exam2
Basics of metabolism and energy
Define energy. What are the 2 main types of energy? How do they differ? Which one is “stored” energy (Slide 2)
energy is the ability to do work
kinetic energy
in motion
potential energy
stored energy
List 2 forms of potential energy in cells and how they are released (Slide 3)
Energy in Concentration gradients across membranes.
Energy is released when concentrations equalize on both sides of the membrane
Energy stored in chemical bonds.
Energy is released when bonds break
Give an example of how potential energy can be transformed into kinetic energy in living systems (Slide 4)
The potential energy stored in the chemical bonds of ATP can be transformed into the kinetic energy of muscle movement
Define G or Gibbs Free energy. What is ΔG of a chemical reaction? How is it determined?
(Slide 5)
Gibbs Free Energy (G) = amount of energy available to do work (aka usable energy)
This change in G of a reaction system is depicted by ΔG
ΔG of a reaction R P is given by
ΔG = GP - GR
Free energy
Based on changes in G (i.e., ΔG) of a reaction, be able to determine if a reaction is exergonic or endergonic (Slide 6, 7)
exergonic
If energy is released in a chemical reaction, then
ΔG<0.
Products of these reactions will have less free energy
than the reactants
endergonic
If a chemical reaction requires an input of energy,
then ΔG>0.
Products of these reactions will have more free energy
than the reactants
Which type of reaction (hydrolysis or dehydration biosynthesis) is exergonic? Which is endergonic? (Slide 8)
Dehydration synthesis, that is, builds molecules up – at the expense of energy (endergonic reaction)
hydrolysis breaks molecules apart, liberating energy (exergonic reaction).
Define metabolism. Define “metabolic pathway” and provide 2 examples (Slide 9)
Metabolism refers to the sum total of all chemical reactions of a
cell or organism.
A metabolic pathway is series of biochemical reactions that
converts one or more chemicals into a final product.
for example, energy from the sun is captured during photosynthesis to convert CO2 and H2O into glucose
(C6H12O6). The energy stored in glucose is released during cellular respiration, regenerating CO2 and
H2O. (We will discuss in subsequent lectures
What are the 2 main types of metabolic pathways? Overall, which type is exergonic and which one is endergonic? (Slide 10)
anabolic
endergonic
catabolic
exergonic
How are anabolic and catabolic pathways related? What is the role of ATP in coupling anabolic and catabolic pathways? (Slide 11)
Catabolic pathways provide the energy and bldg. blocks for anabolic
pathways
Anabolic and Catabolic pathways are coupled via ATP
ENZYMES
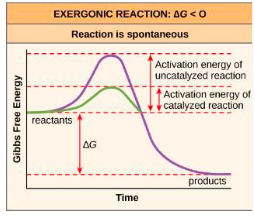
Define “activation energy.” Do exergonic reactions /endergonic reactions /both require activation energy? (Slide 12)
Activation energy is the energy required for a reaction to get started.
Typically, both exergonic and endergonic reactions need an input of activation energy to get started.
Define “transition state.” (Slide 12).
When the reactant absorbs the activation energy and reaches a highly reactive “transition state.” This can then form the product
What are catalysts? How do they speed up chemical reactions? What happens to them once the reaction is completed? (Slide 13)
Catalysts are substances that speed up chemical
reactions by lowering the activation energy requirement
to generate a transition state.
Catalysts are regenerated once the reaction is completed
Know how to interpret a reaction time course chart and recognize the differences between catalyzed and uncatalyzed reactions (Slide 13)
Define “enzyme.” To which category of biological macromolecules do enzymes typically belong? In addition to speeding up reactions, what major feature do enzymes have? (Slide 14)
Enzymes (biological catalysts) are proteins that bind with reactant molecules (aka substrates) and help them reach a transition state with less activation energy
Enzymes are very specific because each enzyme can only bind very specific reactants
Enzymes are regenerated once the reaction is complete
Define “substrate.” Define “active site.” What determines the specificity of enzyme- substrate interactions. (Slide 15)
Reactants of enzyme-catalyzed reactions are called substrates
Enzymes bind very specific substrates
The part of the enzyme where the substrate binds is the enzyme’s “active site.”
The 3D shape of the enzyme and substrate determines this specificity
Interpret the enzymatic mechanism shown in the figure on Slide 15 (what is happening to the substrate here?)
List 3 ways by which enzymes may help the substrate reach a transition state with lower activation energy (Slide 17)
position two substrates so they align perfectly for the reaction
provide an optimal environment, i.e. acidic or polar, within the active site for the reaction
contort the substrate so it is less stable and more likely to react
What is the benefit of enzyme regulation? List 3 ways by which enzymes may be regulated (Slide 18)
Regulation of enzyme activity helps cells control their
environment to meet their specific needs.
For example, digestive cells in your stomach work harder after
a meal than when you sleep.
Enzyme activity can be regulated by
Making more or less of the enzyme
Activators or inhibitors that enhance or inhibit enzyme
function
Adding or removing functional groups to its amino-acid side chains (post-translational modification)
Photosynthesis
Define autotrophs and photoautotrophs. Give 3 examples of photoautotrophs (Slide 1)
Autotrophs are organisms that can make complex, energy-rich
organic molecules (e.g., glucose) from simple molecules such
as CO2.
Photoautotrophs capture light and use it as the energy source
to “fix” carbon into complex organic molecules.
They include
(a) plants, (b) algae, and (c) cyanobacteria
How do heterotrophs get energy? (Slide 2)
Heterotrophs, including animals, fungi, and most bacteria, rely
on the complex, energy-rich organic molecules produced by
autotrophs for their energy needs
What are the components that photosynthesis uses to produce sugars? How do plants obtain each of the components that it needs for photosynthesis? What is the key by-product? How does it exit? (Slides 3)
Photosynthesis uses light energy to produce carbohydrates from carbon dioxide and water.
Oxygen is a by-product.
In land plants:
H2O is absorbed by the roots from
the soil.
CO2 is acquired from the air as a
result of gas exchange through the
stomata (small pores on the leaf
underside).
Oxygen exits through the stomata
What are stomata? What are guard cells? How do guard cells control gas exchange and protect a plant from desiccation (drying out) (Slide 4)
Stomata are pores on the surface of leaves. Each stoma (singular) is
flanked by a pair of guard cells.
Gas exchange occurs through stomata. Water vapor escapes through stomata too
Guard cells can sense and balance the need for CO2 and H2O by
expanding (to open) or contracting (to close) stomata. This helps
prevent desiccation (Drying out)
Memorize the overall reaction of photosynthesis (Slide 6)
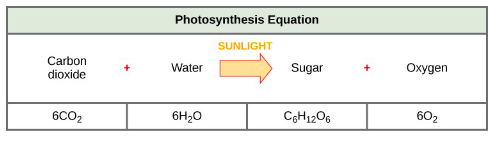
During photosynthesis light energy is converted to
chemical energy.
During photosynthesis, recognize what is being oxidized and what is being reduced. (Slide 6)
Water gives up electrons and is oxidized to oxygen.
CO2 gains electrons and is reduced to sugars.
What type of energy conversion occurs during photosynthesis? (Slide 6)
During photosynthesis, the energy conversion that occurs is from light energy to chemical energy in the form of glucose.
chemical
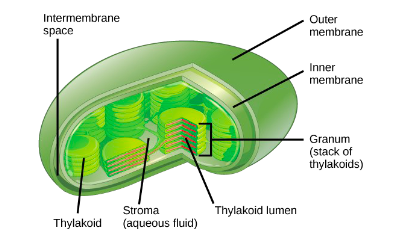
Be able to draw a chloroplasts and label the following parts: Outer membrane, Inner membrane, Stroma, Grana, Thylakoids, Lumen (Slide 7)
What are the 2 stages of photosynthesis? Where in the chloroplasts does each stage occur?
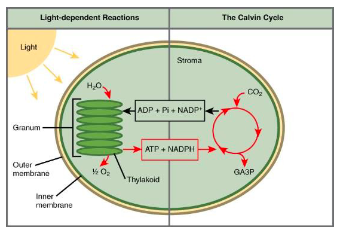
Light-dependent reactions:
Make ATP and NADPH (an e- carrier).It occurs in the thylakoid membranes of chloroplasts.
(2) The Calvin cycle
Uses the ATP and NADPH to make sugar (food)
It occurs in the stroma of chloroplasts.
What happens during each stage? How are the 2 stages connected? (Slide 8)
The light-dependent reactions:
What are the components that conduct light-dependent reactions? Where are they located (Slide 10)?
Located in the thylakoid membranes:
Photosystems II and I: contain chlorophylls, absorb light.
Two Electron Transport Chains (ETC)
Two enzyme complexes – ATP synthase and NADP
reductase
What is the name of the 2 main pigments that capture light? Where are they located? What colors of light are absorbed by chlorophylls? What color is reflected? (Slides 10 and 11)
the main pigments of thylakoid membranes are:
chlorophyll a (a)
chlorophyll b (b)
Chlorophyll a and b capture light for photosynthesis;
reason leaves are green (green wavelengths are reflected)
List the steps of the process of light reactions. What is the role of water in this process? (Slide 12)
PS II absorbs light. It also extracts electrons from water* and energizes them with energy obtained from light.
Energized electrons travel down an electron transport chain to PS I.
This energy is used to make ATP via chemiosmosis** and ATP synthase.
PS I also absorbs light, re-energizes electrons. Electrons travel down another electron transport chain to NADP. This turns NADP into NADPH.
ATP and NADPH will now go to Calvin Cycle
Calvin cycle
What is the relationship between the Light-dependent reactions and the Calvin Cycle (Slide 14)
The ATP and NADPH from the Light-Dependent Reaction are
made/deposited in the stroma for use in the Calvin Cycle.
When the light-dependent reactions make ATP and NADPH, where in the chloroplast are these molecules deposited (Slide 14)?
STROMA
Where in the chloroplast does the Calvin Cycle occur (Slide 14)?
The Calvin Cycle occurs in the stroma of the chloroplast.
For each glucose produced by the Calvin Cycle, how many CO2, ATP and NADPH are required (Slide 14)?
6CO2 ,12 ATP, and 12 NADPH
Detour: Oxidation and Reduction reaction basics
Define “Redox reaction.” (Slide 15)
One reactant is oxidized; other is reduced
What are the hallmarks of becoming oxidized, i.e., to become oxidized is to: ____, _____, _____ (Slide 15 table)
give up electrons or H+, lose potential energy, function as reducing agent
What are the hallmarks of becoming reduced, i.e., to become reduced is to: ____, ____, _____ (Slide 15 table)
gain electron or H+, gain potential energy, function as an oxidizing agent
Know the names of the 3 stages of the Calvin Cycle and the order in which they occur (Slides 16 and 17)
carbon fiaxation
6 CO2 (1-Carbon each) added to 6 RuBP (5 carbons each) by enzyme RUBISCO to generate 12 molecules of 3-PGA (3 carbons each)
Reduction- reduction of 3-PGA
All 12 molecules of 3-PGA are reduced to form 12
molecules of GA3P (3 carbons each).
This reduction process uses electrons from 12 NADPH and energy from 12 ATP.
Regeneration - regeneration of RuBP
10 molecules of GA3P (3 carbons each) recombine to regenerate 6 molecules of RuBP (5 carbons each).
Name the two reactants and the product of Carbon Fixation. How many carbons do each one of these molecules have? Per glucose made, how many of each reactant and product are involved? What is the name of the enzyme that catalyzes Carbon Fixation (Slide 17)?
6 CO2 (1-Carbon each) added to 6 RuBP (5
carbons each) by enzyme RUBISCO to generate 12 molecules of
3-PGA (3 carbons each)
Name the main reactant and product of Reduction. How many of each are there per glucose produced? Energy from how many ATPs and electrons from how many NADPH are used up
(Slide 17)?
All 12 molecules of 3-PGA are reduced to form 12
molecules of GA3P (3 carbons each).
1. This reduction process uses electrons from 12 NADPH and
energy from 12 ATP
Name the main reactant and product of Regeneration. How many molecules of the reactant make how many molecules of the product? So, what exactly is regenerated (Slide 17)?
10 molecules of GA3P (3 carbons each) recombine
to regenerate 6 molecules of RuBP (5 carbons each).
Two molecules of GA3P are left over after Regeneration. What happens to them? (Slide 17)
The 2 left over molecules of GA3P (3 carbons each) combine to
form 1 molecule of glucose (6 carbons)
Not a testable objective: Be fascinated by RUBISCO, its flaws, and the possibilities of increasing food production by getting around its flaws? (Slide 18)
Metabolism review and Glucose oxidation
What is the order in which macromolecules are broken down by cells of our body for ATP production (Slide 2)?
First use carbohydrates
Then fats
And finally proteins
What is the most important energy carrier molecule in cells? Why does it need to be made all the time? (Slide 3)
ATP
Cells generally contain enough ATP to sustain from
30 seconds to a few minutes of activity
What is the most important energy storage molecule in cells? What is the main purpose of this stored energy? (Slide 3)
The most important energy storage molecule in cells is ATP (adenosine triphosphate).
The main purpose of this stored energy is to power various cellular processes and activities.
Recall the basic structure of a polysaccharide (e.g., starch, glycogen). What happens when they are ingested through our digestive system? What is the product of polysaccharide digestion? What happens to this product? (Slide 4)
When you eat a polysaccharide:
O-O-O-O-O-O-O (polysaccharide) —> hydrolysis
O O O O O O O (individual glucose monomers) —>
Glucose molecules are absorbed by our cells and
metabolized by oxidation. Energy is released and
ATP is made with the energy.
Memorize the overall aerobic glucose oxidation equation (Slide 5)
Glucose oxidation equation
C6H12O6 + 6 O2 → 6 CO2 + 6 H2O
glucose oxygen carbon dioxide water
What is the overall goal of glucose oxidation (Slide 5)?
What are the 2 types of glucose catabolism? Under what circumstances does each one occur? (Slide 6)
What are the 4 stages of aerobic glucose catabolism? Which stage also occurs in anaerobic fermentation? (Slide 7 with an assist from 6)
In brief, what happens during each of the 4 steps of aerobic glucose catabolism? (Slide 7)
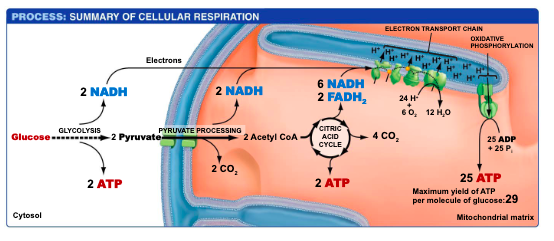
For glycolysis, answer the following questions: (Slides 9 and 10)
a. Where in the cell does it occur?
b. What goes in?
c. What comes out?
d. In which molecules is the released energy carried? How many of these are made?
e. Which molecules carries electrons? How many of these are made?
f. (BANK YOUR ATPs and NADHs).
Review the structure of mitochondria (Know the location of the outer membrane, inner membrane, intermembrane space and mitochondrial matrix. (Slide 11)
Pyruvate processing: (Slides 12, 13 and 14)
a. Where in the eukaryotic cell does it occur?
b. Name the enzyme complex that catalyzes the reactions of Pyruvate processing
c. What goes in?
d. What comes out?
e. In which molecules are the released electrons carried? How many of these are made in total?
f. ADD these them to your BANK.
Citric Acid cycle (Slides 16, 17, 18, 19):
a. Where in the cell does it occur?
The citric acid cycle occurs in the mitochondria of the cell.
b. What goes in?
2 Acetyl CoA + 2 oxaloacetate
c. What comes out?
4 CO2 + 2 oxaloacetate + 2 CoA
d. In what molecules is the released energy stored (BANK YOUR ATPs NADHs, and FADH2’s ).
The released energy in the citric acid cycle is stored in ATP, NADH, and FADH2 molecules.
e. Why is it called a “cycle”? What is the name of the molecule that is regenerated?
The citric acid cycle is called a "cycle" because it is a series of chemical reactions that regenerate citric acid. The molecule that is regenerated is oxaloacetate.
Summarize what happened to the glucose molecule that came into cellular respiration and the gain of ATP and electron carriers (Slide 20)
The glucose molecule undergoes glycolysis, producing pyruvate. Pyruvate enters the citric acid cycle in the mitochondria, generating ATP and electron carriers NADH and FADH2. These carriers then enter the electron transport chain to produce more ATP through oxidative phosphorylation.
Each molecule of glucose is completely oxidized to 6 CO2 , which are expelled when you exhale
4 ATP—directly used as fuel
Oxidative phosphorylation (Electron transport chain, Chemiosmosis and ATP synthesis):
a. What are the 3 processes that comprise oxidative phosphorylation? (Slide 21)
Electron Transport Chain
Chemiosmosis
ATP synthesis
b. Where in the cell does it occur? (Slide 21)
inner mitochondrial membrane
c. In what form is most of glucose’s original energy contained?(Slide 22)
contained in the electrons-carriers NADH and FADH2
d. What happens to the electrons contained in the electron carriers? (Slide 22)
ultimately
transferred to oxygen to form water
e. What is the role of oxygen in this process? (Slide 22)
FINAL ELECTRON ACCEPTOR.
f. What is the released energy used for? (Slide 22)
used to make a large number of ATP
What is the electron transport chain and what does it do? How many protein complexes constitute the electron transport chain? Where are they located? (Slides 23 and 24)
The proteins that transfer electrons from NADH and
FADH2 to oxygen are called the electron transport
chain (ETC)
The proteins of ETC are organized into four protein
complexes
Called complexes I, II, III and IV
located : inner mitochondrial membrane
Know the steps of the entire electron transport process (Slides 24 and 25).
Electrons are removed from NADH and FADH2
These electrons move through the complexes, and eventually end up in oxygen to form water.
Energy is released during this electron movement
Released energy is used to pump H+ (hydrogen ions aka protons) across the inner membrane into the intermembrane space
Forms a strong concentration gradient
Most of the chemical energy from glucose is now accounted for by a proton concentration gradient
At the end of electron transport, where is glucose’s energy stored? (Slide 25).
Glucose's energy is stored in ATP molecules at the end of electron transport in the mitochondria during cellular respiration.
What is ATP synthase? (Slide 26)
the enzyme that synthesizes ATP
What is chemiosmosis? Besides mitochondria, where else does chemiosmosis occur? (Slide 26)
The movement of hydrogen ions down its concentration gradient through ATP synthase.
light-dependent reactions when electrons move from PS II to PS I
Be able to connect how the energy from the H+ concentration gradient is used to make ATP (chemiosmosis and ATP synthesis) (Slides 26 and 27)
The energy from the H+ concentration gradient is used in chemiosmosis to drive ATP synthesis in the mitochondria during cellular respiration.
Summarize oxidative phosphorylation (Slide 28)
Oxidative phosphorylation: The entire process of
oxidizing NADH and FADH2 to gain energy, and use
that energy to make ATP
Summarize the entire process of cellular respiration of glucose (Slide 29)
Glycolysis: Glucose, a 6-carbon molecule, is broken down into two molecules of pyruvate, a 3-carbon molecule. This process occurs in the cytoplasm and produces a small amount of ATP and NADH (nicotinamide adenine dinucleotide).
Citric Acid Cycle/ Krebs Cycle: Pyruvate is transported into the mitochondria and converted into acetyl-CoA. Acetyl-CoA enters the Krebs cycle, a series of reactions that oxidize acetyl-CoA to produce NADH, FADH2 (flavin adenine dinucleotide), and some ATP. Carbon dioxide is released as a byproduct.
Oxidative Phosphorylation: NADH and FADH2 generated in glycolysis and the Krebs cycle donate electrons to the electron transport chain located in the inner mitochondrial membrane. As electrons pass through the chain, they release energy, which is used to pump protons (H+) across the membrane, creating a proton gradient. This gradient drives ATP synthesis as protons flow back into the mitochondrial matrix through ATP synthase, a process called chemiosmosis. Oxygen serves as the final electron acceptor, forming water when it combines with electrons and protons.
Attribute Aerobic respiration Photosynthesis
Occurs in cells of
heterotrophs YES NO
(Yes or No)
Occurs in cells of
photoautotrophs YES YES
(Yes or No)
Overall reaction per
glucose Glucose (C6H12O6) + 6O2 → 6CO2 + 6H2O + Energy (as ATP)
6 CO2 + 6 H2O + light energy → C6H12O6 + 6 O2
Stages Glycolysis Light-Dependent Reactions:
Pyruvate Oxidation Light-Independent Reactions (Calvin Cycle):
Citric Acid Cycle (Krebs Cycle)
Electron Transport Chain
and Oxidative Phosphorylation:
Organelle and/or cell
region where each
stage occurs
Glycolysis:
Location: Cytoplasm (cytosol)
Pyruvate Oxidation:
Location: Mitochondrial matrix (inside the mitochondria)
Citric Acid Cycle (Krebs Cycle):
Location: Mitochondrial matrix (inside the mitochondria)
Electron Transport Chain and Oxidative Phosphorylation:
Location: Inner mitochondrial membrane and mitochondrial matrix (inner membrane for the electron transport chain, matrix for oxidative phosphorylation)
Light-Dependent Reactions:
Location: Thylakoid membrane within chloroplasts
Light-Independent Reactions (Calvin Cycle):
Location: Stroma of chloroplasts
Is oxidation of glucose
happening (Yes / No)?
yes
no
Is reduction of CO2
happening?
(Yes / No)
no
yes
Role of water
Electron Transport Chain (ETC)
Oxygen Transport
Maintaining Cellular Homeostasis
Source of Electrons
Oxygen Production
Generation of Protons (H+)
Stabilization of Reaction Centers
Role of Oxygen
Final Electron Acceptor
Maintenance of Proton Gradient
Regeneration of Electron Carriers
Byproduct of Water Splitting
Supports Aerobic Respiration
Contributes to Atmospheric Oxygen
Electron carriers
NAD+ (Nicotinamide adenine dinucleotide)
FAD (Flavin adenine dinucleotide
Coenzyme Q (Ubiquinone)
Cytochromes
NADP+ (Nicotinamide adenine dinucleotide phosphate)
NADPH (Reduced Nicotinamide adenine dinucleotide phosphate)
ATP (Adenosine triphosphate)
DNA structure:
Name the 2 types of nucleic acids, and their overall functions (Slide 1),
DNA (deoxyribonucleic acid)
DNA codes for the genome of cell - entire genetic content
RNA (ribonucleic acid)
RNA is primarily involved in protein synthesis
What are the building blocks of DNA called? (Slide 2).
deoxyribonucleotides
What are the parts of each deoxyribonucleotide? Which ones are common to all 4 deoxyribonucleotides? Which one is different among the 4? (Slide 2).
Phosphate Group (PO4): This is a phosphorus atom bonded to four oxygen atoms, forming a negatively charged phosphate group. It is attached to the 5' carbon of the sugar molecule.
Deoxyribose Sugar: Deoxyribose is a five-carbon sugar that forms the backbone of DNA. It has a hydrogen atom (-H) attached to the 2' carbon instead of a hydroxyl group (-OH), which distinguishes it from ribose, the sugar found in RNA.
Nitrogenous Base: The nitrogenous base is attached to the 1' carbon of the deoxyribose sugar. There are four types of nitrogenous bases found in DNA:
Adenine (A)
Guanine (G)
Cytosine (C)
Thymine (T) (this one is diff and can be replaced w U uracil in RNA)
(All Slide 3)
What is the natural structure of DNA?
double helix - made of 2 polynucleotide strands
Name the bond that holds deoxyribonucleotides within each polynucleotide strand of a DNA double helix.
phosphodiester bonds
Name the bond that is formed between the 2 polynucleotide strands of the double-helix? Which specific part of the deoxyribonucleotides are involved in this bonding?
phosphodiester bonds
The 2 strands connected by hydrogen bonds between specific nucleotides (A-T, G-C). This is also called complementary base-pairing
What is the meaning of complementary base-pairing?
a-t ; g-c
What is the meaning of anti-parallel orientation of the polynucleotide strands?
Strands are also anti-parallel.
One goes in 5’ 3’, other in 3’ 5’
What were the 2 contributions of Erwin Chargaff to figuring out DNA structure? What is Chargaff’s rule? (Slide 4)
the amounts of adenine and thymine in DNA were roughly the same, as were the amounts of cytosine and guanine.
% adenine = % thymine
% cytosine = % guanine
(Activity) Observe the DNA double helix on Slide 6. Complete the following:
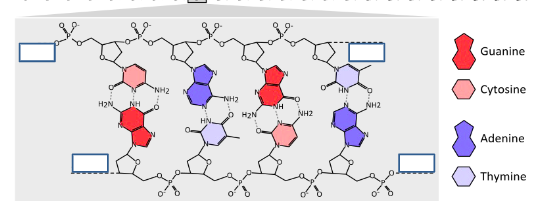
a. Write the sequence of the top strand of the double helix. Be sure to include direction (5’, 3’)
5’ c a g t 3’
b. Write the sequence of the bottom strand of the double helix. . Be sure to include direction (5’, 3’)
3’ g t c a 5’
c. Label the 5’ and 3’ ends correctly in the boxes next to each strand on the picture
d. Label the phosphodiester linkages
e. Label the hydrogen bonds that hold the base-pairs together.
f. Label the purines and pyrimidines.
Purines: Adenine, Guanine
Pyrimidines: Cytosine, Thymine, Uracil
Why is it advantageous to have all base pairs to be purine – pyrimidine pairs (Slides 6 and 7)?
it allows for equal distance between the 2 strands
Which base-pair (A-T or G-C) is harder to break up via heat? Why? (Slides 6 and 7)
A-T base pairs are linked by 2 H-bonds each, G-C base pairs
are linked by 3 H-bonds. Thus G-C pairs are harder to break
up
What are the parts of each ribonucleotide? Which ones are common to all 4 ribonucleotides? Which one is different among the 4? (Slide 8).
Ribonucleotides consist of a phosphate group, a ribose sugar, and a nitrogenous base.
The phosphate group and ribose sugar are common to all 4 ribonucleotides.
The nitrogenous base is the part that differs among the 4 ribonucleotides.
Slide 9) What is the natural structure of RNA? Name the bond that holds ribonucleotides together within the polynucleotide strand?
How are secondary RNA structures formed sometimes? What are the base-pairing rules here?Can RNA base-pair with DNA?
Natural structure of RNA is a single polynucleotide strand
phosphodiester bonds
Sometimes, there is internal base-pairing (A- U and C-G) within an RNA strand, creating secondary structures.
RNA can also base-pair with DNA
DNA replication:
What is DNA replication? Why does DNA need to be replicated? When is DNA replicated? (Slide 1)
What is DNA replication?
• The doubling of one DNA double-helix into 2 double-helices
Why does DNA need to be replicated?
• Cell division
• Development
• Repair
• Replacement
When is DNA replicated?
• Just before cell division
What are the 3 possible models of DNA replication? How would the newly replicated DNA look in each of the 3 models? (Slide 2)
Conservative replication – original parent double strands remain intact and two completely new double helix strands are synthesized
Semi-conservative replication– the two strands come apart – each acts as a template for synthesis of a new complementary strand.
Dispersive replication– both original parent strands are broken up into small pieces and incorporated into newly synthesized strands

How did Meselson and Stahl experimentally determine the model of DNA replication?(Slides 3 – 5)
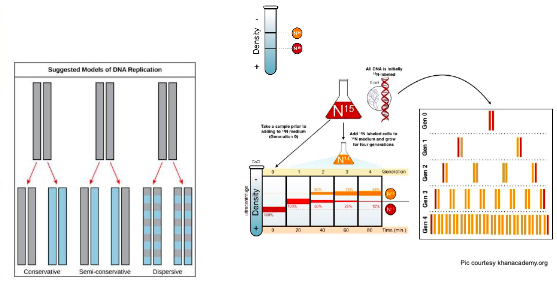
Meselson and Stahl experimented with E. coli grown first
in heavy nitrogen (15N) then in 14N.
DNA grown in 15N (red band) is heavier than DNA grown
in 14N (orange band), and sediments to a lower level in a
centrifuge.
When DNA grown in 15N is switched to media containing
14N, after one round of cell division the DNA sediments
halfway between the 15N and 14N levels, indicating that it
now contains fifty percent 14N.
In subsequent cell divisions, an increasing amount of
DNA contains 14N only. This data supports the semi-
conservative replication model
Key points to keep track of
- Growing bacteria with 15N and 14N nitrogen
- Extracting DNA
- Centrifuging DNA
- Observing relative positions of 15N DNA and 14N DNA in centrifuge
- Based on the relative positions, concluding what the model of DNA replication was?
Activity: If DNA replication was conservative, what would the band pattern look like after 1, 2 and 3 generations?
List the 3 steps in the process of DNA replication (Slide 6)
Separation of the strands of the helix
Synthesis of the new strand
Removing primers and tying the segments together
How are the strands separated? Name the enzyme that is responsible for this (Slide 7)
The DNA strands are separated during replication by the enzyme helicase.
Break hydrogen bonds that hold the 2 strands together
Replication bubbles are created.
How is the new DNA synthesized? Name the 2 enzymes that are most responsible for this step. What does each enzyme do? (Slide 8)
Primase and DNA polymerase III
Primase lays down the RNA primer
DNA Pol III bring the correct nucleotide by using each strand as a “template.”
DNA Pol III then creates the new phosphodiester linkages to hold the adjacent nucleotides together.
DNA Pol III can only place new nucleotides in the 5’ 3’ direction.
In what direction are nucleotides placed during DNA replication? (Slide 8)
During DNA replication, nucleotides are added in the 5' to 3' direction on the growing DNA strand.
Why is one strand replicated continuously and the other strand replicated in fragments (discontinuously) (Slides 9 and 10).
the newly synthesized strand can only grow in ITS 5’ 3’
direction
Therefore, as the replication fork opens, one strand can be
replicated by continuous growth. The other strand has to be
made in the opposite direction, in segments
What happens during the third step? Name the enzymes responsible for this step? What does each enzyme do? (Slide 11)
DNA pol I and DNA ligase
• Break down RNA primer, replace with DNA nucleotides
• Ties the segments together
10. Be able to summarize the process of DNA replication (Slide 12)
A replication fork is formed when helicase separates the DNA
strands at the origin of replication.
Primase synthesizes an RNA primer. DNA polymerase III uses
this primer to synthesize the daughter DNA strand.
DNA polymerase I replaces the RNA primer with DNA.
DNA ligase seals the gaps between the fragments, joining the
fragments into a single DNA molecule.