BIOL 310 - Ch 18 Lecture Notes
Mutations
a change to the genetic code
categories — somatic vs germ-line
types
base substitutions
insertions & deletions
expanding repeats
Somatic vs Germ-line
somatic
every cell that is not a gamete
a portion of the cells are mutant
germ-line
all cells carry the mutation or none of the cells carries mutations
Mutations Types
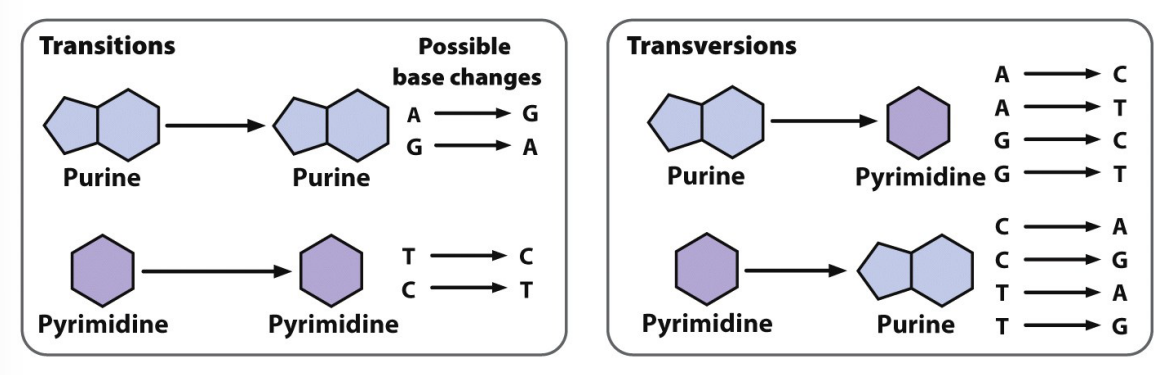
base substitutions
transitions — purine to purine or pyrimidine to pyrimidine
transversions — purine to pyrimidine (vise versa)
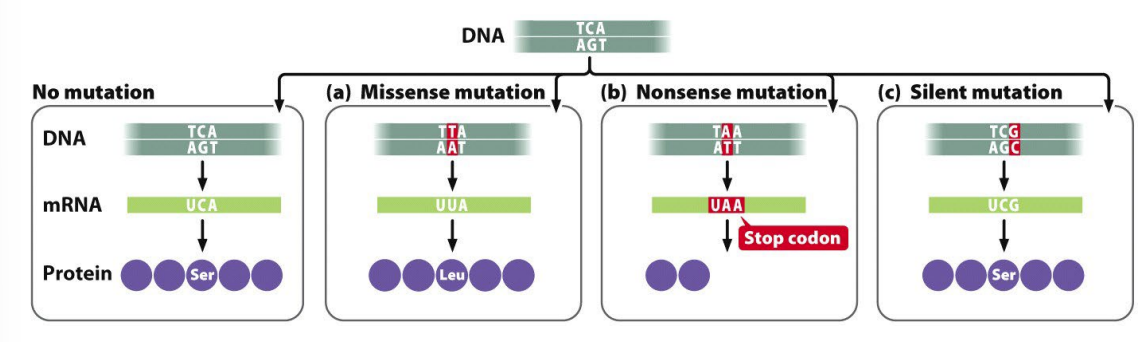
missense mutation - new amino acid replaces
nonsense mutation - stop codon generated
silent mutation - new codon still generates original amino acid
insertions & deletions
loss or gain of codon into genetic code which throws the three base pairs off
expanding nucleotide repeats
DNA polymerase can fall off & the repeats have a tendency to form hairpin loops
when another nuclease attaches onto the hairpin section to elongate — extra five repeats in the loop
next time the strand serves as a template will have thirteen repeats from an original eight
the offspring will also have thirteen repeats
phenotypic effects of mutations
neutral mutation — no effect
loss of function — forward mutation (wild type to mutant state)
gain of function — reverse mutation (mutant to wild type state)
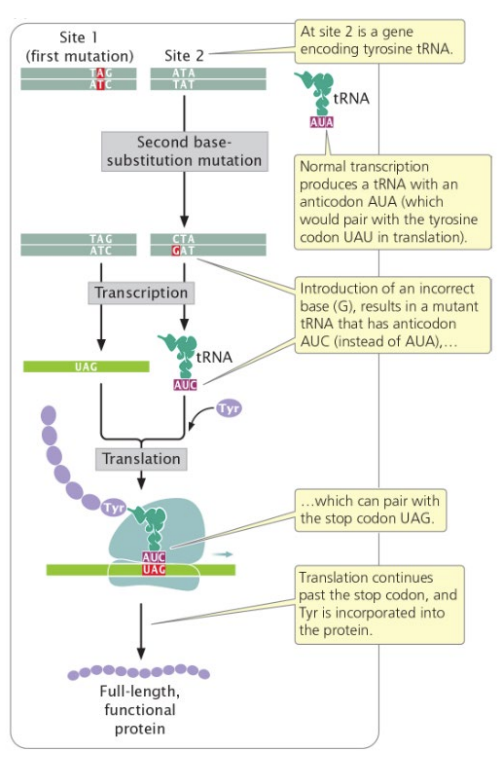
intergenic suppressor
second mutation on second gene reverts phenotype
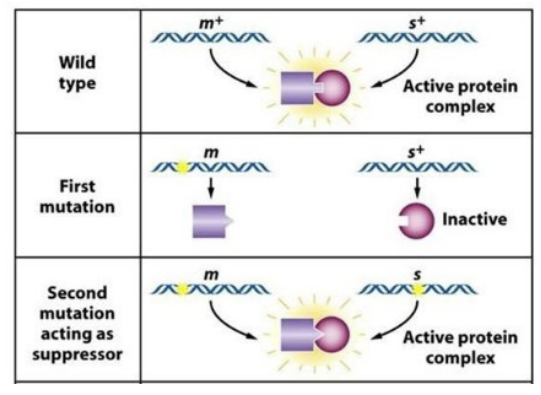
intragenic suppressor
second mutation within the original gene reverts phenotype
mutation rates
mutations per unit (cell division, gamete, genome replicated)
the rate can be “affected” by
probability of a mutation: induced or spontaneous
likelihood of repair
likelihood of detection
generalizations
spontaneous mutations rate is low — 1 to 100 per 10 billion bacterial cells & 1 to 10 per million gametes in eukaryotes
varies by gene & organism
adaptive mutation — some areas of the genome sustain mutation; genetic diversity at that locus can be beneficial (improve at population level)
areas of the genome differ between high or low mutation rate; some areas might be significant to conserve
causes of mutations
what is happening - Bromouracil
spontaneous
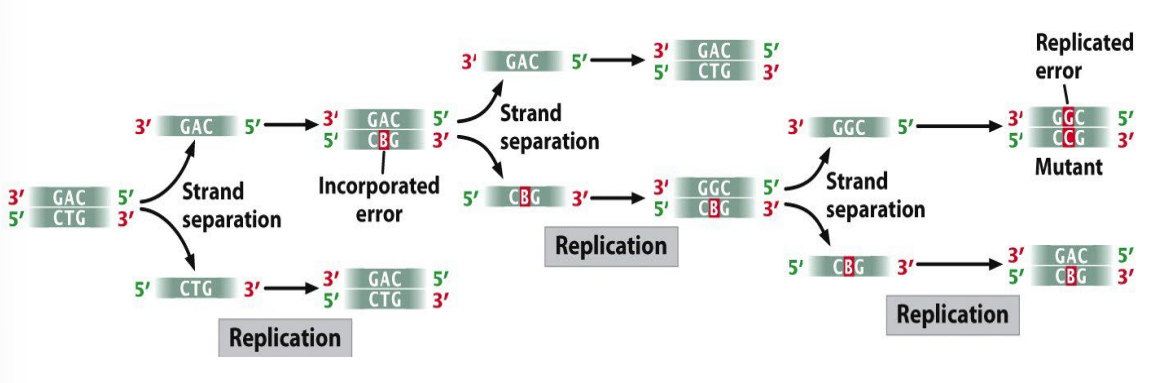
mispairing
replication errors
deletions & insertions — strand slippage
unequal crossover
depurination
deamination
induced
chemically induced
base analogs — bases that look similar to the wild-type bases but have different hydrogen bonding properties
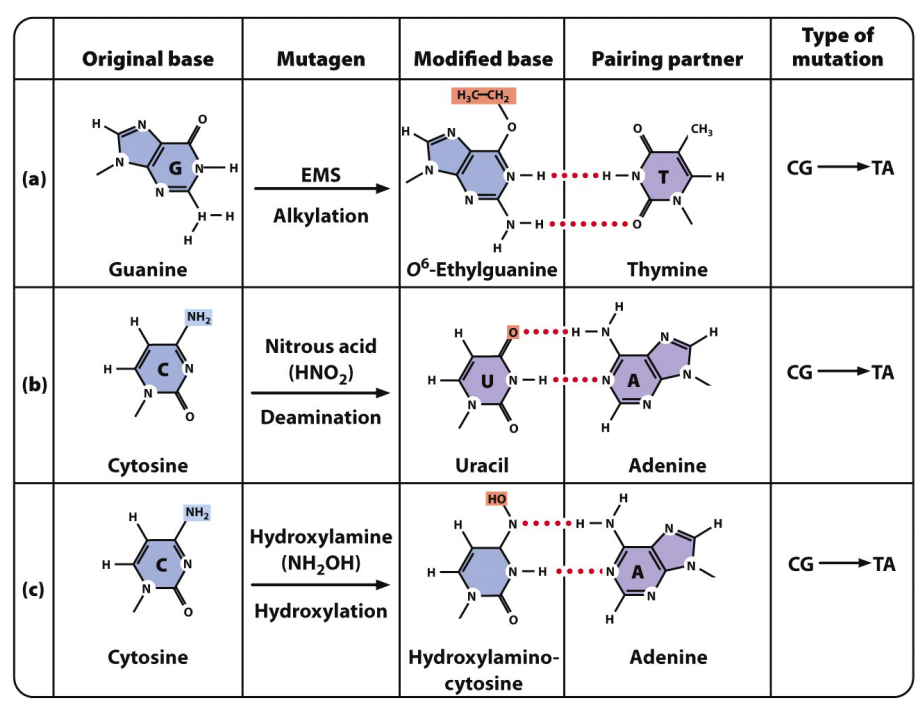
alkylation
altered bases such as CG → TA
deamination
altered bases such as CG → TA
oxidation
hydroxylation
altered bases such as CG → TA
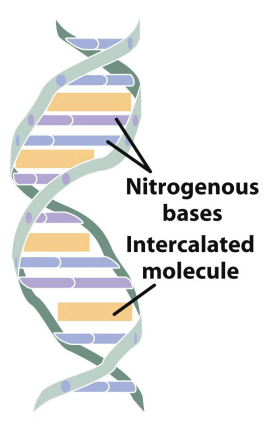
intercalation
dyes that attach themselves to the DNA & act as bases, can pair with wild-type bases
shoved between nitrogenous bases
a type of insertion mutation - frame shift
radiation
ionizing radiation (x-rays)
breaks chromosomes and can cause deletions, duplications, inversions, and translocations
homologous recombination: two copies of chromosome I if one has a break the cell can use the chromosome that is complete to fill in the other with gaps
non-homologous end joining: multiple chromosomes present and both sustain breaks; the cell shoves them together (last ditch effort)
UV — hydrolysis of cytosine: mispairing during replication
thymine dimers block replication & transcription
polymerases stall at the dimers and fall off
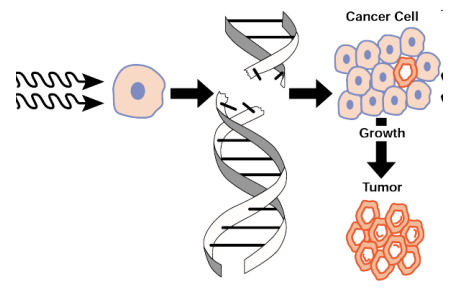
radiation & mutation
cumulative effect
large dose all at once = accumulated dose over time
radiation measured in Roentgen units
determining mutagenicity
the ames test
histidine auxotrophs — bacteria that cannot produce their own supply of histidine due to a frameshift in a gene of Histidine operon
putative mutagen
media with histidine traces for DNA replication by the bacteria culture
treatment plate with more colonies suggest that the chemical added is more mutagenic than the control
radiation & humans
lethal levels are highly damaging
sub-lethal levels
increased somatic mutations
effectively no germline mutations
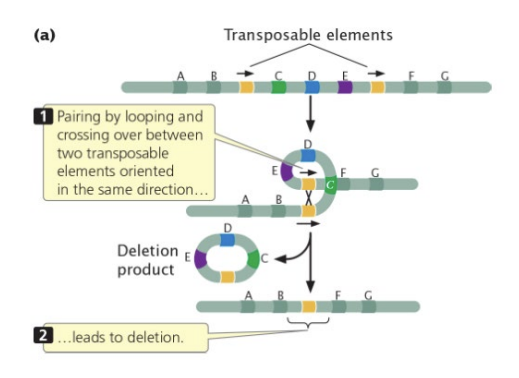
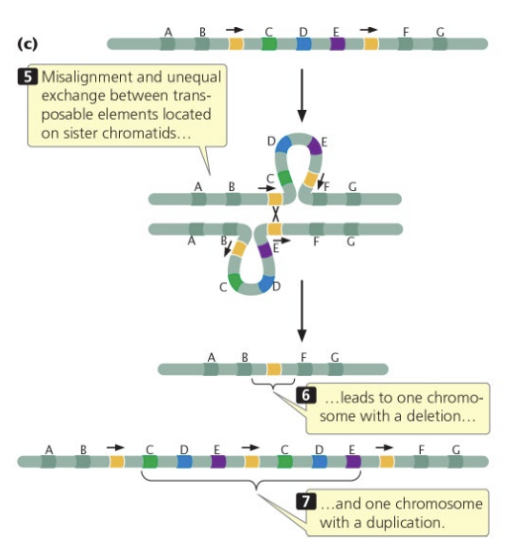
transposable elements
~45% of human genome is transposons
mutation effects
deletions, inversions, duplications
during replication recombination event between different regions of the same chromosome with the same transposon yields deletion product
transposons in inverse orientation (facing towards each other) yields an inversion product
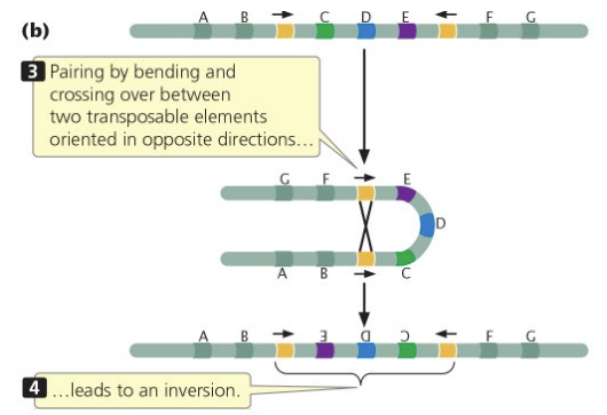
two colinear transposons & two homologous chromosomes have a recombination event to yield one chromosome with a deletion and one with a duplication
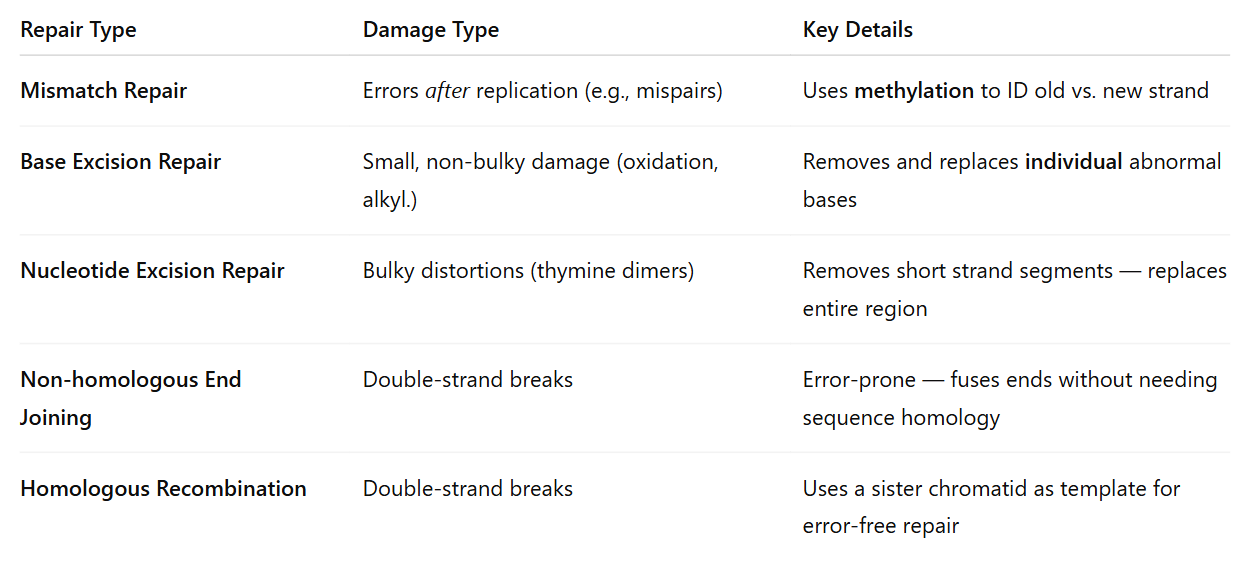
DNA repair mechanisms in E. coli
DNA replication error is 1 error per 10,000 nucleotides
after proofreading it becomes 1 per billion nucleotides
mismatch repair — enzymes that can key in problems to errors not caught by proofreading; recruit enzymes to fix any bumps in the DNA
recruit mismatch repair complex to problem region; fill gap by ligase and polymerase
excision repair
base excision pair — removing abnormal or chemically modified bases
nucleotide excision repair — remove larger defects such as thymine dimers
DNA repair endonuclease — binds to & excises the damaged base(s)
DNA polymerase fills in the gap
DNA ligase seals the nick left by DNA polymerase
nucleotide excision repair
proteins recognize damaged bases
nicks are produced and the ‘damaged’ section is removed
gaps are filled in by polymerase & ligase seals any remaining nicks
faulty repair and genetic disease
failure to repair
increase standing mutation load
increase likelihood of disease phenotype
associated with increase cancer incidence