L5: Protein Folding Landscapes, Kinetics, And Misfolding
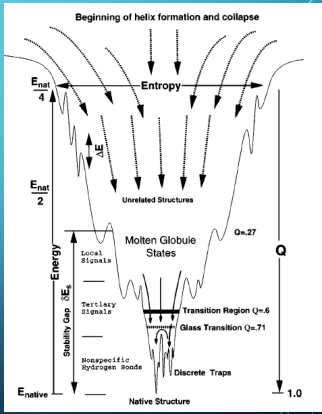
Q = fraction of protein folding
horizontal = entropy - number of possible states
slopes - steeper towards N - incentivises proetin to ‘leave’ kinetic traps/transition states
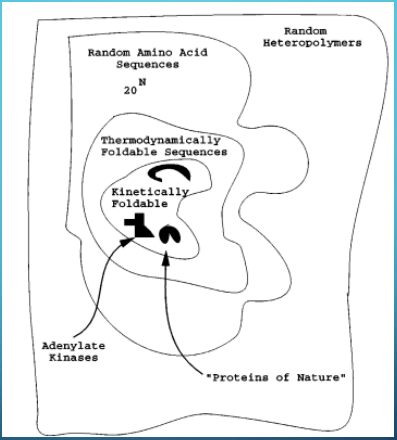
Why do Statistical Distributions Matter in Folding?
Proteins fold stochastically due to molecular fluctuations.
always fluctuation in the system
cooperativity
Different distributions describe different aspects of folding dynamics.
local minima/transition state
free energies
end to end distance
Binomial Distribution - probability of protein being in folded/unfolded state @ equilibrium
Poisson Distribution - rare folding/unfolding events over time, single-molecule kinetics
Gaussian Distribution - distribution of native-state fluctuations and energy variations
Exponential Distribution - waiting times between folding transitions in an energy landscape
Energy Landscapes
probability distributions of folding pathways
diffusion folllows statistical laws
Random walk models - protein confformational exploration
Biased random walk - energy-driven folding
enthalpic contribution
folded vs unfolded - thermodynamic
Mean Squared Displacement
efficiency of exploration
kinetics
‘useless’ pathways/entropic conformations
slope of t vs MSD - lower slope = higher efficiency
Levinthal’s Paradox
100 residue polypeptide; 2 different ɸ and Ψ bond angles; each angle can be in one of 3 stable conformations.
Total conformations - 3200
If the protein tried each conformation in 1 femtosecond (10−15 sec), it would take longer than the age of the universe to find its native structure.
But in reality, proteins fold within milliseconds to minutes!
This paradox suggests that protein folding must be guided rather than purely random.
SOLVED - Anfinsen’s experiment (RNAase)
Folding is not random but hierarchical.
Early-stage interactions limit the search space.
Folding energy barriers control rate-limiting steps.
How does nature solve the paradox?
physical and energetic constraints
Energy Landscapes Guide Folding
biased energy funnel
reduce explorable conformations
restrict movements to lower-energy regions
Directed Search via Local Interactions
Secondary structures (α-helices, β-sheets) - form early, limit search space
Hydrophobic collapse - reduces conformational freedom
local native-like interactions
Parallel pathways
different parts may fold independently before assembling
flexibility
Molecular Chaperones
Hsp70, GroEL, Hsp90
help prevent kinetic traps
stabilise partially folded states
create an isolated environment, preventing aggregation
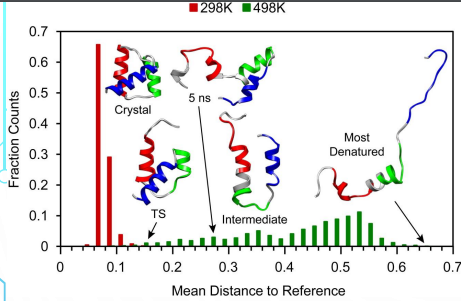
x axis - kinetics
y axis - thermodynamics
Kinetics of Protein Folding & Transitions State Theory
Biochemistry - how far and how fast?
Why study kinetics? Folding time varies from microseconds to minutes, depending on solution conditions
What determines folding speed? Number of residues, number of intermediate conformations, Energy barriers and transition states, Chaperones and cellular crowding effects
Two-State vs. Multi-State Folding
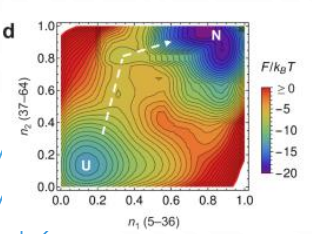
ambient temperature + energetically favourable

larger energy difference = slower folding rate
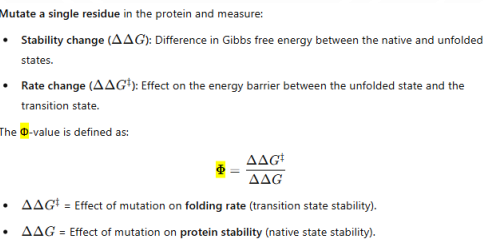
how far? how stable will the resulting strcuture be?

Deep local minima → Kinetic traps → Misfolding risks.
Anfinsen’s experiment
trace amount of beta-mercaptoethanol - dissolves incorrect S-S bonds
 Knowt
Knowt