Bacteria 1
Classification of bacteria
determined using microscopic, macroscopic and biochemical characteristics
size
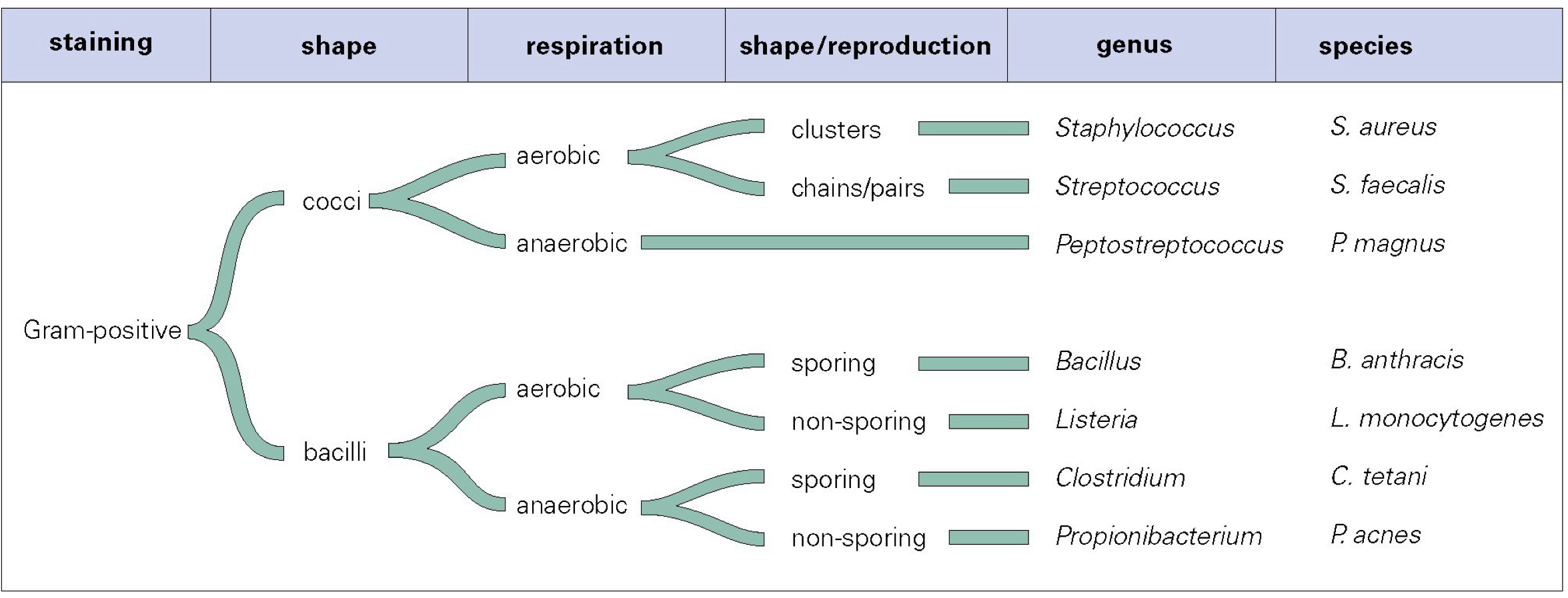
staining property
gram-positve
gram-negative
shape
cocci (round)
bacilli (rod)
spirilla (helical)
fusiform (pointed rod)
cluster
chains or pairs
respiration
aerobic
anaerobic
reproduction
sporing
non-sporing
important subspecies groups (biotype, strain, group) are identified based on
immunological properties
how they react with the immune system
serogroups and serotypes
defined by the bacteria cell wall, flagellar and capsule antigens (test with specific antisera——blood serum containg antibody)
biochemical characteristics
example: certain strain of Staphylococcus aureus release a β-haemolysin (toxin that causes RBC to lyse)
antibiotic susceptibility
phage typing
identifies bacterial strains based on their susceptibility to specific bacteriophages
example: differentiate between isolates of Vibro cholerae and Salmonella enterica serovars
Direct genetic approach
using PCR and probes to detect organism-specific sentinel (disease-causing) DNA sequence
Structure of bacteria
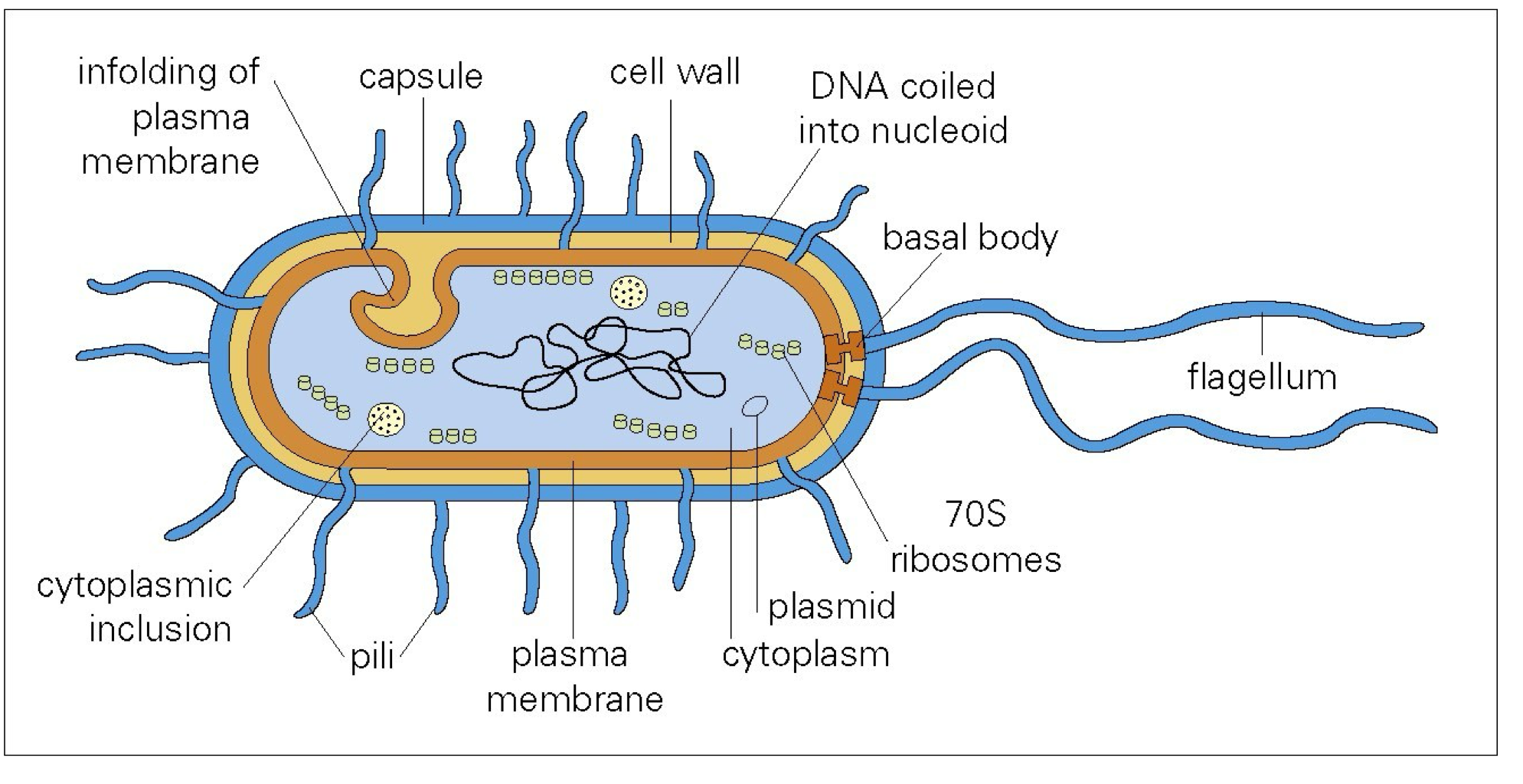
Cell wall
General Characteristics
peptidoglycan——a mixed polymer of hexose sugars and amino acid: the main component of cell wall
can be digested by lysozyme
cell wall is the major contributor to the ultimate shape of an organism
synthesis of peptidoglycan is distrupted by beta-lactam and glycopeptides antibiotics
all bacteria except mycoplasmas are surrounded by cell wall
might hv an addtional capsule
high molecular weight of polysaccharides → slimy suface
✔︎ protection against phagocytosis
important in deteremining virulence
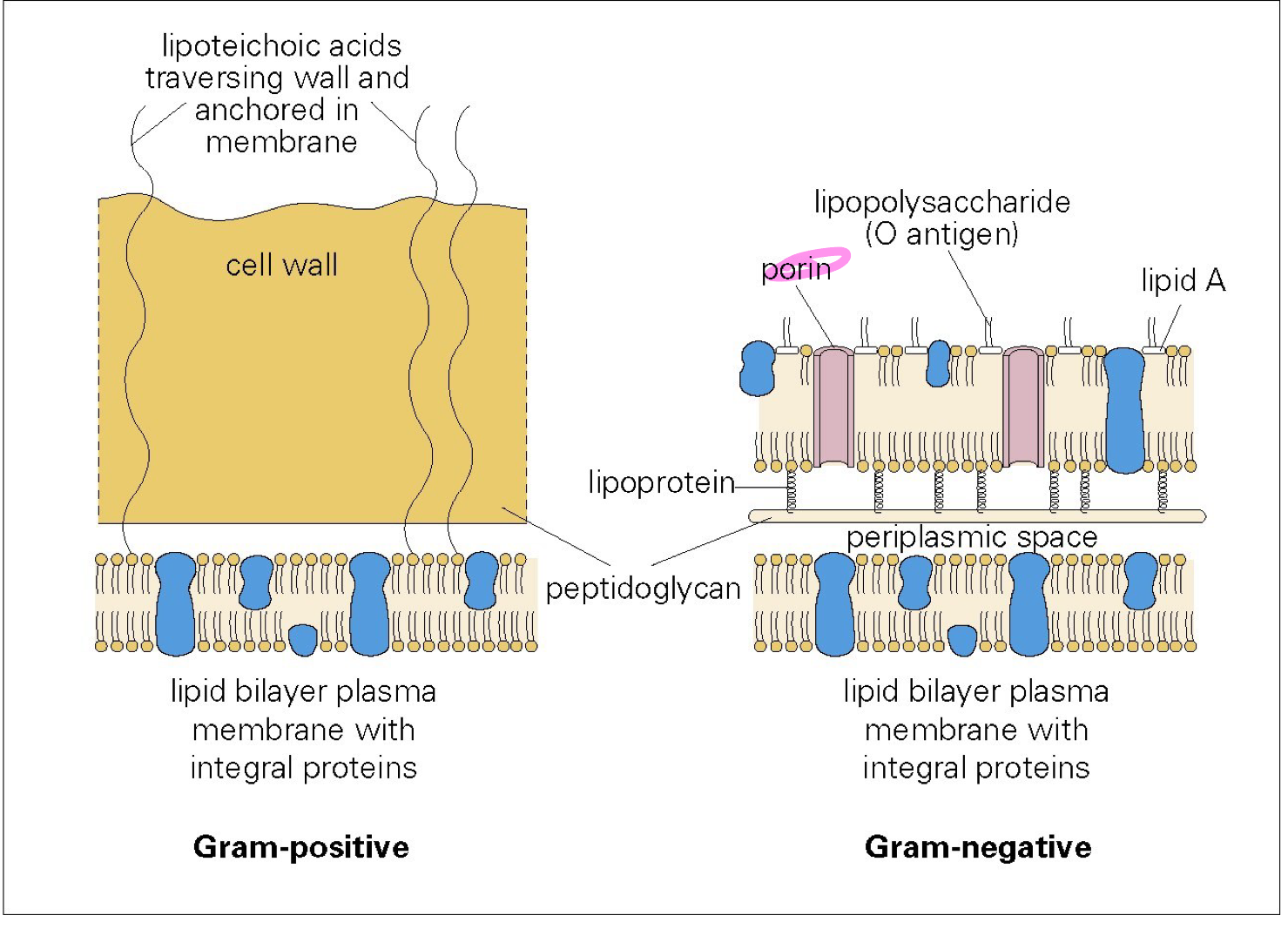
Gram staining
Gram-positive
thick layer (20-80nm) of peptidoglycan layer
external to cell membrane
contain other macromolecules
lipoteichoic acids (LTA)
★Mycobacteria
diff chemical basis for cross-linking to lipoprotein layer
outer layer envelop contain various complex lipids (mycolic acid)
cell wall compenents hv a pronounced adjuvant activity——promote immunologic responsiveness (can be used to enhance vaccine)
Gram-negative
thin peptidoglycan layer (5-10nm)
overlaid by an outer membrane
lipopolysaccharides(LPS)
O antigen (carbohydrate chain): antigenic property
endotoxin (lipid A component): toxic property
lipoprotein
porin
for entry of hydrophilic molecules
anchored to lipoprotein molecules
Flagella
log helical filaments extending from the cell surface
enable motility of bacteria
╳ utilising ATP
built of flagellins (protein): strongly antigenic
H antigen: important targets of protective antibdy responses
Pili (fimbriae)
more rigid than flagella
sex pili
attachment to other bacteria
for transfering DNA material
common pili
attachment to host cell
help prevent phagocytosis
antigenic variation: antigen changes
involve recombination of genes coding for ‘constant’ and ‘variable’ regions of pili
avoid immune variation
Nutrition
take up small molecules across cell wall
uptake and transport by cell membrane via
facilitated diffusion by carriers
active transport
e.g. amino acid, oligosaccharides, small peptides
Gram-negative bacteria can take up larger molecules
preliminary digest in periplasmic space (rich in enzyme)
some species require
only minimal nutrients
e.g. E. coli can grow with glucose and inorganic salt only
complex nutrients
e.g. Streptococci grow only in complex media with many organic compounds
General nutritional requirement
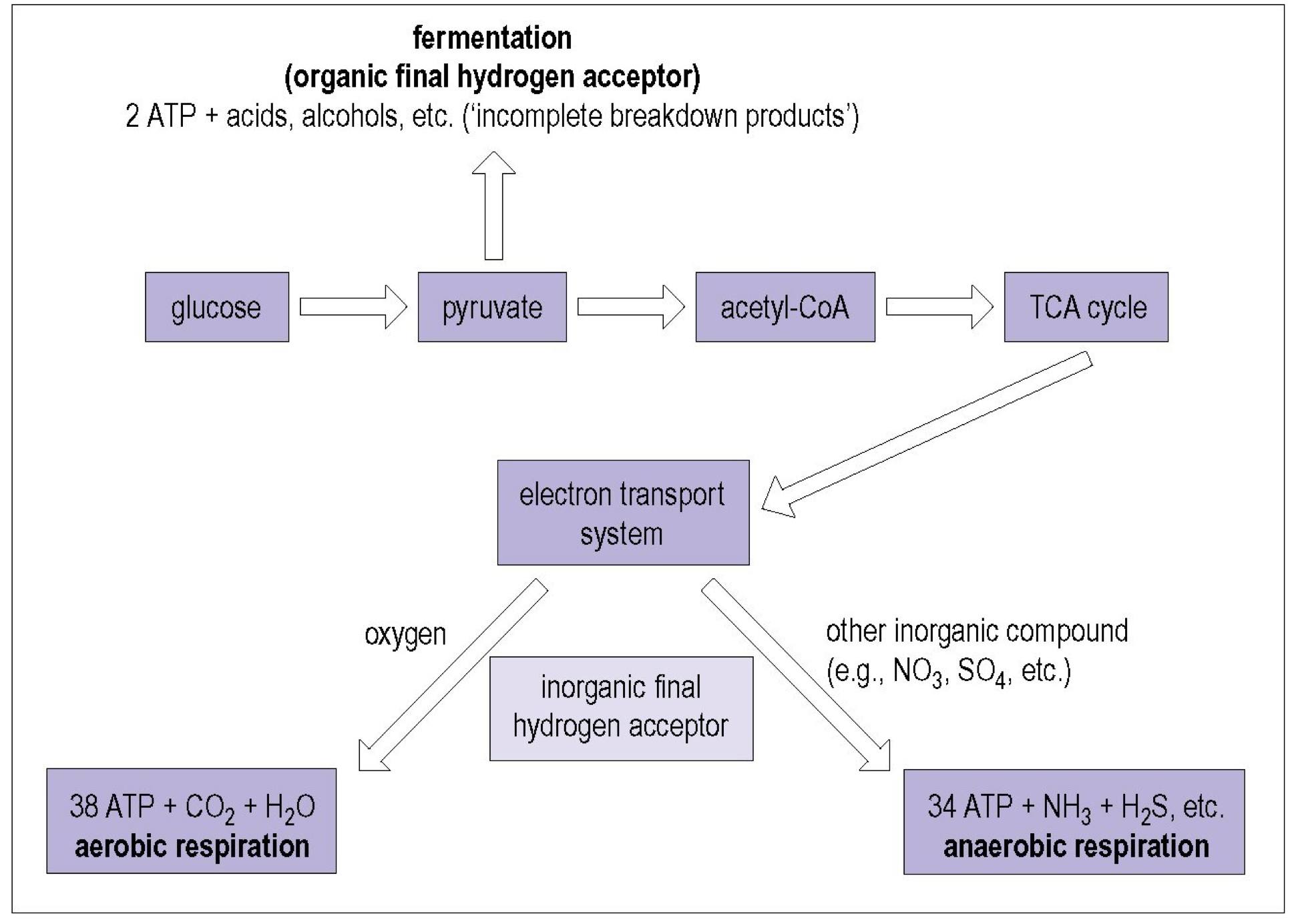
Respiration
requirement of oxygen can be obligate or facultative
aerobic
final product: 38 ATP+CO2+H2O
final electron acceptor: oxygen
anaerobic
final product: 34 ATP+NH3+H2S, etc (less ATP)
final electron receptor: other organic or inorganic molecule
less efficient but useful when absence of oxygen and other substrate are available
typically in host’s body
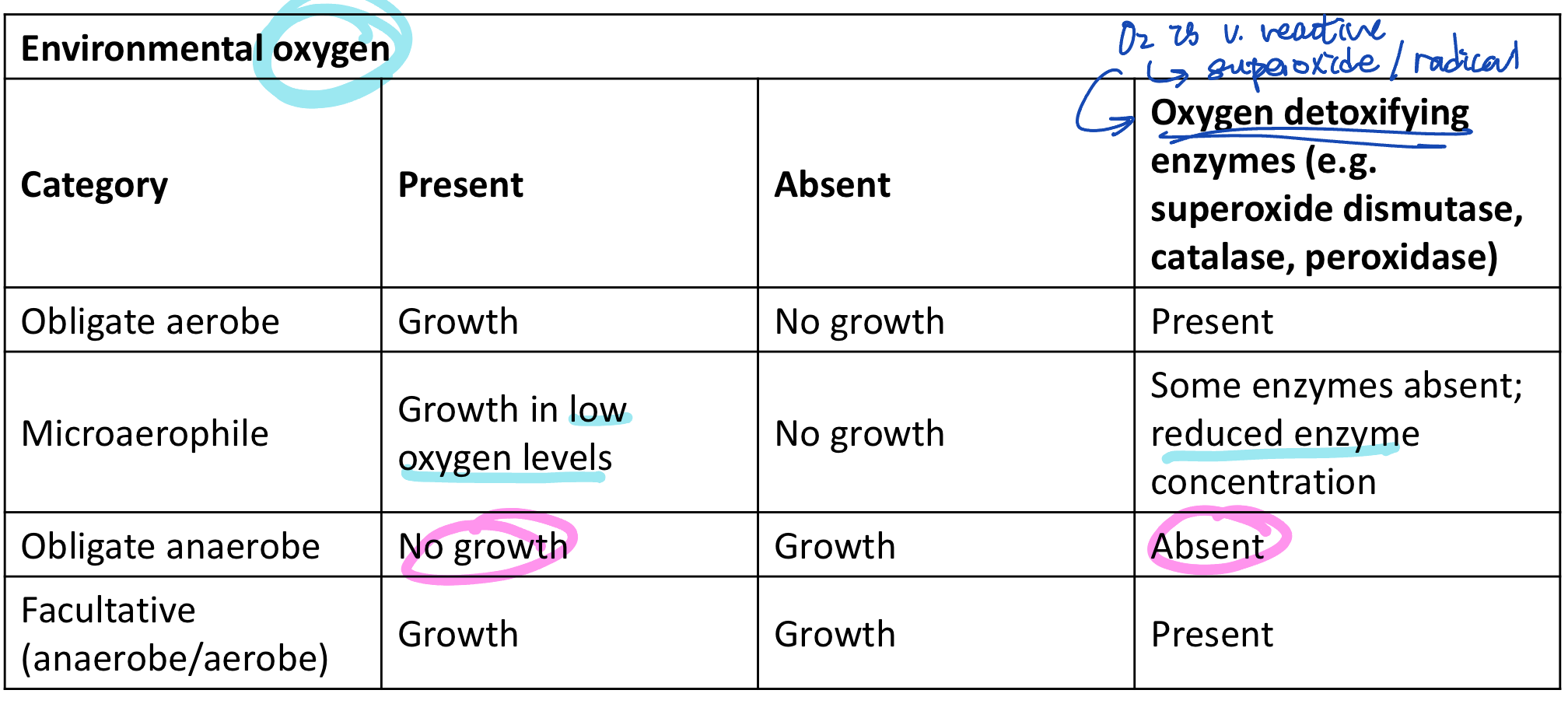
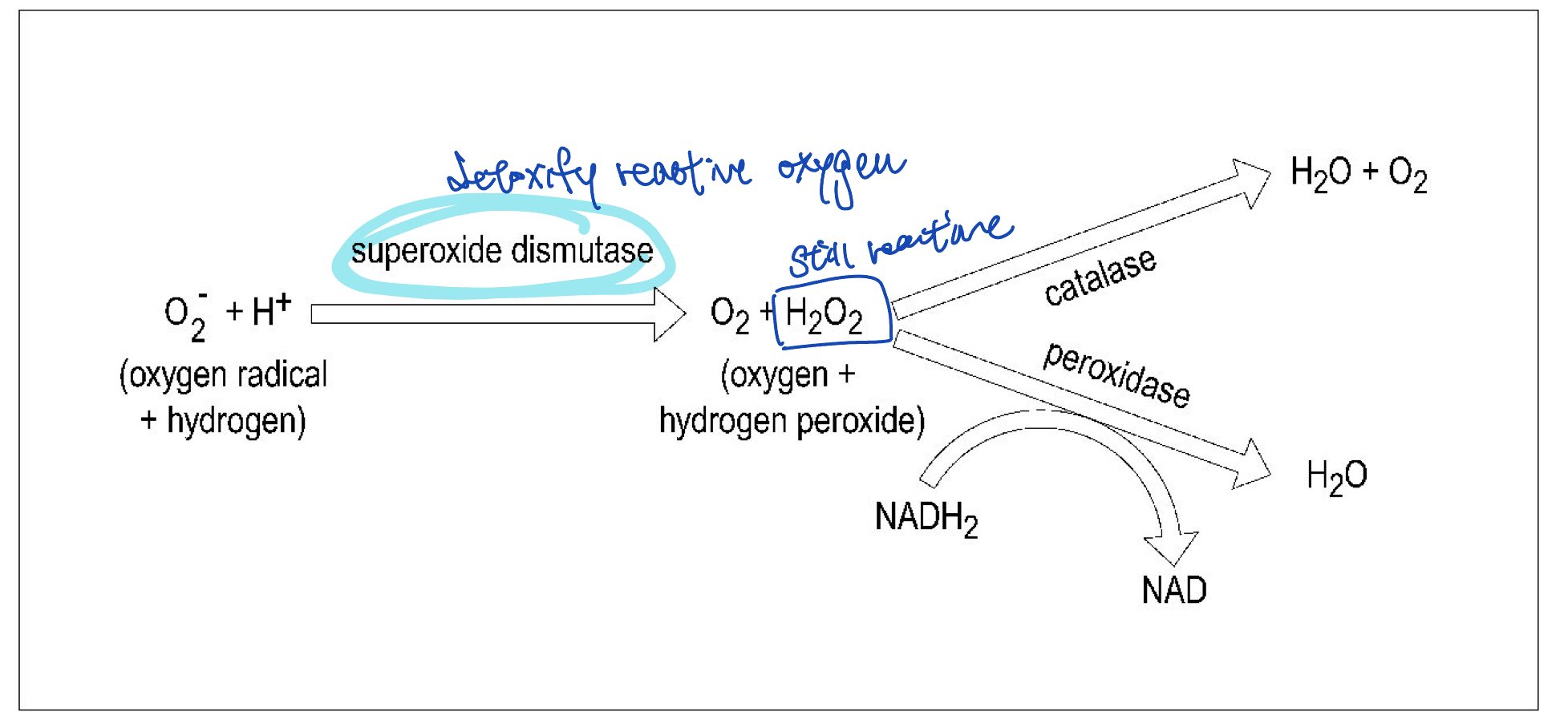
mode respiration and ability of bacteria grow in the presence of O2 relates to their ability to enzymatically deal with potentially destructive intracellular reactive oxygen species
whether it has oxygen detoxifying enzymes
superoxide dismutase
calatase
perodidase
examples of reactive oxygen species
free radicals
anions with oxygen
superoxide
Growth and division
depends large part on the availability of nutrients and genetical factor
Example:
E. coli double every 20min in lab vs 1-2 hours in a nutritionally depleted environment
Mycobacterium tuberculosis divide every 24 hr
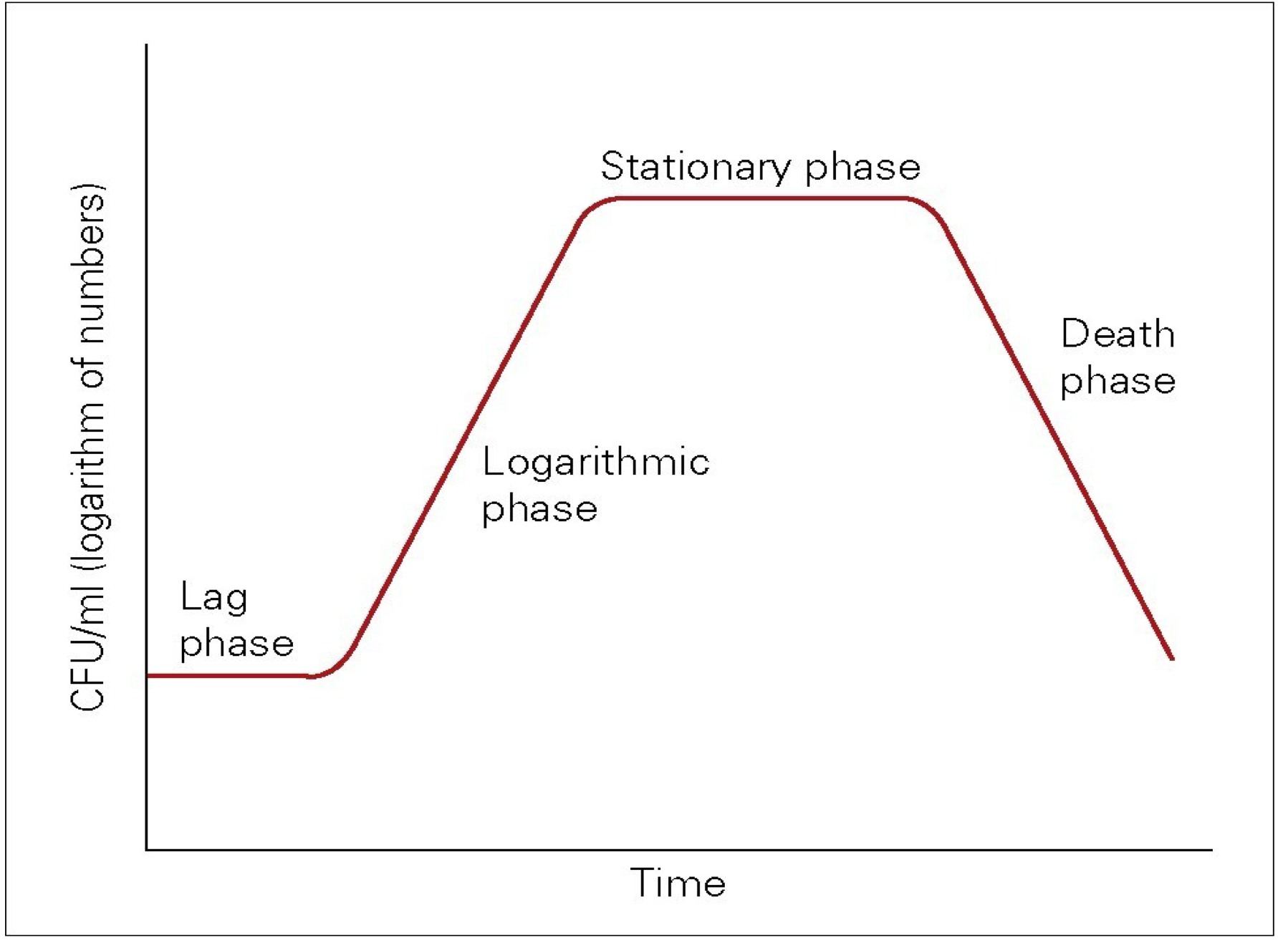
Bacterial growth curve
lag phase
period of adjustment when introduced to a new environment
sense the new environment→alter gene→synthesis of favorable protein for the specific environment to grow
log or exponential phase
population double in a constant rate
stationary phase
nutrients are depleted and toxic product accumulate
→equalise state: cell growth slows down and stops
death phase
Genomic DNA
characteristics
all bacterial genomes are circular
DNA has no introns
╳ nucleus → tightly coiled in to a nucleoid
genetic info can be extrachromosomal: encoded in plasmid (a small circular self-replicating DNA molecule)
replication process
begins at a single site——OriC: origin of replication
multienzyme replication complex bind to OriC → inititate unwinding and separation of DNA strands
helicases (separate DNA strands)and topoisomerases (release DNA from supercoiled form, e.g. DNA gyrase) are used
DNA polymerase helps incorportation of DNTPs
replication must be accurate
importance: DNA has info defining the properties and processes of cells
✔︎ proofreading mechanism
base selection
3’-5- exonuclease
mismatch repair
→ reduce frequency of error ~ 1 per 1010
Gene expression
majority of genes are transcribed into mRNA
e.g. up to 98% in E. coli
some are transcribed to produce ribosomal RNA species
5S, 16S, 23S
✔︎ scaffold (platform) for assembling ribosomal subunit
others are transcribed into transfer RNA
for decoding mRNA into functional proteins
Transcription
frequency of trancription initiation is influenced by
exact DNA sequence of the promoter site
overall topology (supercoiling) of DNA
presence or absence of regulatory protein that bind adj to and may overlap the promoter site
sigma factor
a component of RNA polymerase
important in promoter recognition
esp. in controlling exp of genes inv in spore formation in Gram +ve bacteria
several different factors allow sets of genes to be switched on
by altering level of expression of a particular sigma factor
bacterial arrangement
monocistronic: one promoter and one terminator for one mRNA transcript
polycistronic
a single promoter and terminator flank multiple structural gene——operon
≥ 1 protein is synthesed
importance: ensure protein subunits for particular enzyme complex or specific biological process are synthesised simultaneously and in the correct stoichiometry (ratio)
e.g.
uptake and metabolism of lactose are encoded by lac operon
cholera toxin from vibrio cholerae
pili of uropathogenic (pathogenic to urinary tract) E. coli → mediate colonisation
Regulation of gene expression
importance: affect the ability of teh bacteria to adapt to changes in their environment
highly regulate expression of many virulence determinants
to conserve metabolic energy
make sure virulence determinants are only produced when particular property is needed
Example
enterobacterial pathogens (bacteria in guts): can adapt to environment like
low temp & low nutrients (water)
37°C with rich supply of C and N but low O2 and free Fe (human guts)
adapt by switching on and off a range of metabolic and virulence-associated genes
gene expression can be alter by changing the amount of mRNA transcription
alter the efficiency of binidng of RNA polymerase to promoter site
activation: positive gene regulation——increase rate of transcription
binding of activator protein to operator site (where regulatory protein bind to)
binding of RNA polymerase to promoter
initiation of mRNA transcription
absence of activator protein → RNA polymerase fails to bind to promoter → no transcription
repression: negative gene regulation——inhibit transcription
binding of repressor protein to operator site
inhibition of binding or activity of RNA polymerase
block mRNA transcription
absence of repressor protein → RNA polymerase can bind to promoter → allow inititation of mRNA transcription
regulons: coordinated regulation of multiple genes
multiple genes are controlled by the same regulator protein (can be activator or repressor)
Translation
ribosome bind to specific sequence of mRNA (Shine-Dalgarno sequences——upstream promoter sequence)
begin to translate at AUG start codon
hybridise with specific complementary sequence of initiator of tRNA molecule
polypeptide chain elongation by condensation reaction of ribosome: couple the incoming amino acid to grow
termination at stop codon