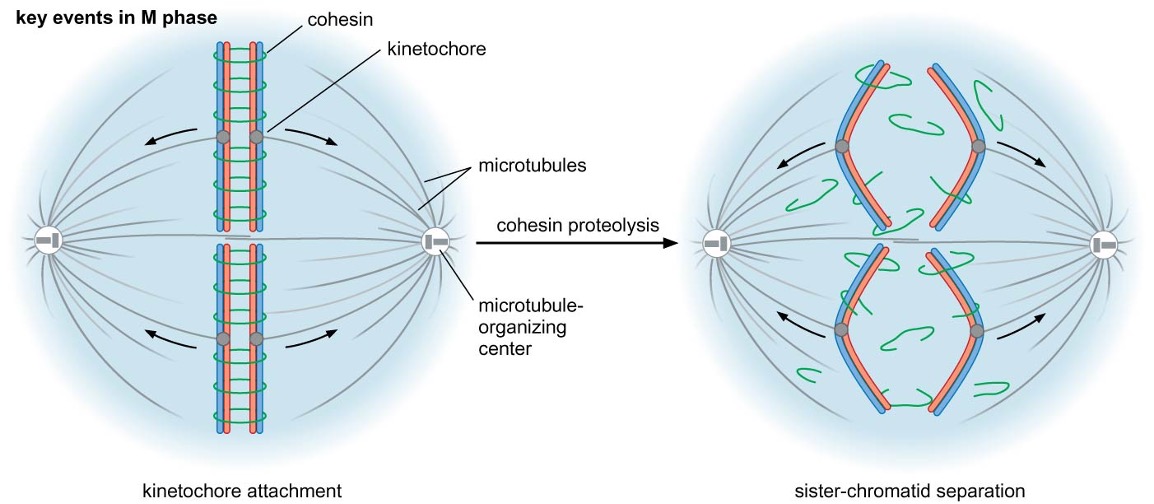
mitotic spindle forms and kinetochores of sister-chromatid pair attach to microtubules emanating from opposite MTOCs (: bivalent attachment) → alignment of chromosomes in middle of cell
When a cell wants to divide into two new cells, it needs to make sure that each new cell gets a copy of all its genetic material. To do this, the cell creates something called a "mitotic spindle," which is like a scaffold made of tiny tubes called microtubules.
The genetic material of the cell is organized into structures called chromosomes, and each chromosome has a copy of itself that we call a "sister-chromatid pair." These pairs need to be separated so that each new cell gets one copy of each chromosome.
To make sure this happens correctly, the sister-chromatid pairs attach to the mitotic spindle using special structures called "kinetochores." Each pair attaches to microtubules that come from opposite ends of the spindle, so that they are pulled apart in opposite directions.
When all the sister-chromatid pairs are attached to the spindle, they line up in the middle of the cell. This makes sure that each new cell gets one copy of each chromosome.
Anaphase
proteolytic destruction of cohesin → chromosome segregation
Telophase
nuclear envelope reforms
chromosome becomes decondensed
Level of cyclin is the checkpoint for a separase to break down cohesin
5) Meiosis
Interphase
after DNA replication, homologous chromosomes pair with each other and recombine
Meiosis I
the two kinetochores of each sister-chromatid pair bind to one pole of the spindle (: monovalent attachment)
homologs are physically connected at chiasma (crossing over regions)
sister-chromatid cohesion along the arms is eliminated during anaphase I → separation of recombined homologs, but that near the centromere is maintained
Meiosis II
cohesion remaining at centromere is eliminated
four haploid cells are formed
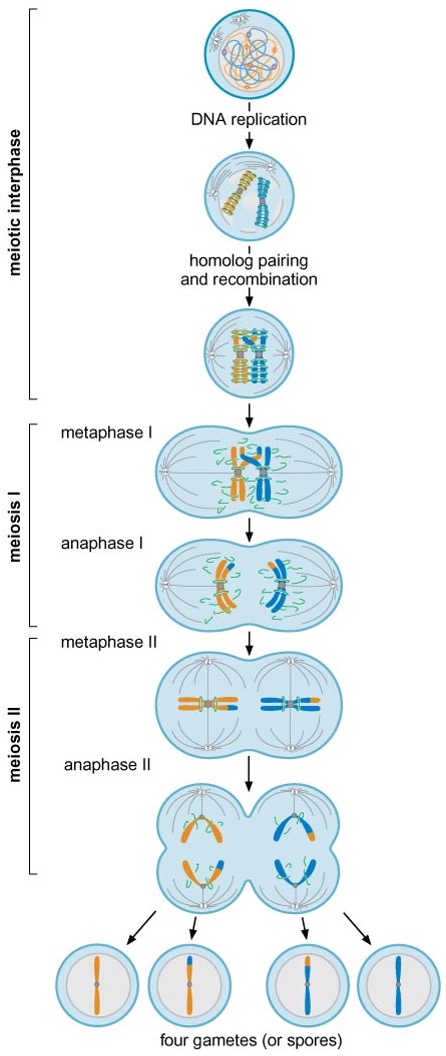
6) Observation of chromosome structure by microscopy
condensed chromosomes are easy to visualize by simple light microscopy
in interphase, 30 nm or 10 nm chromatin fibers are observed
2. The Nucleosome
1) Nucleosomes are building blocks of chromosomes
each nucleosome is composed of a core of 8 histones and the DNA wrapped around them in the left-handed direction
nucleosome assembly compacts DNA ~6 fold
linker DNA: DNA between each nucleosome
core DNA: ~147 bp DNA that is most tightly associated with nucleosome
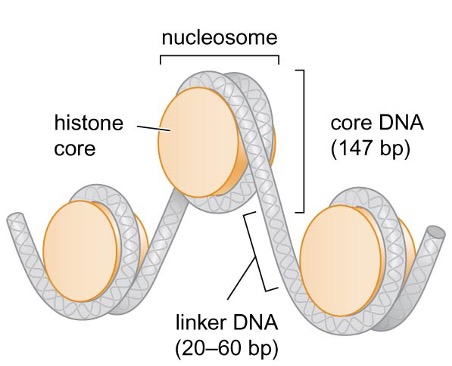
2) Nucleosome assembly
Histones
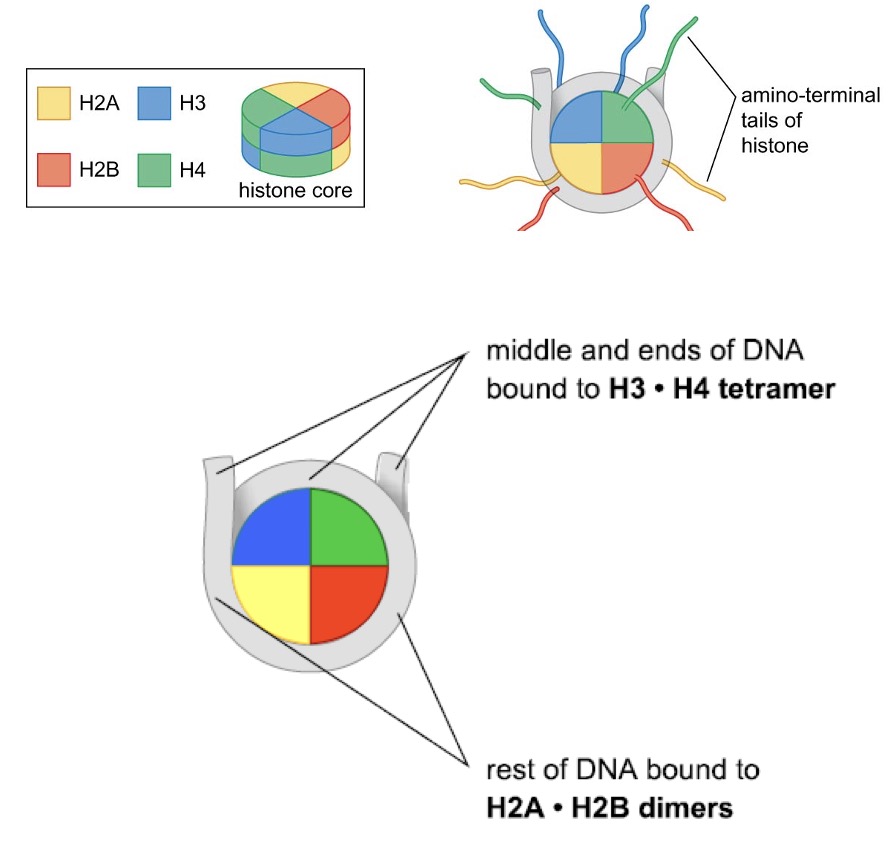
2 copies of each of the core histones (H2A, H2B, H3, H4, all 11~15 kDa) form the histone octamer core of the nucleosome
linker histone, H1, binds to linker DNA
histones have high content of (+) charged amino acids
the histone-fold domain found in every core histone mediates the assembly of histone-only intermediates → H3 and H4 form H3∙H4 tetramers, H2A and H2B form H2A∙H2B dimers (in solution)
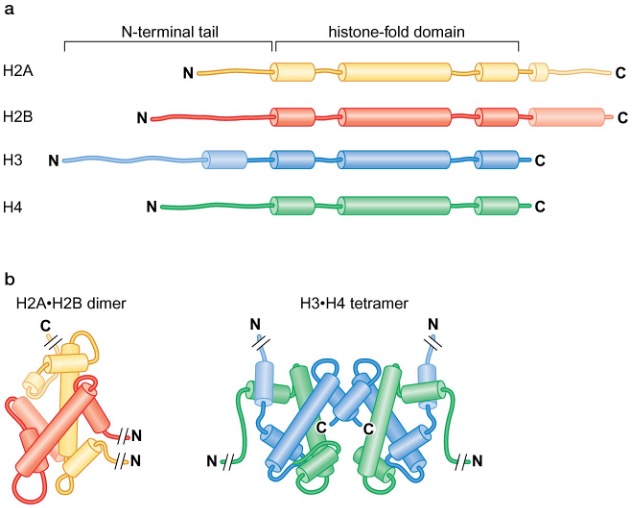
Nucleosome assembly
H3∙H4 tetramer binds to DNA = make the structure → 2 x H2A∙H2B dimers join H3(2)∙H4(2)–DNA complex
N-terminal tails lack a defined structure and are accessible within the intact nucleosome → act as sites of posttranslational modifications that alter function of individual nucleosomes
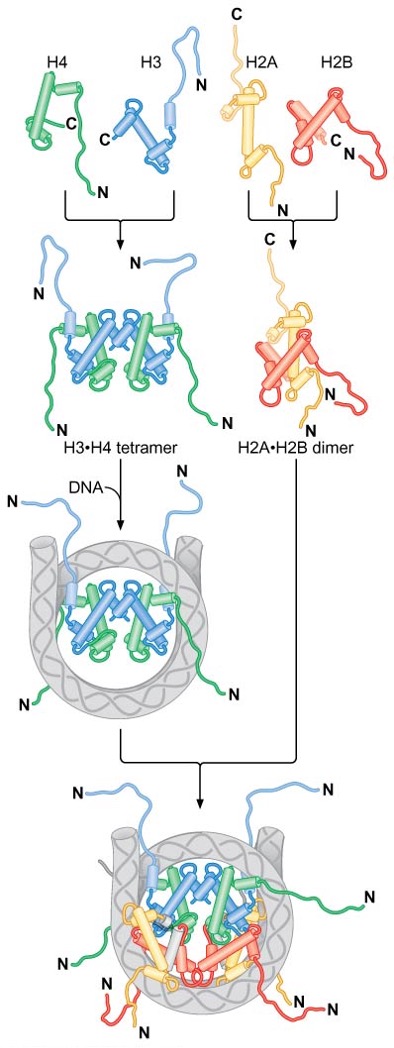
3) Histones bind characteristic regions of DNA within the nucleosome
nucleosome has an approximate twofold axis of symmetry (: dyad axis)
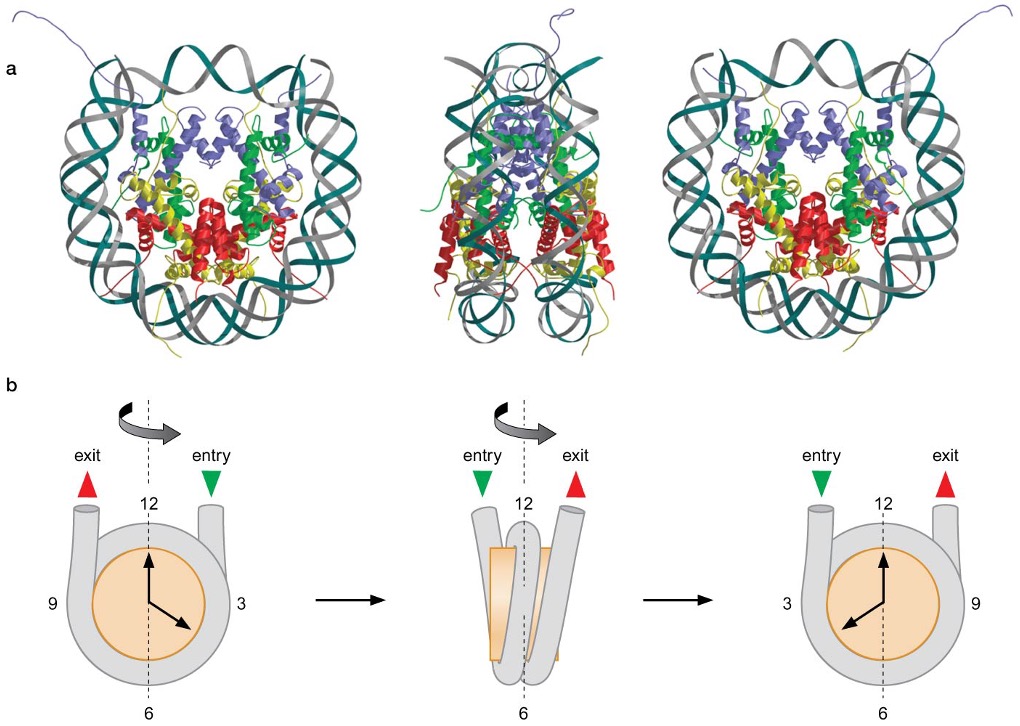
H3∙H4 tetramers and H2A∙H2B dimers each interact with a particular region of DNA within the nucleosome → H3(2)∙H4(2) tetramer binds to the central 60bp and 13 bp-ends of both DNA, while H2A∙H2B dimers associate with ~30 bp of DNA on either side of the central 60 bp bound by H3 and H4
Sequence-independent interaction between core histone and DNA
DNA and core histone have 14 contact sites, 1 for each time minor groove of DNA faces histone octamer (147 bp/10.5 = 14) → large number of H bonds (~40) provide driving force for DNA bending, basic nature of histone masks (-) charge of phosphates that can resist DNA bending and facilitates close juxtaposition of two adjacent DNA helices wrapped around the histone octamer
most H bonds are between proteins and O atoms in the DNA backbone near the minor groove, 7 H bonds are between proteins and parts of DNA bases that are not distinguishable between different base pairs → histone – DNA contact is non-sequence specific
4) Histone N-terminal tails stabilize DNA wrapping
H2B, H3 N-terminal tails emerge from between the 2 DNA helices where path of exit of H2B and H3 tails is formed by 2 adjacent minor grooves
H4 and H2A tails emerge either above or below the DNA helices → histone tails direct DNA to wrap around the histone octamer in a left-handed manner (torroidal writhe) → induces (-) supercoiling
DNA wrapping stores (-) superhelicity
each nucleosome changes the linking number of associated DNA by ~1.2 → acts as storage of (-) superhelicity → initiation of DNA replication, transcription, recombination, ... can all unwind DNA by removing nucleosome
topoisomerases decrease the linking number in DNA not bound to nucleosomes and reduce (+) superhelicity generated by nucleosome assembly → enables formation of more nucleosomes
in bacterial cccDNA, gyrase maintains a (-) supercoiled state
bacteria that grow at very high temperatures (>80°C) have reverse gyrase that helps keep the genome (+) supercoiled to prevent DNA from unwinding
3. Higher-order Chromatin Structure
Heterochromatin
limited gene expression (too compact to be expressed)
associated with particular chromosomal regions (~telomere, centromere,...)
nucleosomal DNA is assembled into higher-order structures
Euchromatin
higher gene expression
nucleosomal DNA is in much less organized assemblies
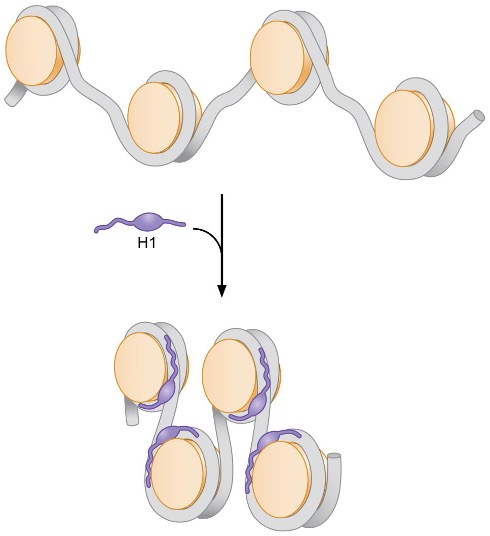
1) H1 binding
after nucleosome assembly, one H1 interacts with DNA at 2 regions, linker DNA at one end of nucleosome and middle region of associated 147 bp → tightens association between 2 regions and increases length of DNA wrapped around histone octamer
if assume that angles of DNA entry and exit are ~20° relative to dyad axis, this would result in nucleosomes alternating on either side of central region of linker DNA bound by H1 → H1 binding results in a distinct zigzag appearance of nucleosomal DNA
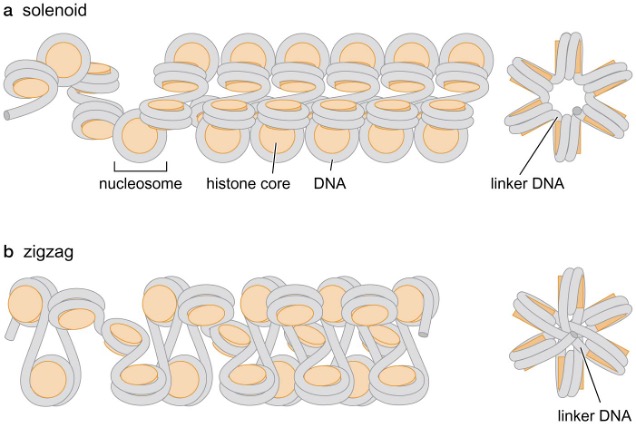
2) 30-nm fiber
binding of H1 stabilizes higher-order chromatin structures
Models for 30-nm fiber structure
Solenoid model
nucleosomal DNA forms a superhelix containing ~6 nucleosomes per turn
flat surface of histone octamer discs are adjacent to each other
linker DNA never passes through axis of fiber
Zigzag model
linker DNA passes through central axis in a relatively straight form → longer linker DNA favors this conformation
Stabilization of 30-nm fibers by histone N-terminal tails
histone N-terminal tails most likely stabilize the 30-nm fiber by interacting with adjacent nucleosomes
interaction between (+) charged N-terminus of H4 and (-) charged, conserved region of histone-fold domain of H2A is particularly important for 30-nm fiber formation
3. Nuclear scaffold
packaging of DNA into nucleosomes and 30-nm fibers compacts length of linear DNA by ~40-fold
further folding is thought to be achieved by forming loops of 40-90 kb held together by a nuclear scaffold
topoisomerase II is abundant in scaffold preparations and may be part of the mechanism that holds DNA at the base of loops
SMC (Structural Maintenance of Chromosome) proteins are also abundant in scaffolds, and may serve a role by interacting with chromosomal DNA
Histone variants alter nucleosome function
histone variants can replace standard histones to form alternate nucleosomes
histone variants are highly similar to core histones in amino acid sequence but are found at different locations in chromatin where they play special functions
Examples:
In double-strand DNA breakage, H2A.X located adjacent to breakage is phosphorylated at a Ser that is not present in H2A (H2A.X replaces H2A) → specifically recognized by DNA repair enzymes
In centromeric DNA, where nucleosomes are incorporated into the kinetochore, CENP-A, an H3 variant that has an extended N-terminal tail, is associated with nucleosomes → acts as binding site for components of the kinetochore
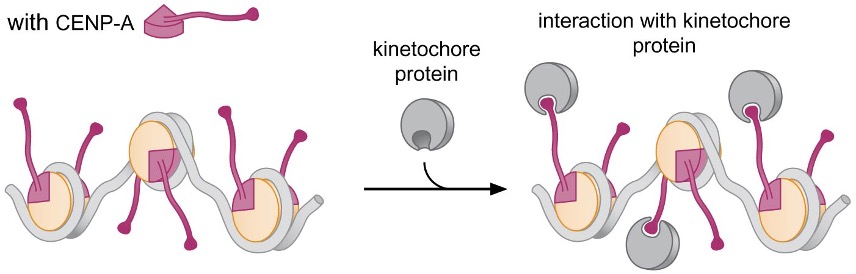
4. Regulation of Chromatin Structure
1) Nucleosome positioning
nucleosomes are not fixed in their locations due to non-sequence-specific interactions with DNA
nucleosome positioning allows DNA-binding site for regulatory proteins to remain inaccessible linker DNA region
Modes for nucleosome positioning
Protein-dependent positioning
competition between nucleosomes and DNA-binding proteins
preferential nucleosome assembly adjacent to nucleosome-bound DNA- binding protein’s binding site
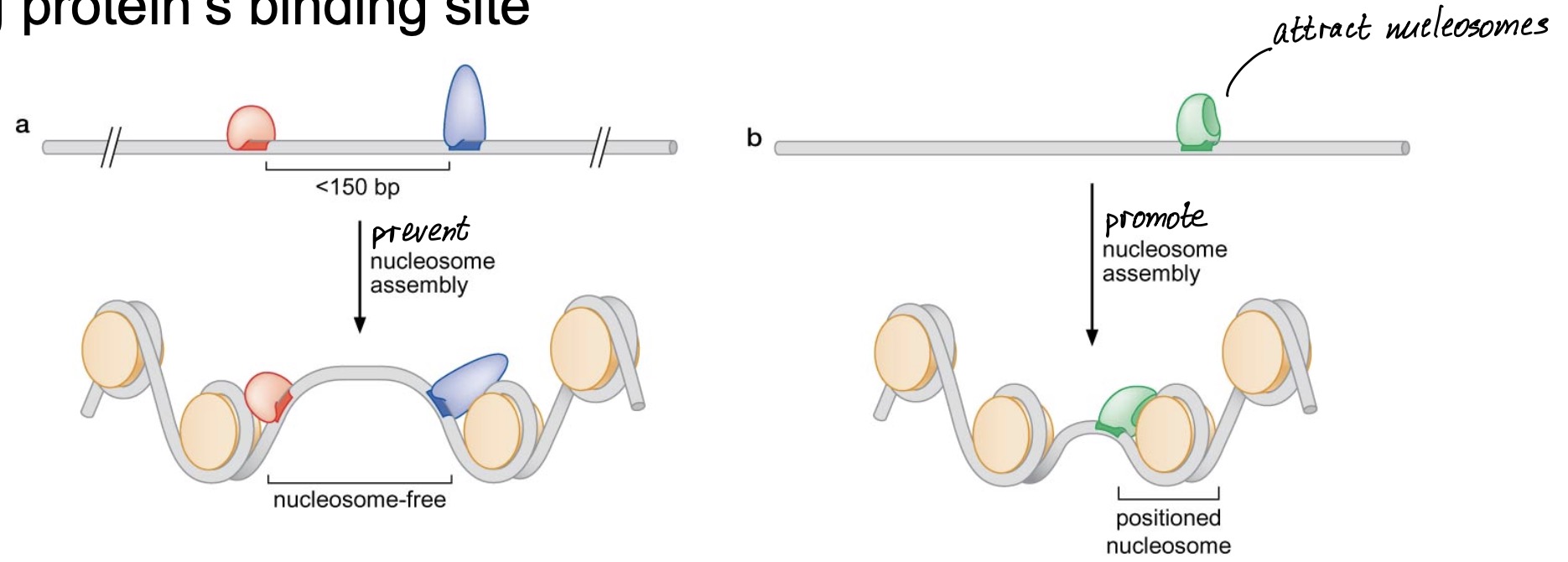
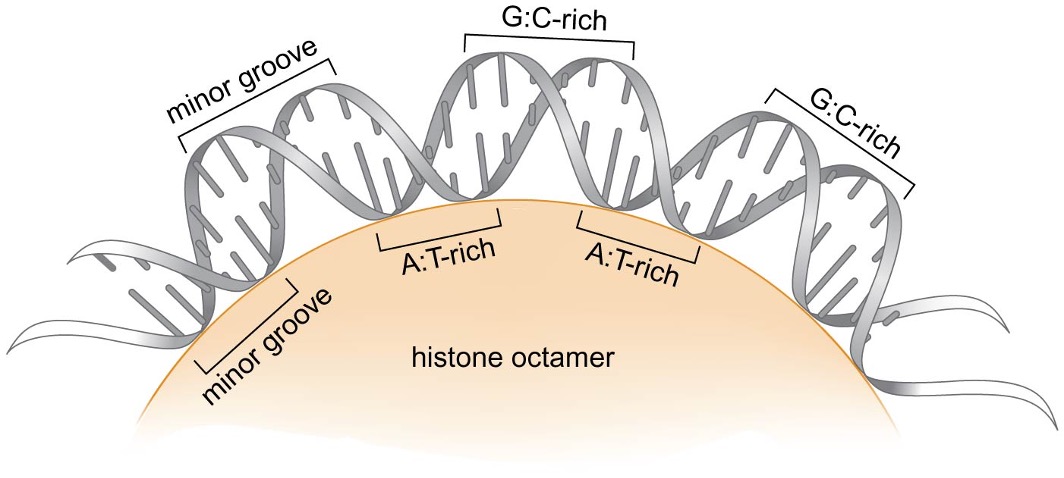
Sequence-dependent positioning
DNA wrapped around the histone octamer is smoothly bent in most parts but sharper bends occur at some locations
nucleosomes preferentially form on DNA sequences that bend easily
minor groove narrowing is energetically more favored in AT-rich sequences → favored nucleosome binding region sequence: AT-rich sequence in minor groove facing histone octamer, and GC-rich sequence in minor groove facing away from histone octamer
2) Modification of histone N-terminal tails
Histone N-terminal tail modifications (resulting in regulation)
acetylation, methylation, phosphorylation, ubiquitination are modifications that are commonly found in histones → can affect chromatin structure and function
Example: Lys acetylation neutralizes and reduces (+) charge of histone → can reduce affinity for DNA and affects the ability of nucleosome arrays to form more repressive higher-order chromatin structure
Effect of histone modifications
most histone modifications are reversible via specialized enzymes (histone acetyltransferase - add, histone deacetylase - remove, histone methyltransferase - add, histone demethylase - remove, kinase - add phosphate, phosphatase - remove phosphate)
alter the function of chromatin and act as on/off switches used for regulation
Example: euchromatin contains highly acetylated histones
histone methylation can be associated with either active/inactive chromatin (depending on which residue is methylated) via other proteins that determine the functional result of methylation
Example: methylation of Lys9 in H3 → transcriptionally silent chromatin
Example: methylation of Lys4 in H3 → transcriptionally active chromatin
vast majority of possible histone modification combinations can lead to various functional outcomes (: histone code)
Protein domains that recognize modified forms of histone tails
bromodomain ↔ acetylated histone tails (Bind to acetylated tails)
chromodomain-TUDOR domains, PHD (plant homeodomain) fingers ↔ methylated histone tails
SANT domain ↔ unmodified histone tails
in many cases, proteins containing these domains recognize a single modified site of a specific histone component
some proteins include many of these domains → specialized for recognizing multiply-modified histone tails
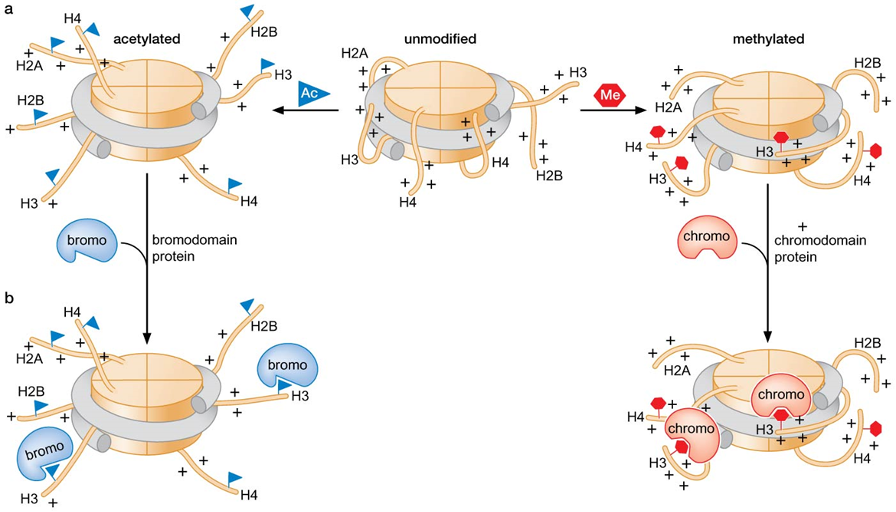
Alteration of function by domains that recognize histone tail modifications
modified histones can recruit enzymes that will further modify adjacent histones
Example: histone acetyltransferase (: includes bromodomains) → can modify histones in nucleosomes adjacent to acetylated histone
modified histones can recruit proteins that act on DNA
Example: HP1 contains chromodomain → becomes recruited to methylated Lys9 on H3 → initiates heterochromatin formation
many nucleosome-remodeling complexes and transcription regulators include domains that recognize histone tail modifications
Example: TFIID contains bromodomain → transcription machinery is directed to sites of histone acetylation → increased transcriptional activity
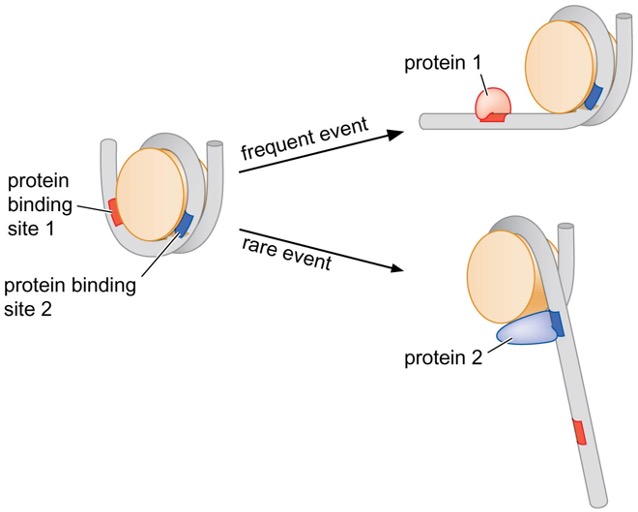
3) Interaction of DNA with the histone octamer is dynamic
nucleosomes exist in various structures
specialized proteins can increase/decrease dynamic association of the histone octamer and DNA → allow changes in DNA accessibility
many DNA-binding proteins can recognize and bind to DNA only when DNA is exposed by transient unwrapping from the histone octamer
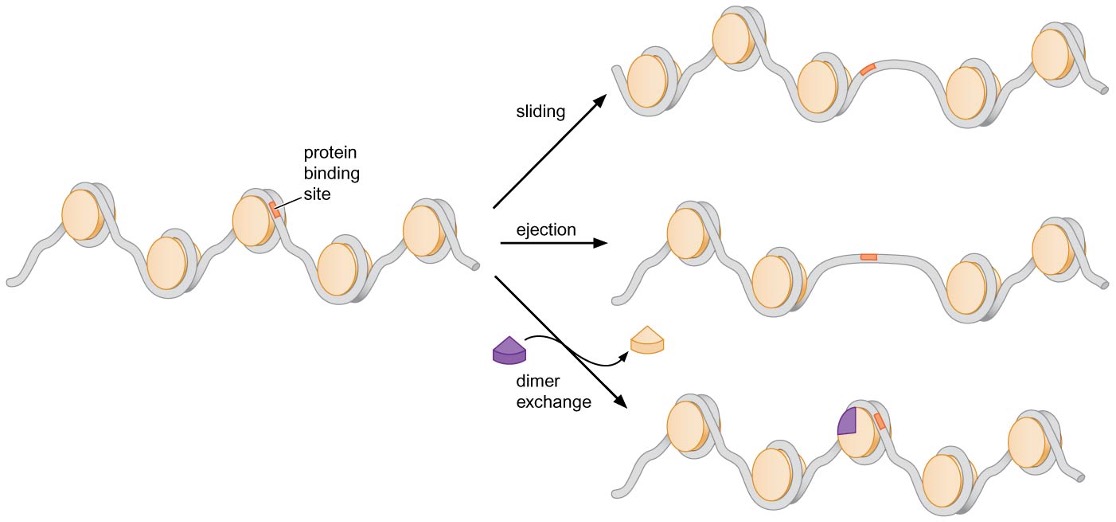
4) Nucleosome-remodeling complexes facilitate nucleosome movement
Nucleosome-remodeling complexes facilitate changes in nucleosome location or interaction with DNA using energy of ATP hydrolysis
Changes mediated by nucleosome-remodeling complexes
Sliding of DNA along surface of histone octamer ~ catalyzed by all nucleosome-remodeling complexes
Transfer / ejection of histone octamer from one DNA helix to another
Exchange of H2A∙H2B dimer with variants of the dimer
Model for nucleosomal DNA sliding
DNA translocating domain of ATP- hydrolyzing subunit binds nucleosomal DNA while other subunits bind tightly to histones
→ ATP-dependent DNA translocating activity breaks 5 histone-DNA contacts while pulling DNA from the nearest linker domain into the nucleosome and creates a loop of DNA
→ broken contacts re-form with translocated DNA and loop disrupts another histone-DNA contact
→ loop moves like a wave across the surface of histones, breaking contacts until all contacts have re-formed with DNA between them
nucleosome-remodeling complexes differ in any given cell, but each complex contains a related ATP-hydrolyzing subunit that catalyzes DNA movement
other subunits associated with each complex can modulate their function
some complexes contain subunits that bind to chemically modified histones
Targeting of nucleosome remodeling complexes
nucleosome remodeling complexes can be targeted to specific sites by interaction with other proteins → can become recruited to a precise location on DNA where needed
nucleosome remodeling enzymes that contain specialized domains that bind to histone tail modifications may become targeted to specific sites and alter DNA accessibility
5. Nucleosome Assembly
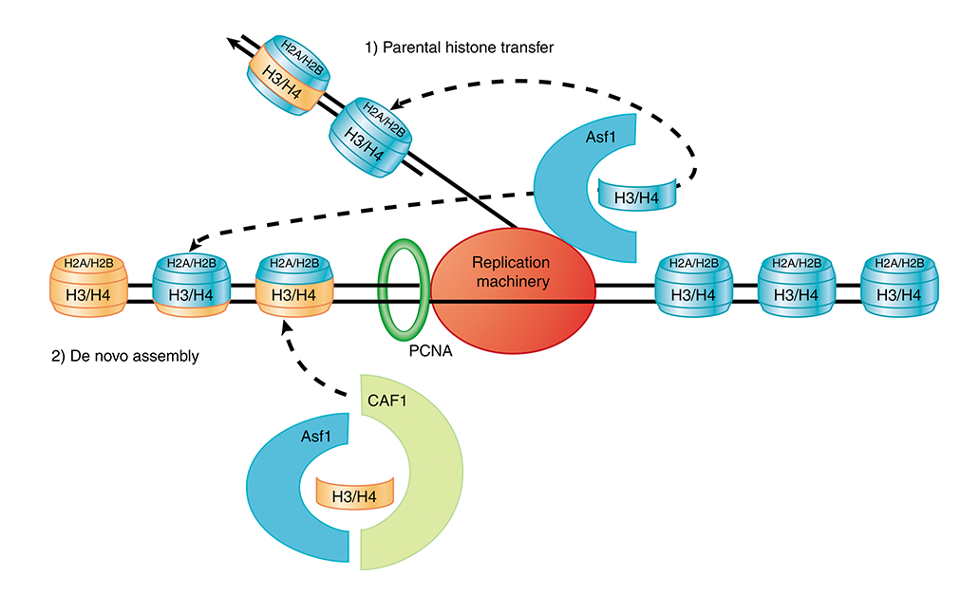
1) Nucleosomes are assembled immediately after DNA replication
old histones tend to rebind one of the daughter chromosomes at a position near their previous position on the parental chromosome → localized inheritance of modified histones ensures that a subset of modified histones is located in similar positions on each daughter chromosome
H3·H4 tetramers appear to remain bound to one of 2 daughter DNA duplexes (not released into free pool)
H2A·H2B dimers are released and enter free pool of histones
modification is further maintained by recruitment of enzymes that add similar modification to adjacent nucleosomes
2) Histone chaperones
replicated DNA is immediately incorporated into nucleosomes → accessory proteins assist histone to associate with DNA, acting as ‘histone chaperones’
CAF-1 (chromatin assembly factor-1) binds to newly synthesized H3 and H4, and is recruited to PCNA (sliding clamp)
ASF1 (anti-silencing function 1) interacts with helicase and may be involved in transferring parental nucleosomes to newly synthesized DNA
CAF-1 and ASF1 also appear to interact with each other
Model for histone assembly/disassembly during DNA replication
DNA replication displaces histone octamers into H3∙H4 tetramers and H2A∙H2B dimers
→ H2A∙H2B dimers enter free pool of histones,
H3∙H4 tetramers are assisted by CAF-1 or ASF1 such that they are directly transferred to daughter DNA
→ two random H2A∙H2B dimers bind to H3∙H4 tetramer
→ nucleosomes assemble ~600 bp behind replication fork
Summary
Within the cell, DNA is organized into large structures called chromosomes. Although the DNA forms the foundations for each chromosome, approximately half of each chromosome is composed of protein. Chromosomes can be either circular or linear; however, each cell has a characteristic number and composition of chromosomes. We now know the sequence of the entire genome of thousands of organisms. These sequences have revealed that the underlying DNA of each organism’s chromosomes is used more or less efficiently to encode proteins. Simple organisms tend to use the majority of DNA to encode proteins; however, more complex organisms use only a small portion of their DNA to encode proteins. The increased complexity of regulatory sequences, the appearance of introns, and the presence of additional regulatory RNAs (e.g., miRNAs) all contribute to the expansion of the non-coding regions of the genomes of more complex organisms. Cells must carefully maintain their complement of chromosomes as they divide. Each chromosome must have DNA elements that direct chromosome maintenance during cell division. All chromosomes must have one or more origins of replication. In eukaryotic cells, centromeres play a critical role in the segregation of chromosomes, and telomeres help to protect and replicate the ends of linear chromosomes. Eukaryotic cells carefully separate the events that duplicate and segregate chromosomes as cell division proceeds. Chromosome segregation can occur in one of two ways. During mitosis, a highly specialized apparatus ensures that one copy of each duplicated chromosome is delivered to each daughter cell. During meiosis, an additional round of chromosome segregation (without DNA replication) reduces the number of chromosomes in the resulting daughter cells by half to generate haploid gametes. The combination of eukaryotic DNA and its associated proteins is referred to as chromatin. The fundamental unit of chromatin is the nucleosome, which is made up of two copies of each of the core histones (H2A, H2B, H3, and H4) and ~ 147 bp of DNA. This protein - DNA complex serves two important functions in the cell: it compacts the DNA to allow it to fit into the nucleus, and it restricts the accessibility of the DNA. This latter function is extensively exploited by cells to regulate many different DNA transactions including gene expression. The atomic structure of the nucleosome shows that the DNA is wrapped about 1.7 times around the outside of a disc-shaped, histone protein core. The interactions between the DNA and the histones are extensive but uniformly base-nonspecific. The nature of these interactions explains both the bending of the DNA around the histone octamer and the ability of virtually all DNA sequences to be incorporated into a nucleosome. This structure also reveals the location of the amino-terminal tails of the histones and their role in directing the path of the DNA around the histones. Once DNA is packaged into nucleosomes, it has the ability to form more complex structures that further compact the DNA. This process is facilitated by a fifth histone called H1. By binding the DNA both within and adjacent to the nucleosome, H1 causes the DNA to wrap more tightly around the octamer. A more compact form of chromatin, the 30-nm fiber, is readily formed by arrays of H1-bound nucleosomes. This structure is more repressive than DNA packaged into nucleosomes alone. The incorporation of DNA into this structure results in a dramatic reduction in its accessibility to the enzymes and proteins involved in the transcription of the DNA. The interaction of the DNA with the histones in the nucleosome is dynamic, allowing DNA-binding proteins intermittent access to the DNA. Nucleosome-remodeling complexes increase the accessibility of DNA incorporated into nucleosomes by increasing the mobility of nucleosomes. Two forms of mobility can be observed: sliding of the histone octamer along the DNA or complete release of the histone octamer from the DNA. In addition, these complexes facilitate the exchange of HA/H2B dimers. Nucleosome-remodeling complexes are recruited to particular regions of the genome to facilitate alterations in chromatin accessibility. A subset of nucleosomes is restricted to fixed sites in the genome and is said to be "positioned." Nucleosome positioning can be directed by DNA-binding proteins or particular DNA sequences. Modification of the histone amino-terminal tails also alters the accessibility of chromatin. The types of modifications include acetylation and methylation of lysines, methylation of arginines, and phosphorylation of serines, threonines, and tyrosines. Acetylation of amino-terminal tails is frequently associated with regions of active gene expression and inhibits the formation of the 30-nm fiber. Histone modifications alter the properties of the nucleosome itself, as well as acting as binding sites for proteins that influence the accessibility of the chromatin. In addition, these modifications recruit enzymes that perform the same modification, leading to similar modification of adjacent nucleosomes and facilitating the stable propagation of regions of modified nucleosomes/chromatin as the chromosomes are duplicated. Nucleosomes are assembled immediately after the DNA is replicated, leaving little time during which the DNA is unpackaged. Assembly involves the function of specialized histone chaperones that escort the H3.H4 tetramers and H2A-H2B dimers to the replication fork. During the replication of the DNA, nucleosomes are transiently disassembled. Histone H3•H4 tetramers and H2A H2B dimers are randomly distributed to one or the other daughter molecule. On average, each new DNA molecule receives half old and half new histones. Thus, both chromosomes inherit modified his-tones that can then act as "seeds" for the similar modification of adjacent histones.