Cycle 6
+ calculate allele and genotype frequencies
Keratinocyte: skin cells, don’t make pigment
Melanocytes: produce pigment in keratinocytes (skin & hair cells)
Melanin: the pigment produced in the melanosomes inside the melanocytes
each melanosome makes one type of melanin (different types of melanin make differencolours, Eumelanin-black, Pheomelanin-red) black & red make brown
melanocytes transfer melanosomes to keratinocytes to give them pigment
Allele Dominance
the B allele makes MC1R always on, therefore it is dominant over both red and brown
even if you have a W allele (sometimes on, sometimes off) the B allele produces too much cAMP therefore the levels never drop low enough to produce pheomelanosomes
the W allele makes MC1R sensitive to ASP which means it turns on and off
when it is on it produces eumelanosomes, and when it is off it produces pheomelanosomes, the build up of both causes the pigment to be brown
the R allele makes the MC1R always off but when heterozygous, the other allele is dominant because it is producing eumelanosomes at least some of the time, this makes the cAMP levels high enough that pheumelanosomes are NOT produced
dominant alleles do not increase in frequency unless they are selected for
If there is selection against a dominant allele, the allele can be “weed”ed out of the population, whereas recessive alleles can hide in the heterozygous individuals
Mendelian Pigs Spotted Coat Colour
the S allele is a mutant version of the B allele that causes MC1R to lose function completely
the S allele is highly unstable, therefore some cells in organisms with SS genotypes mutate to be SB and the B allele causes the MC1R to regain function and create black fur on these cells, therefore the pig will have red and black fur
if the organism is heterozygous for S then it will have a fewer black spots because each cells only has one S allele, therefore half as many chances to get the black mutation
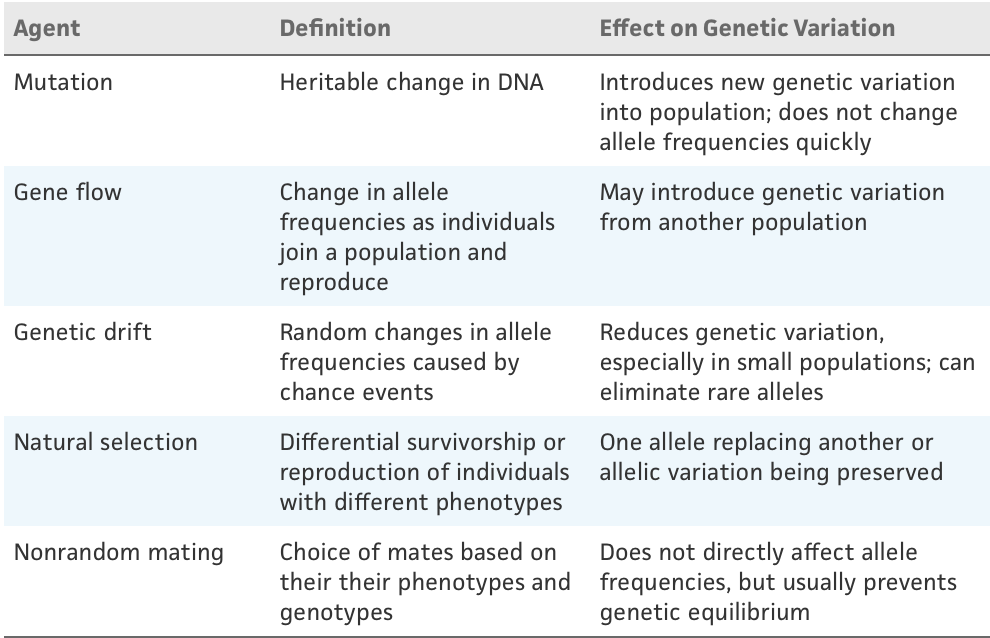
Hardy-Weinberg Equilibrium
if the populations follow the equation p2+2pq+q2 then the population is at equilibrium
Conditions:
No mutations
No migration from other populations.
The population is infinite in size (i.e., there is no genetic drift).
No selection
Random mating
Quantitative Variation: individuals differ in small, incremental ways - continuous spectrum
Qualitative Variation: they exist in two or more discrete states, and intermediate forms are often absent
Phenotypic Variation: caused by genetic differences between individuals, differences in the environmental factors that individuals experience, or by an interaction between an individual’s genetics and the environment
Gene pool: the set of all genes in a population
Genotype frequencies: % of individuals possessing each genotype (p, pq, q)
Allele frequencies: frequency of individual alleles
frequencies stay the same if there isn’t selection
frequency is based on the fitness of the allele
population is evolving if the frequencies are changing
Genetic equilibrium ![]() m: when allele frequencies do not change between generations
m: when allele frequencies do not change between generations
Heterozygote Advantage: both allele frequencies are maintained at an equal amount
WSS < WRS > WRR
Heterozygote Disadvantage: Whichever allele starts off as being more rare will decrease until it reaches 0 frequency, the common allele increases in frequency
Genetic variation is eliminated
WSS > WRS < WRR
Homozygous Advantage: The phenomenon where individuals with two identical copies of a gene have a higher fitness than those with two different copies of the same gene.
Homozygous Disadvantage: High homozygosity rates can also be problematic for a population as they can unmask recessive deleterious alleles generated by mutations, reduce heterozygote advantage, and be detrimental to the survival of small, endangered animal populations.
Genetic Drift
generally leads to the loss of alleles and reduced genetic variability; it therefore causes allele
and genotype frequencies to differ from those predicted by the Hardy–Weinberg model