W3 L4: Cell signalling
Signal transduction:
Principles of signalling:
- For any signal to be received or sent you need the following:
- signal itself - can be protein, short peptide, ion, small molecule e.g. nucleic acid
- means of signal getting to desired destination e.g. systemic circulation
- Something to receive signal - a receptor e.g. tyrosine kinase receptor ~ insulin receptor
- something to interpret signal - second messengers
- Response to signal - outcome for cell e.g. transcription of particular genes or changes in enzyme activity
 Method of interpretation: second messengers
Method of interpretation: second messengers
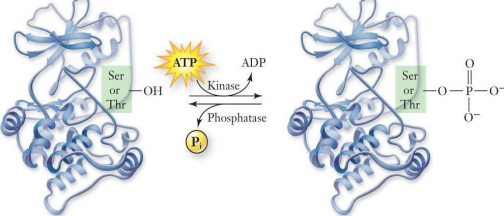
- Proteins (inc. enzymes) are modified
- Modification Δ shape or charge of molecule inc:
- Phosphorylation (add phosphoryl gp to AA → more -vely charged)
- Acetylation (prevents other things activating that protein)
- Methylation (make it bulky & stop something from binding)
- Cleavage (cutting of longer AA seq. to generate activates protein)
- Δ enables either the activation of enzymatic function or recognition by another protein
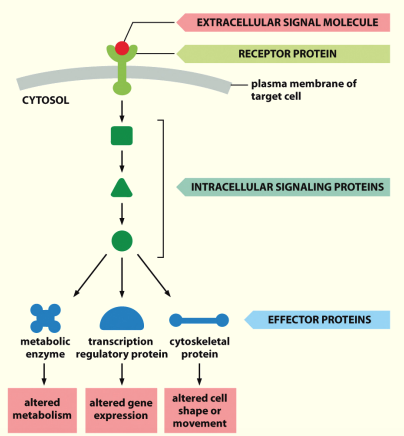
Signal transduction
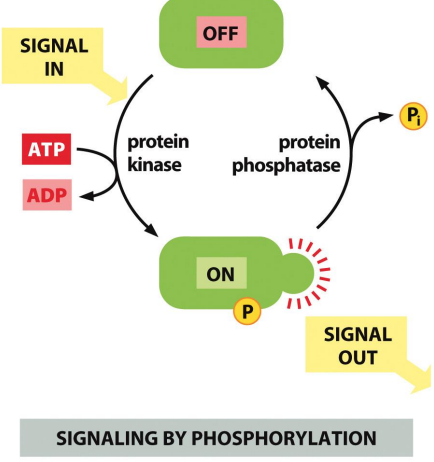
Phosphorylation
Protein kinases & phosphatases are employed in virtually all signalling pathways
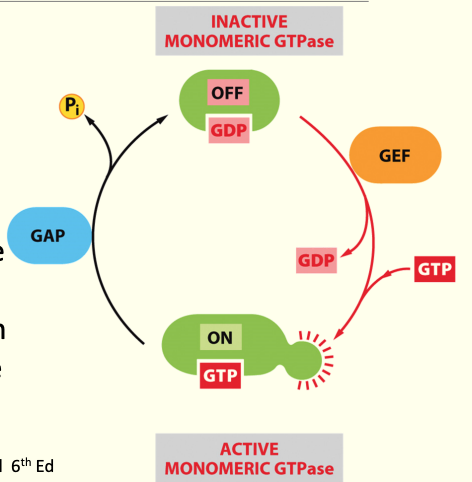
GTPases
GTP-binding proteins used in signal transduction as on/off switches
GDP is bound to GTPase→ has no activity
GDP associates w/ GEF (guanine exchange factor) → GDP is removed & GTP able to bind→ shape Δ & ↑ in activity (swaps GDP for GTP)
GAP (guanine activating protein) proteins aid removal of a Pi from GTP (to generate GDP), rendering the GTPase as inactive again
Signalling Cascades
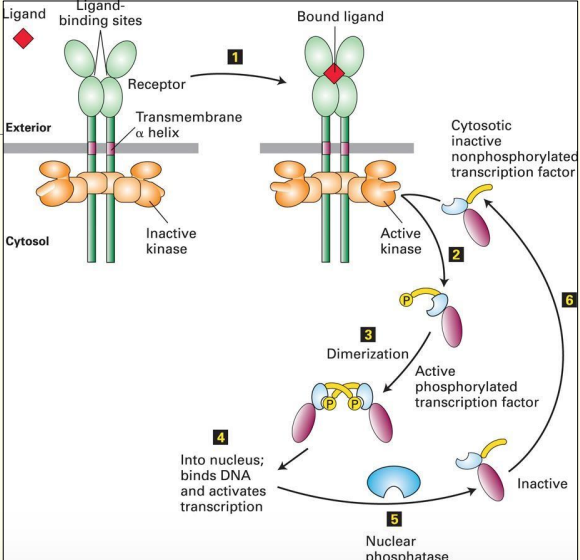
A simple signal transduction pathway involving 1 kinase & 1 target protein.
Phosphorylation & dephosphorylation are required to ensure that transduction only occurs in the presence of a stimulus
Ligand will only bind to receptor is both parts of binding site are present
When ligand binds - causes shape Δ of binding site & activates kinase domain → can now phosphorylate
- this activates another protein (TF) & causes dimerisation of 2 proteins which enter the nucleus - bind DNA & activate transcription
How can we ensure transmission is effective?
- Use lipid rafts to keep components of the receptors together:
- Using accessory for ligand binding
- Using adapter proteins for signal transmission
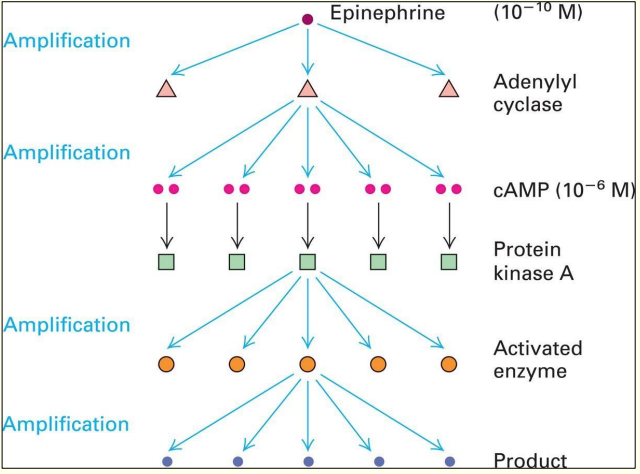
The domino Effect
Amplification of extracellular signal, enables either:
- Activation of multiple pathways to induce multiple effects simultaneously, OR
- Convergence of multiple pathways on a single target
Ensuring transmission
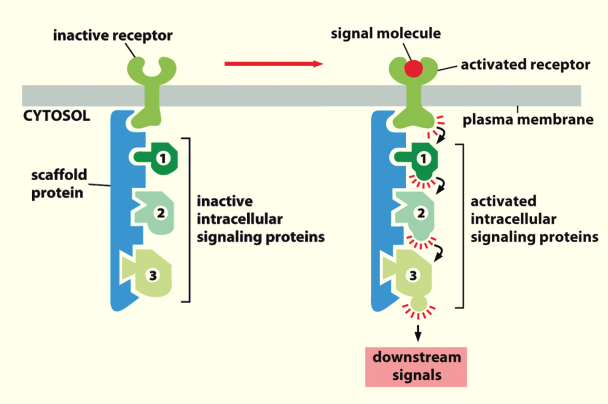
Scaffolding
- Inactive intracellular signalling proteins bind to scaffold protein through specific **‘structural motif’**→ whole complex associated w/ inactive receptor
- Ligand/signalling molecule binding causes the receptor to become activated & enables communication of signal from 1 protein to another as proteins are in close proximity to each other (proximity induced activity)
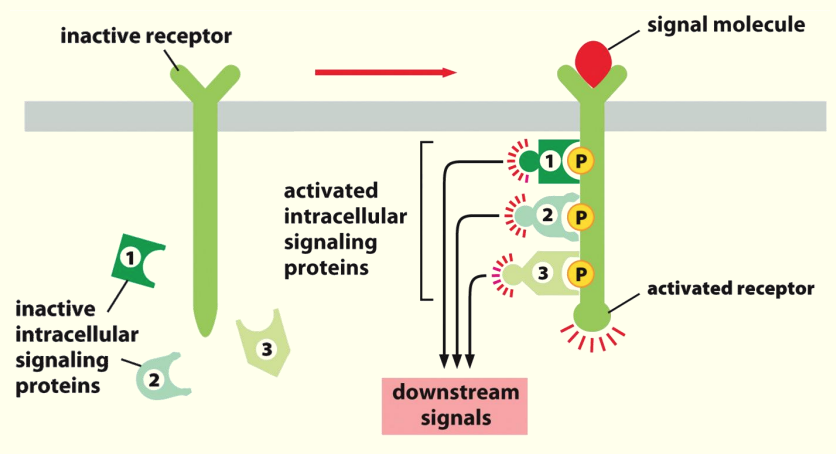
 Complex formation
Complex formation
- Ligand/signalling molecule causes receptor to become activated through phosphorylation
- Inactive intracellular signalling proteins bind to phosphorylated AAs on receptor
- Binding causes activation of proteins which start the signalling cascade
 Docking sites
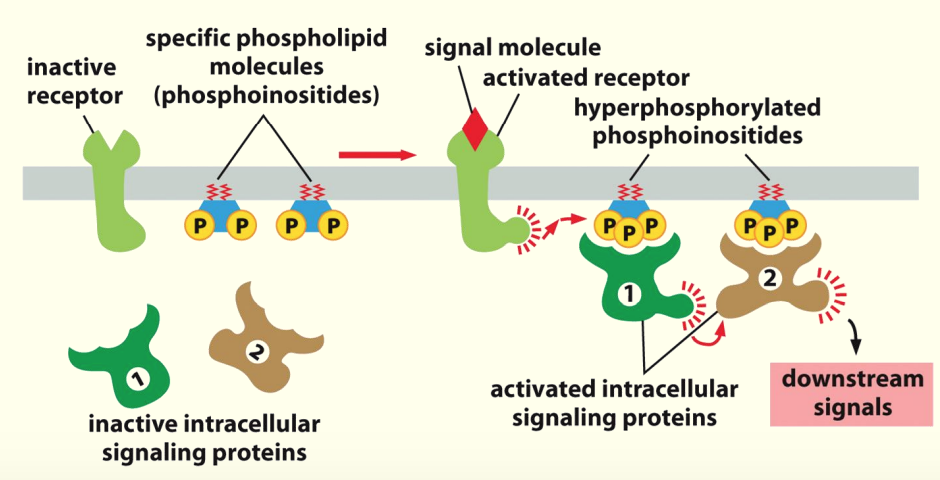
Docking sites
Membrane lipids used as docking sites for proteins in cascade
Activation of the receptor by ligand/signal causes phosphorylation of membrane lipids ~ phosphoinositides (phosphorylated PIP2 to make PIP3)
Hyperphosphorylated phosphoinositides recognised by intracellular proteins - then activated by induced proximity or direct modification
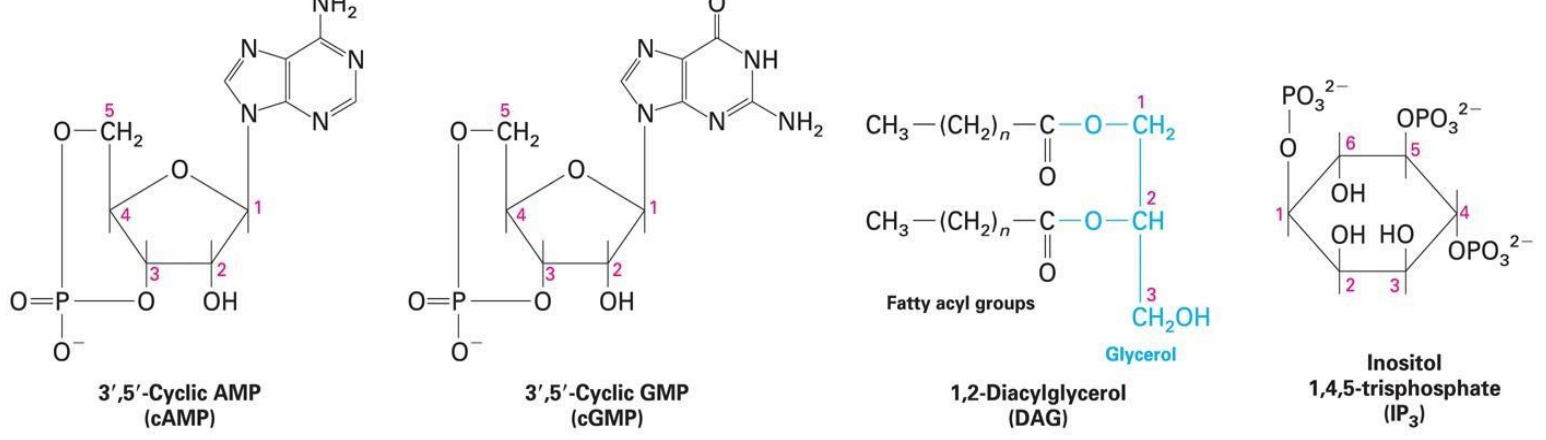
4 main 2º Messengers:
3’,5’- Cyclic AMP (cAMP)→ activates protein kinase A (PKA)
3’,5’- Cyclic GMP (gAMP)→ activates protein kinase G (GKA) & opens cation channels in rod cells
1’,2’- diacylglycerol (DAG)→ activates protein kinase C (CKA)
Inositol 1,4,5- trisphosphate (IP3)→ opens Ca2+ channels in ER
Receptor types:
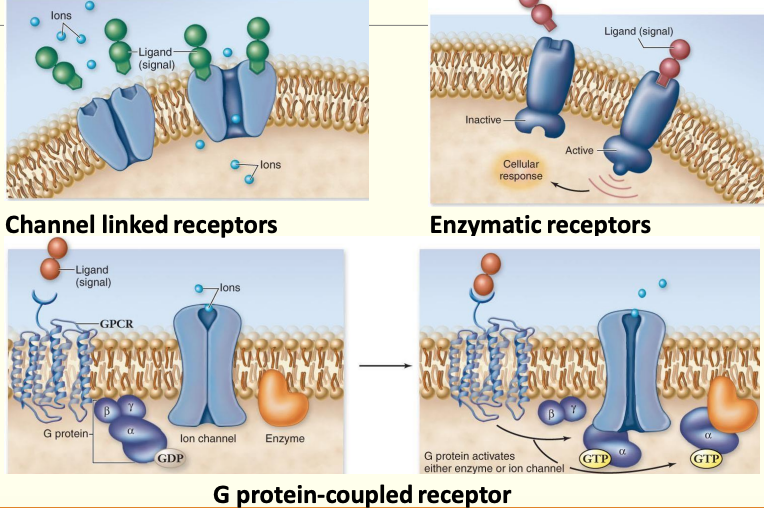
Receptors as detectors
- Receive signal & activate second messengers
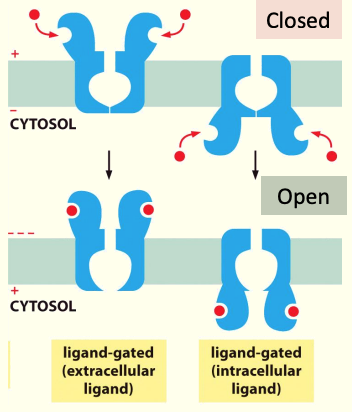
- Ion channels
- Form a “pore” in the membrane
- Only open when signal particle is bound to protein
- Restrict movement of ions from 1 side of membrane to another
- G proteins
- 7 transmembrane domain containing proteins, embedded in membrane
- N terminus on outside of cell & C terminus is on inside of cell
- Associated w/ proteins on inner surface of membrane that determine what type of response occurs
- 4 flavours
- Enzyme coupled receptors
- Single pass transmembrane receptors that ‘dimerise’ in membrane in response to ligand binding
- Activate each other to recruit intracellular proteins
- Result in use of second messengers
Ion Channel-Coupled Receptors
- Transiently Δ permeability of plasma membrane when ligand binds to receptor & enables it to open briefly
e.g. GABAₐ & Nicotinic Ach receptors
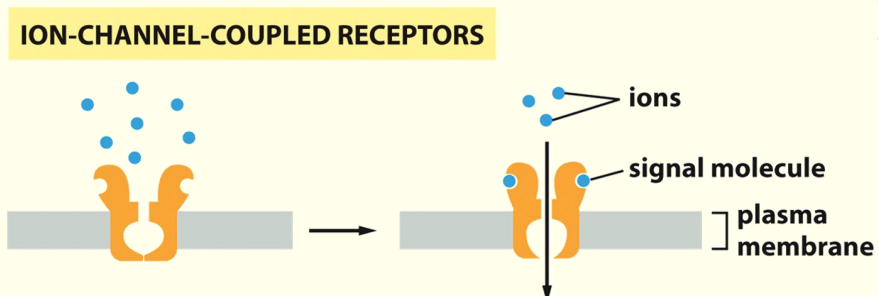
Specialised transmembrane proteins
Protein generates a pore in the membrane
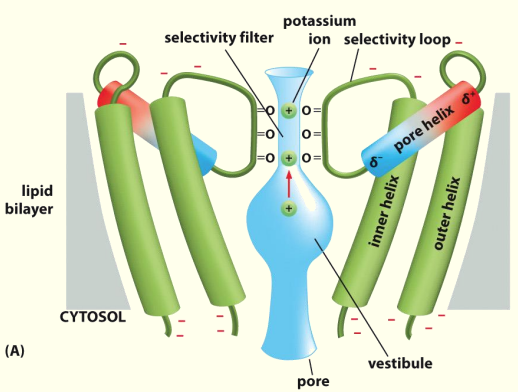
AAs protruding into channel work as selectivity filter to limit entry into the pore
When ligand binds to receptor it Δ shape so that pore can open to allow movement of appropriate ions through the pore
Examples:
Nicotinic acetyl choline receptors
- Diff. subunits in diff. locations,
- Cation channels - attracting +vely charged ions e.g. K + and Na+
GABAₐ →
- Inhibitory neurotransmitter
- 21 diff genes for subunits
- Chloride channel
- hyperpolarises the cell
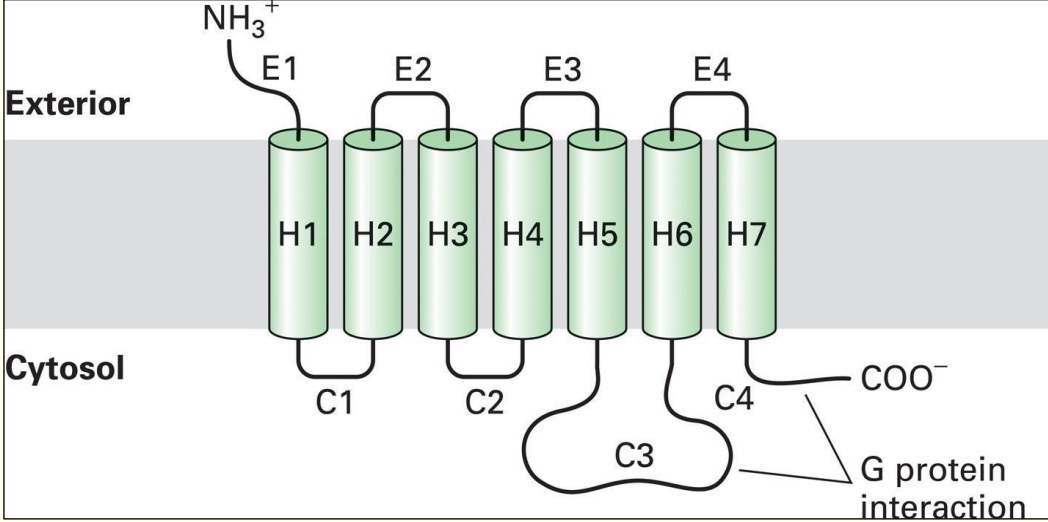
G Protein-Coupled Receptors
Also known as GPCRs or G-proteins
N-terminus (NH3+ end) on outside & carboxylic acid end (C-terminus) is inside
General structure of G Protein–Coupled receptors→ have 7 transmembrane domains (crosses membrane 7 times)
Have a loop e.g. C3 which is larger than the rest → loop & COO- end interacts w/ trimeric G proteins
Trimetric (α, β and ɣ subunits) G proteins relay signals from GPCRs
Anchors used to keep G-proteins close to surface & to the ligand
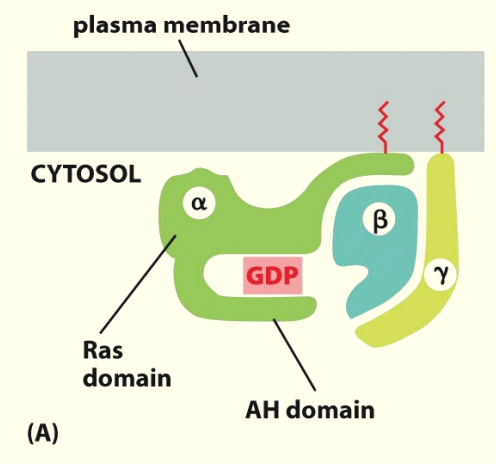
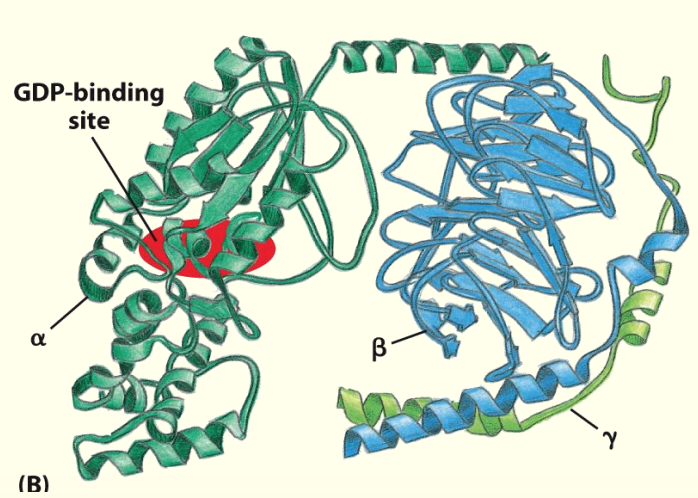
Binding of ligand to receptor recruits trimeric G-proteins
Association of the G-protein w/ receptor leads to activation & consequently activation of enzyme partner
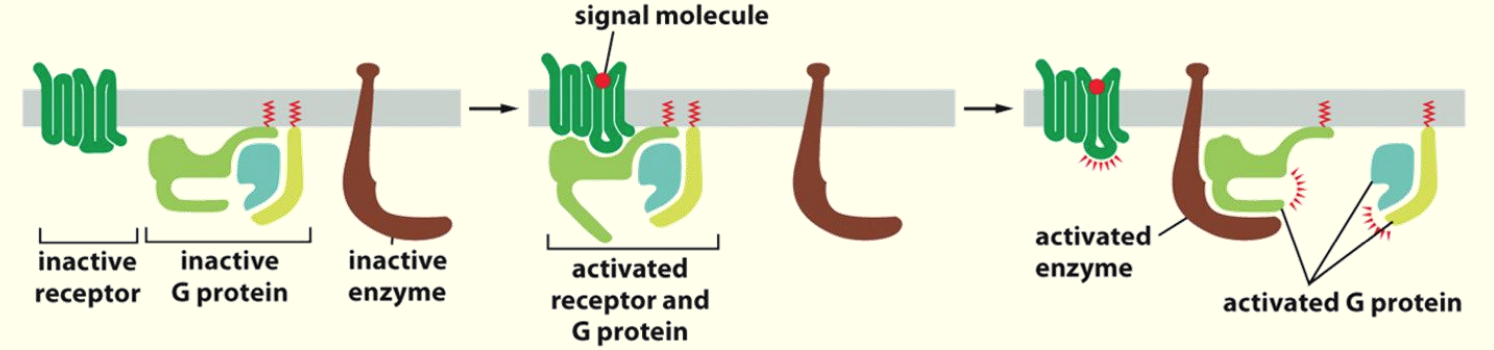
Activation of G proteins by activated GPCRs
Binding of ligand to GPRC leads to displacement of GDP from inactive trimeric G protein complex
Binding of GTP to α subunit of trimeric G protein leads to its activation & dissociation into α, β and ɣ subunits
- β & ɣ are able to have effects elsewhere
α subunit is now active & can activate an effector protein
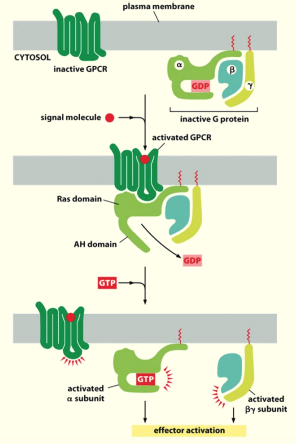
Activation of effector proteins associated with GPCRs
General mechanism of the activation of effector proteins associated with GPCRs
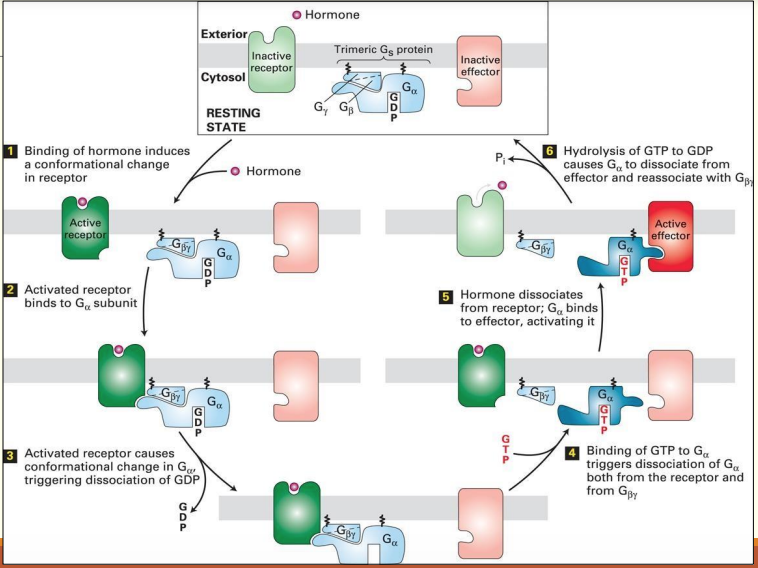
Diff. G proteins are activated by diff. GPCRs & in turn regulate diff. effector proteins
All effector proteins in GPCR pathways are either membrane bound ion channels or enzymes that catalyse the 2nd messengers
Calcium signalling
Activation of Phospholipase C
Phospholipase C metabolises phosphorylated lipids such as Phosphatidylinositol 4,5-bisphosphate (PIP2 )
- PIP2 needed for adaptor proteins to bind → to get clathrin binding
β & ɣ come in close prox. to activated phospholipase C-β & cause it to become activated
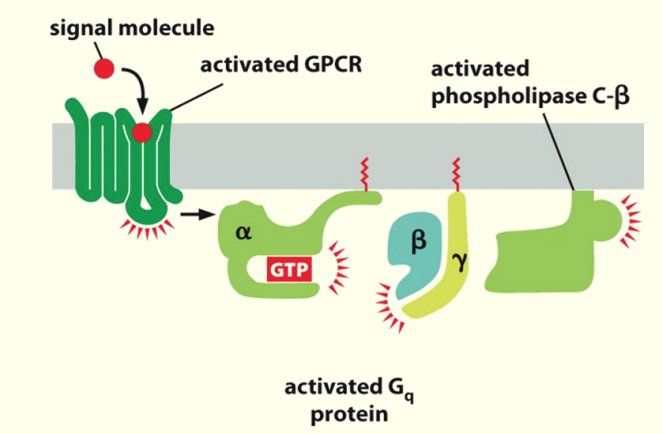
Hydrolysis of PI(4,5)P2
Activation of 2 different effects BUT with a common goal
DAG → activates protein kinases C - adds phosphate gps to proteins
IP3 → cause release of Ca2+ from ER
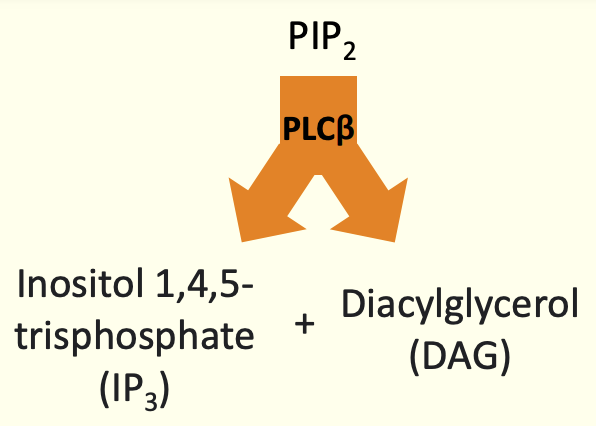
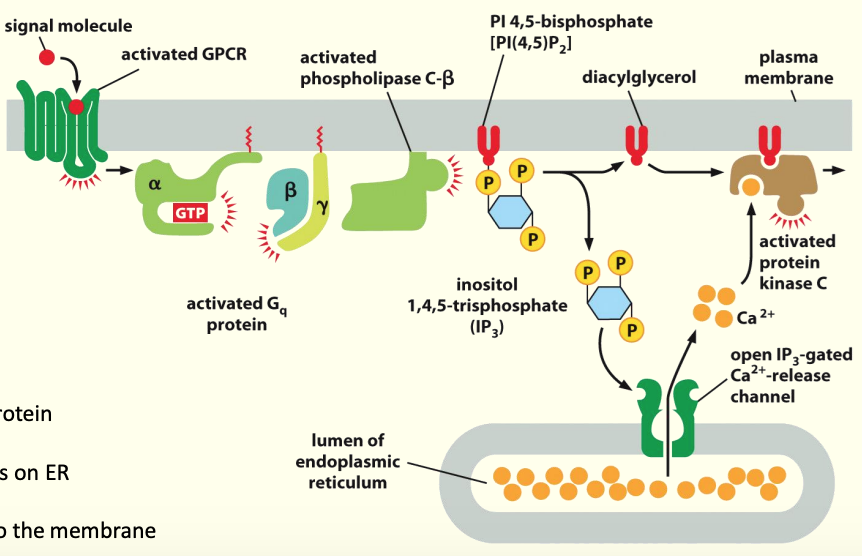
How this relates to intracellular Calcium:
Activation of PLC-β by a Gαo or Gαq protein
Cleavage of PIP2 into DAG & IP3
IP3 interacts w/ opens Ca2+ channels on ER
Release of stored Ca2+ into cytosol
Ca2+ recruits Protein kinase C (PKC) to membrane
DAG activates PKC
Active PKC phosphorylates substrates
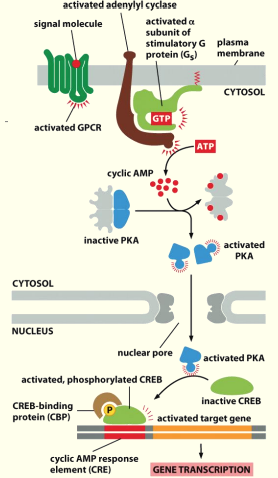
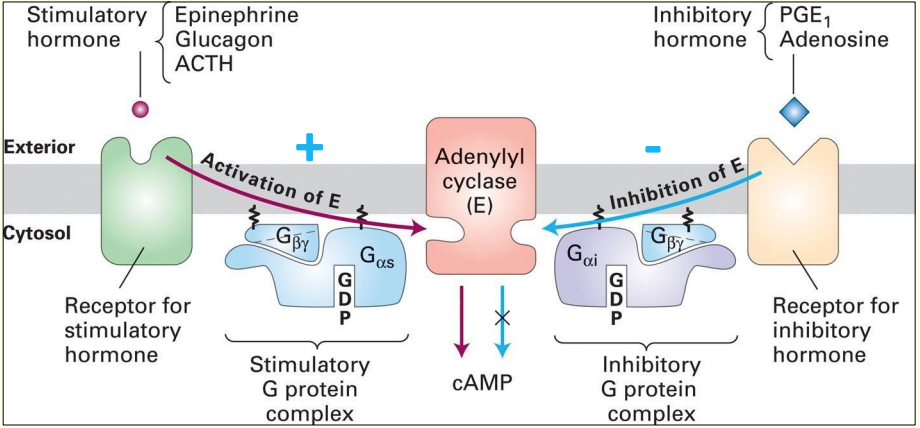
GPCRs that activate or inhibit Adenylyl Cyclase
Binding of ligand e.g. epinephrine to receptor causes stim. of Adenylyl cyclase
Adenylyl cyclase converts ATP into the secondary messenger cyclic AMP (cAMP)
Inhibitory hormone e.g. Adenosine - binds to a diff. type of G protein-coupled receptor
- swapping out of GDP for GTP, causes release of βɣ subunit which associate w/ Adenylyl cyclase & inhibit it
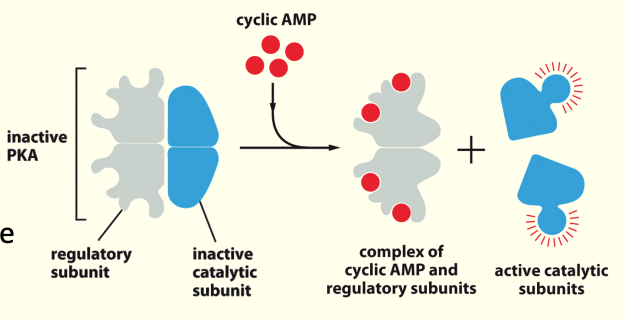
Adenylyl cyclase, cAMP and Protein Kinase A (PKA)
When low levels of cAMP are present- Protein Kinase A (PKA) is inactivated by binding of regulatory subunit.
When adenylyl cyclase is activated & produces cAMP, [cAMP] ↑
PKA has greater affinity for cAMP than regulatory subunit so cAMP binding then releases the inhibitory subunit & PKA becomes active
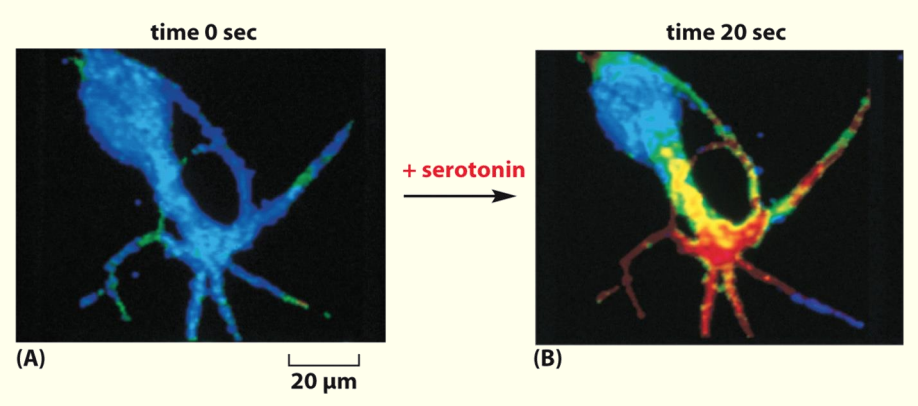
An increase in cyclic AMP in response to external signal
In this nerve cell the neurotransmitter serotonin activates a GPCR causing a rapid increase in cAMP levels
Serotonin binds to GPCR on cell surface & causes rapid ↑ in cAMP (shown by red area)
What happens after…
Activation of adenylyl cyclase by a Gαs protein
Conversion of ATP into cAMP
cAMP concentration in the cytoplasm rises
The regulatory subunit has greater affinity for cAMP
cAMP binds to regulatory (inhibitory) subunit of PKA, displacing the catalytic subunit.
Activated PKA can phosphorylate targets
Activated PKA can move into the nucleus and phosphorylate CREB, which controls transcriptional activation of gene promoters