Bacterial Gene Regulation
Vocabulary:
Gene regulation: The mechanisms and systems that control the expression of genes, can occur at many levels including transcription, mRNA processing/stability, translation, and posttranslational modifications
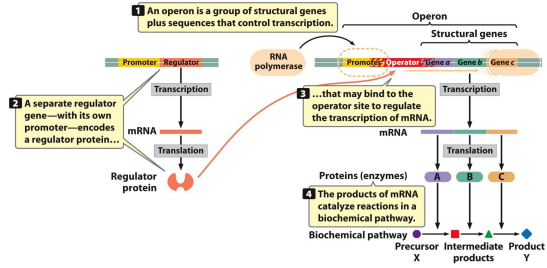
Operon: A group of genes that is transcribed as a unit
Constitutive expression: Involves gene information continually flowing from DNA → protein, proteins are constantly being produced under normal cellular conditions
Structural genes: Encode proteins used for enzymatic purposes of have a structural role in the cell
Regulatory genes: Encode products that interact with other sequences and control the expression or structural or regulatory genes. Typically upstream of the operon
Regulatory elements: Regions of DNA that are used by the products of regulatory genes to regulate gene expression
Positive control: Mechanisms that stimulate gene expression, transcription only occurs when the regulator molecule stimulates RNA production
Negative control: Mechanisms that inhibit gene expression, expressed unless shut off by a regulator molecule
Inducible enzymes: Produced by bacteria to adapt to the environment, present when specific substrates are present
Constitutive enzymes: Enzymes continually produced regardless of environmental conditions
Repressible system: The presence of a specific molecule will inhibit gene expression, abundance of the end product in an environment will repress gene expression
Lactose: Will repress gene activity when absent and induce when present, will cause more enzymes that break down galactose and glucose to be made
Cis-acting elements: Regulatory regions on the same strand as the gene they regulate, typically DNA elements
Trans-acting elements: Molecules that bind to cis-acting sites, typically proteins
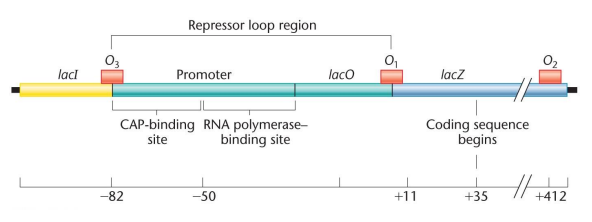
Lac operon: Within the regulation of lactose, this operator overlaps the promoter and the 5’ end of the first structural gene
lacZ: The first structural gene of the lac operon, encodes B-galactosidatse, which converts disaccharide glucose and galactose into monosaccharides
lacY: The second structural gene of the lac operon, specifies the primary structure of permease
lacA: The third structural gene of the lab operon, encodes transacetylase, which aids in removal of toxic byproducts of lactose digestion
LacI: Located close to the lac operon, has its own promoter, produces a repressor molecule which regulates transcription of structural genes
Allosteric repressor: Interacts reversibly with another molecule, causes conformational changes in 3D shapes and change in chemical activity
Cistron: Part of a nucleotide sequence that codes for a single gene
Permease: Part of an operon, actively transports lactose across the cell membrane
Jacob and Monod: Proposed the operon model in 1960, which involved negative control. Thought the lacI gene regulates transcription of structural genes by producing an allosteric repressor molecule
lacP: The operon promoter for lactose
lacO: The operon operator for lactose
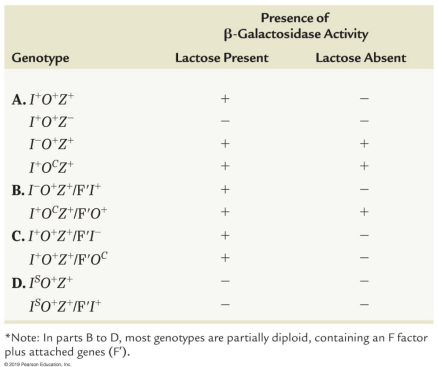
I- mutant: A mutation of the lactose gene where the repressor protein is altered or absent and cannot bind to the operator region, causing structural genes to always be on, recessive and trans acting
P- mutant: A mutation in the promoter of the lactose gene, cis acting and fails to produce functional B-galactosidase
IS mutant: A mutation in the lactose gene where the operon is super-repressed, and the repressor protein that binds to the operator cannot be removed, so it is always off
OC mutant: A mutation of the lactose gene where the nucleotide sequence of the operator DNA is altered and will not bind a normal repressor molecule, so the structural genes are always on, cis acting and constitutive, dominant over wild type
Partial diploid: Full bacterial chromosome plus an extra piece of DNA of an F plasmid
Merozygotes: Allowed researchers to look at the interaction of mutants in both bacterial and plasmid DNA, using different combinations allowed them to determine each component’s role in regulation, and distinguishing cis from trans
Catabolite-activating protein (CAP): Exerts positive control over the lac operon binds to CAP binding site, and facilitates RNA polymerase binding at the promoter and transcription positively
Catabolite repression: Diminishes expression of the operon and represses the metabolites of all other sugars when glucose is available
Adenyl cyclase: Activated on the cell-surface when glucose levels drop, cleaves 2 phosphates from ATP and reconnects the free end back onto the molecule, making a loop
Cyclic AMP (cAMP): Released by adenyl cyclase, spreads through the cell and stimulates the production of enzymes that produce food molecules, inversely proportional to the level of available glucose
Glucose: The preferred source of energy for bacteria, inhibits CAP binding, inhibits activity of adenylyl cyclase which turns ATP → cAMP
Crystal structure analysis: Determines the crystalline structure of lac repressors bound to inducer/operator DNA
Repression loop: The binding of repressor to operators O1 and O3, prevents access of RNA polymerase to promoter
Tryptophan (trp) operon: A repressible gene system, produces enzymes necessary for the synthesis of tryptophan, has 5 contiguous subunits E-A, O operator, and P promoter
Attenuation: Trp structural genes are preceded by a leader sequence, which contains this regulatory site
Antiterminator hairpin: A structure created by RNA in the absence of tryptophan, stalls the ribosome
Terminator hairpin: A structure created by RNA in the presence of tryptophan, doesn’t stall a ribosome
Leader region: IN the trp gene, contains 2 trp codons
Riboswitches: Gene regulation done by attenuation using alternative forms of mRNA secondary structure, may bind a small molecule of cause a conformational change, creates antiterminator or terminator structures
Aptamer: The part of the riboswitch that binds to a ligand
Expression platform: The part of a riboswitch that can form a terminator structure
For maximum transcription levels, the repressor must be bound to by lactose so operon expression isn’t being repressed, and CAP must bind to the CAP binding site
Lactose is an inducible system, having lactose present increases transcription of the gene