Genetic Engineering Step 1 Study Guide
What characteristics make plasmids easy to genetically modify?
- Plasmids have short sequences of DNA
- Plasmids are easy to cut and paste back together
- Plasmids copy independently
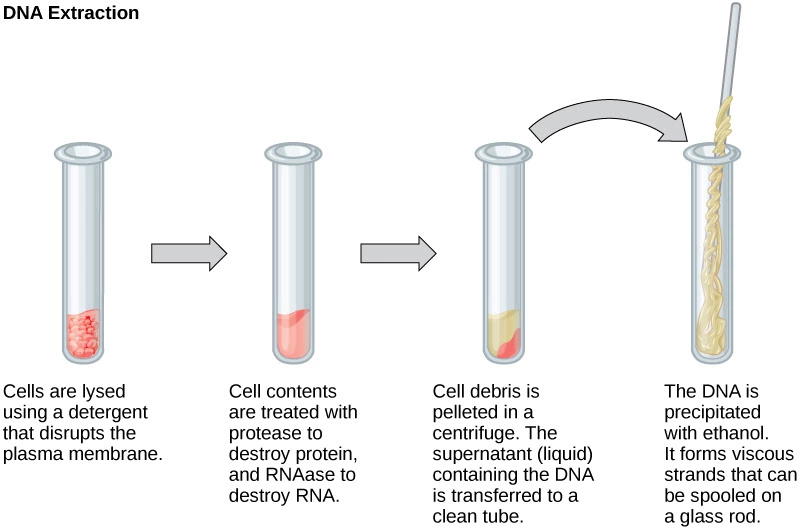
Explain the process and all reagents used to extract DNA.
- Lysozyme is added degrade peptidoglycan in cell walls
- We don’t use lysozyme because we have animal cells
- Detergents are added to lyse (explode) the cell
- Detergents include sarkosyl or sodium dodecyl sulfate (SDS)
- Proteases and salts are added to clump debris
The mix is then centrifuged to create a precipitate and a supernate
The precipitate is at the bottom and is debris
The supernate is at the top and has the DNA
DNA can be extracted with alcohol
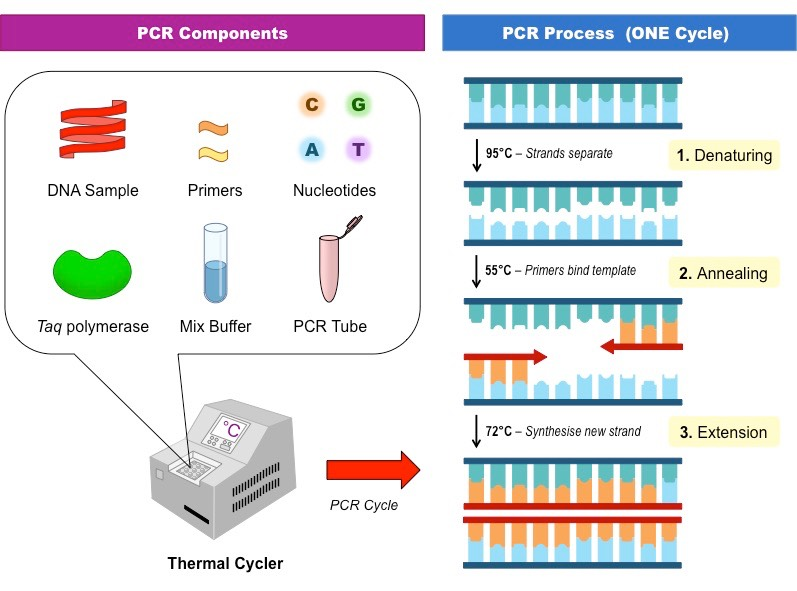
Explain how PCR and Gel Electrophoresis are used in genetic engineering.
PCR- Stands for Polymerase Chain Reaction, used to copy DNA many times
PCR uses primers which target specific sequences in DNA
DNA strands are split apart at 95°
Then primers bind to the split strands at 50°
The polymerase enzyme makes a complementary strand using free nucleotides at 72°
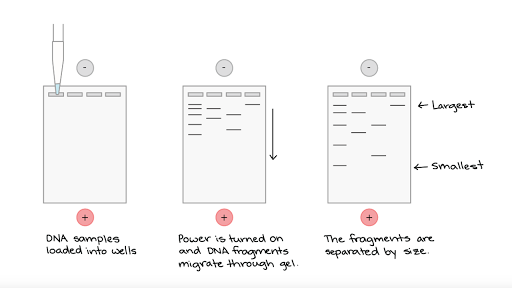
Gel Electrophoresis- Separates DNA fragments based on size
- There is a gel made of agarose that has wells (holes) in it
- DNA is placed into the wells
- An electric charge is run through the gel
- DNA is negatively charged and it will move to the positively charged side
- Smaller fragments will move faster so they will go further
 Vocabulary:
Vocabulary:
genetic engineering- manipulation of genetic information
multiple cloning sites- series of unique restriction enzyme recognition sites
vectors- Vessel to carry genetic information with (often a plasmid)
lysosome- degrades peptidoglycan in cell walls
precipitant- molecular debris, found at bottom of test tube
supernatant- suspends DNA, found at top of test tube
lysed- to explode cells
detergent- Solutions to lyse cells with
protease- Degrades proteins
salts- clumps cellular debris
probes- single-strands of DNA or RNA that bind to a complementary sequence that have a biological marker attached. Shows location of DNA sequences.
hybridization- Process where probe binds to complementary sequence
restriction enzymes- cuts DNA at specific sites
gel electrophoresis- separates DNA fragments based on size
DNA polymerase- Synthesizes new DNA strands in the PCR process
thermal cycler- Used in PCR to take DNA through temperatures best for replication (95°, 50°, 72°)
polymerase chain reaction- used to replicate specific sequence of DNA many times
- We do not replicate all of the DNA
southern blotting- transfers DNA from gel onto membrane with probes. Uses photographic film to identify specific sequences of DNA
exons- coding sequences that leave the nucleus
introns- non-coding sequences of DNA that cannot leave the nucleus