Molecular Biology (BIOB11) Midterm Review
Table of Contents
Genetics and Laws of Heritability
DNA structure
Structure and packaging of Genomes
Structure of bacteria v. human
Packaging DNA mechanisms
Heterochromatin and euchromatin
Epigenetics
Genome maintenance
DNA mutation and structures of replication
DNA replication
Gene expression
Structure and Function of RNA
Transcription and Processing of mRNA
Genetics
Nature of genes
Genes are understood to be a unit of inheritance, one that is passed on.
Chromosomes seemed to be a likely carrier of genes and genetic information. DNA is a vessel for heritability and change, allowing for carrying on and new change.
Central DOGMA is the process of DNA to RNA to protein through transcription, processing, transport out of nucleus, translation, protein folding, and carrying out function.
Regulation is occurring during all of these processes to ensure mutations do not occur, as mutation do not change and they will remain in the cell, meaning that the DNA sequence cannot be changed back.
Mitosis and Meiosis
DNA is replicated once in each process. Mitosis divides once and meiosis divides twice.
Mitosis
Happens in somatic cells. You end up with diploid cells that are genetically identical.
Meiosis
Meiosis occurs in two steps; meiosis 1 and meiosis 2.
Meiosis 1 has two chromosomes (homologous chromosomes) that are pulled apart.
Meiosis 2 looks like mitosis where sister chromatids are pulled apart into two different cells.
End up with 4 haploid cells that are genetically non-identical.
Heritability
Inheritance
Genes are a unit of inheritance discovered by Mendel’s pea plants. Dominant traits are selected over others, second generation exposed recessive traits.
Laws of inheritance
Individuals have two copies of each gene called gametes (a single copy of every chromosome)
Alleles: may be dominant or recessive
Dominant alleles determine phenotype. Homozygous (AA, aa) or heterozygous (Aa)
Law of segregation
An individual’s maternal and paternal chromosomes segregate from one another during gamete formation. One gamete carries one allele for each gene.
Law of independent assortment
Segregation of a pair of alleles for one trait (gene) has no effect on the segregation of alleles of another trait (you’re not going to have all paternal if one whole paternal trait moves into an area)
Happens in meiosis 1. It is important for creating genetic organisms. Increase fitness in population and for change to be selected upon.
Carriers of inheritance
Non-dividable cells move into dividable strands called chromosomes, the carrier of heritable material needed in all cells. Normal cells went through this process
Messing with division causes disease and problems
Homologous chromosomes: Tetrad or bivalent pair in meiosis. Pair together in meiosis 1 and pulled apart. Separates the two sister chromatids into two cells
Law of independent assortment is possible due to crossing over
Breaking and rejoining of pieces of DNA
From a chiasma.
Occurs mostly in homologous chromosomes, although could potentially happen in other stages like heterozygotes.
Genes located close to each other may be linked (not always the case)
Kinetochores (microtubules) attach at centromere and haploid daughter cells post crossing over have diverse genetic makeup.
Issues in meiosis/mitosis
If they couldn’t separate they might go to the same cell
Trisomy: too many chromosomes
Crossing over may not be reciprocated and thus some may end up with too many or too little pieces of regulatory DNA.
Two centromeres happens in cancer, when trying to be pulled one way it ends up being pulled with the other one.
Extra mitotic spindle if you have a third division and mess up meiosis.
Fertilization: leads to a combination of alleles that are not present on either paternal chromosomes.
DNA
DNA carries heritable information. DNA is needed for viruses to transfer. You can separate DNA to find where a negative strain is. DNA was discovered as a combination of building blocks (sugar, phosphate, 4 nitrogenous bases). Franklin’s x-ray diffraction image of DNA, which was the key to figuring out the double helix structure.
What is DNA
Two stands, antiparallel. 5’ with phosphate group and empty 3’. Sugars (deoxyribose), 2’ carbon does not have oxygen only hydrogen.
Right hand spiral with a minor and major groove. Major groove creates protein binding sites (regulation, replication, transcription)
A/T has 2 hydrogen bonds. G/C has 3 hydrogen bonds.
A/G are purines, T/C are pyrimidines.
DNA is storage on genetic information, replication and inheritance, expression of a genetic message. Information is stored as the linear sequence of bases and codes for genes contributing to cellular processes. 2 strands of DNA would come apart and serve as a template (complimentary strand). Break hydrogen bonds to access a single DNA strand.
Changes in DNA mark the passage of time as species form and diverge. Bacteria → archae → eukaryotes. All through, DNA has been the vessel.
****
Structure and Packaging of Genomes
Structure of bacteria v. human genomes
Differ in size and arrangement
Eukaryotes: Animal, plant, fungi. Keeping genomic DNA in nucleus, additional DNA in mitochondria, pathogens, and plastids. They bind new nucleic acid in (the main stuff comes from DNA).
Multiple origins of DNA
Use telomerase to maintain chromosome ends
Nucleosomes
DNA in nucleus
Prokaryotes: Archae and bacteria. No distinct nuclear compartment. Nucleoid region, circular chromosomes. Horizontal gene transfer is responsible for spread of penicillin resistant strains of gonorrhea. Smaller so the faster it can replicate. Keep what is beneficial and tosses what isn’t
Have very little non-essential DNA sequences
Bacteria is a circle while human cells are 21 chromosomes. Both replicate, translate, take in energy (cell metabolism), cell division.
Both Prokaryotes and eukaryotes
Use DNA as a carrier of heritable DNA
Lagging strand DNA with Okazaki fragments
DNA → RNA → Proteins
Genomes represent genes that perform the same functions. When there is small nucleic change, we are given the opportunity for evolution, however this does not affect important processes. Too fast mutation/evolution can result in death, and thus there are mechanisms in place to maintain the genomes. Genome size is variable and not directly correlated to the amount of genes, like how corn have a big genome size, but have fewer base pairs and chromosomes.
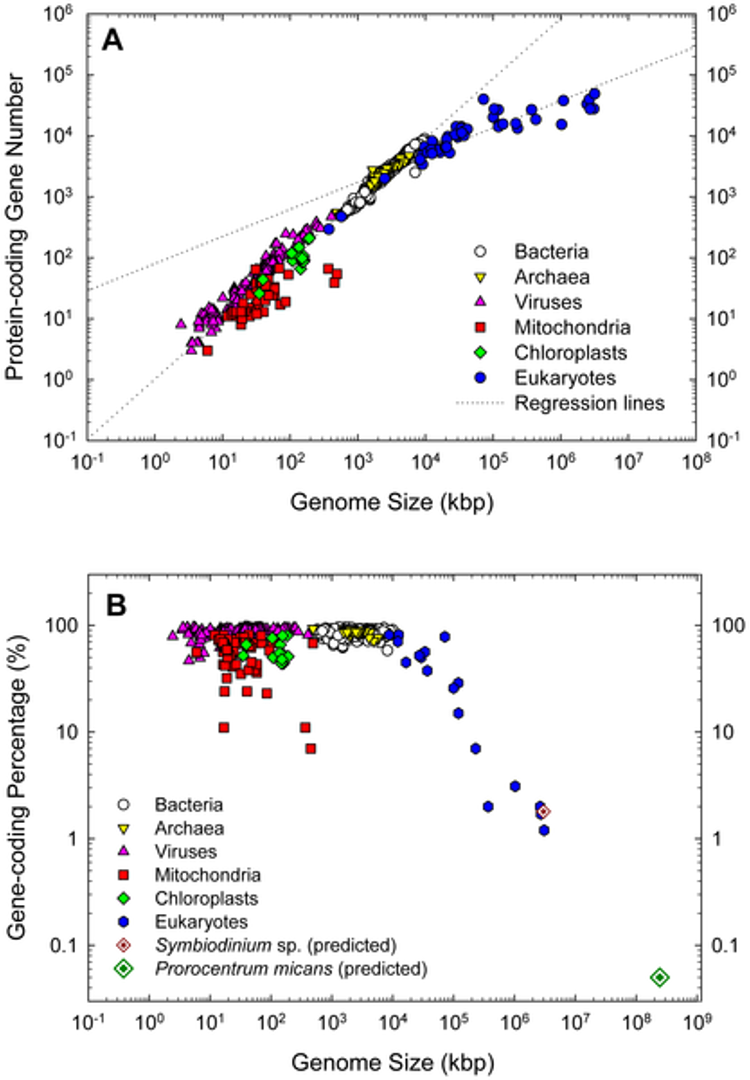
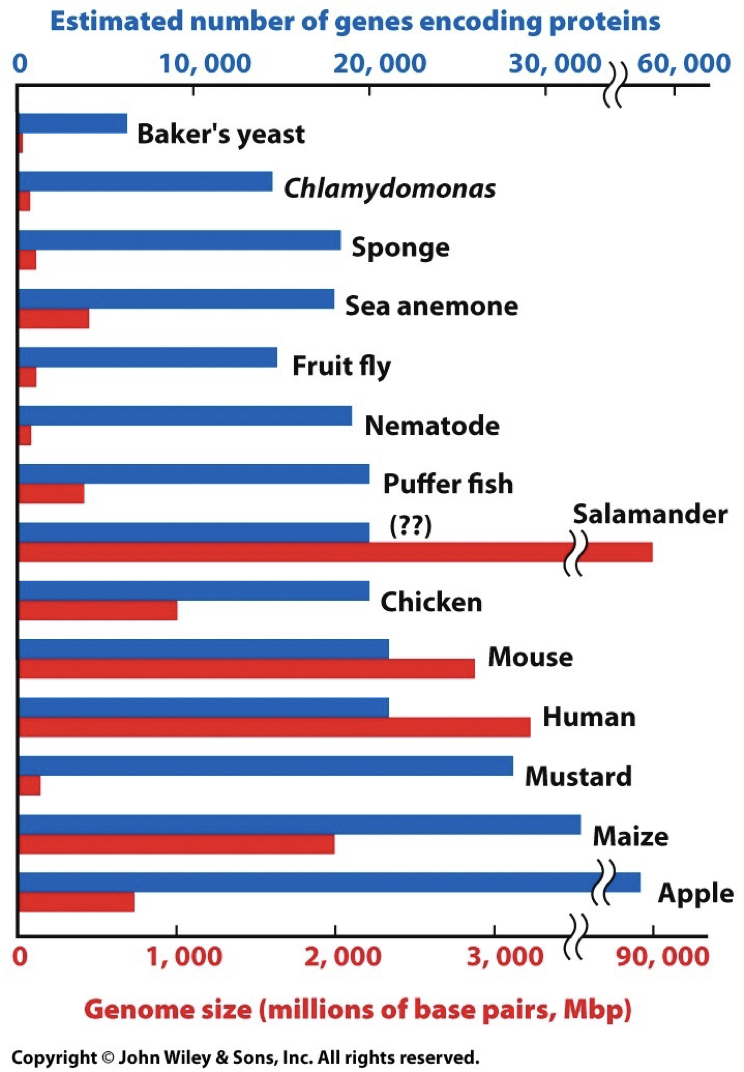
Composition of Human Genes
LINEs: long interspersed nuclear elements (transposons, repeated sequences)
SINEs: short interspersed nuclear elements (transposons, repeated sequences)
Introns: Spliced out and not included in RNA
Exons: Included in RNA, coding sequences
Unique sequences: nonrepetitive DNA that may have meaning
Transposons: Mobile genetic elements that move around and jump around the genome. (LINEs, SINEs)
Repeated sequences: LINEs, SINEs (transposons), simple sequence repeats, segmental duplications.
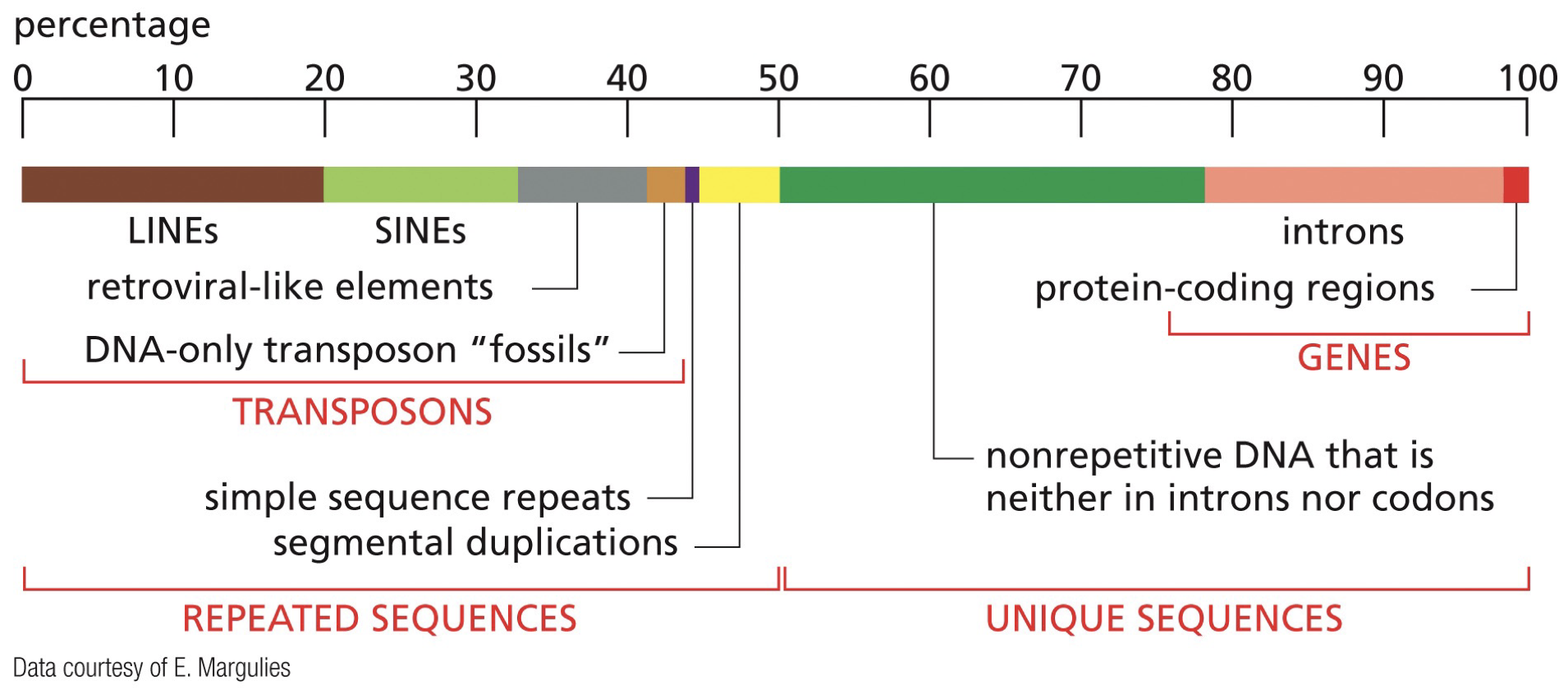 Non-coding sequences exist within genes. Once RNA transcription is completed, the introns are spliced out. Remaining sequence is composed of solely exons, and is then made into proteins, then is made into folded protein.
Non-coding sequences exist within genes. Once RNA transcription is completed, the introns are spliced out. Remaining sequence is composed of solely exons, and is then made into proteins, then is made into folded protein.
Packaging DNA Mechanisms
DNA is highly packaged and organized and going to be associated with many things at a time.
Nucleus: 6.4 billion base pairings in 46 chromosomes. 2 meters of base pairings. 1 base pairing per 6 H2O.
DNA supercoiling
DNA becomes a double helix, and then twists on itself even more to become supercoiled. Very tense and opening causes tension. Increase stability, compact, allows for unwinding of sections, relieves strain.
Positive supercoils: overwound
Negative supercoils: underwound
Topoisomerase: Enzymes that regulate supercoiling of DNA in the cell.
Nucleosomes
Eukaryotic DNA is associated within histones to form chromatin. Histones are highly conserved proteins rich in basic amino acids. They have a high positive charge and interact with the negative charge of DNA (phosphate groups).
2 wraps of DNA and a histone. Histones start with a dimer, then make an octomer.
8 histones form the centre, made up of repeating subunits of DNA and a histone. 200 base pairs per histone.
Histone fold/histone handshake: different sequences in the tail where modifications occur, regulatory modification. This tells the cell how to organize DNA. H2A - H2B, H3-H4.
H1: Linker histone, links DNA together. Amount of H1 in there affects how tightly histones package together. 30 nm fibres is the H1 and core nucleosome. We stack them together which give a 30 fold.
Looped domains occur following condensed chromatin. Interacting with proteins, forming scaffolds 30 nm fibre in loops to position chromatin into 700 nm folds, then 1400 nm fold. Eventually get to 10 000-fold (mitotic chromosomes)
Heterochromatin and Euchromatin
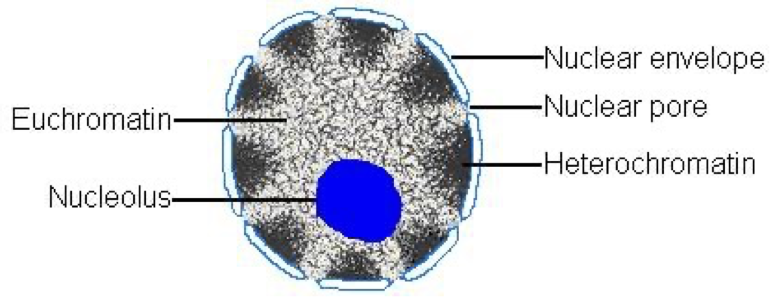
Euchromatin
Less condensed, towards the centre, accessible for protein binding and transcription, this is all due to sections becoming less compact at interphase following mitosis.
Heterochromatin
More condensed, towards periphery, not much functional activity. At least 10%.
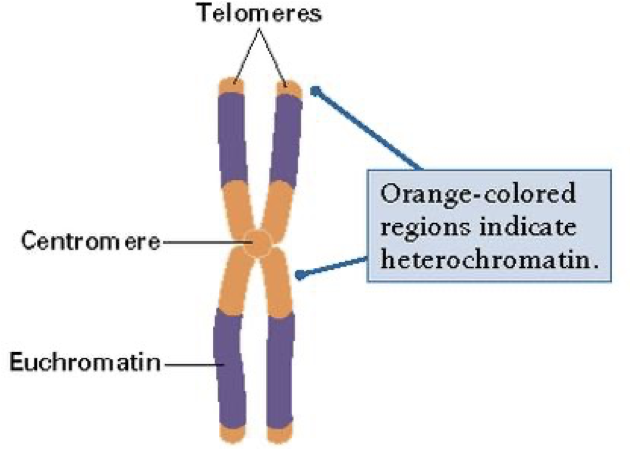
Constitutive heterochromatin
Permanently silent, like the telomere and centromere. Repetitive DNA. Compact at all time.
Facultative heterochromatin
Active only during certain area of an organism’s life, happens as cell and body develops while cell differentiation occurs. Inactivated during certain phases of an organism’s life.
Ex. X-inactivation (doesn’t start inactivated, but is then condensed in embryo)
XX phenotype needs part of one X to be silenced. Two active transcriptionally will be too much DNA. Once silenced it is maintained.
Random whether maternal or paternal is partially condensed. Once fully condensed as heterochromatin, the inactivated x-chromosomes is called a Barr body.
Calico cats: You can see which X-chromosome was turned off based on which cat it took from. Both x are active prior to meiosis. Patch of cells give patches of coat colours. If you clone the cat it will have a different coat because of different random inactivation events. The sequence remains the same.
Importance of facultative and if it all was euchromatin
This has purpose in regulating genes. You need a different set of genes for different cells to specialize cells, like a kidney cell vs. a heart cell.
Things that have similar functions can be put together.
Is able to control cancer better.
Constitutive → facultative → fairly closed and inactive, where it is linker histone bound → weakly transcribed → highly accessible.
Epigenetic inheritance
Epigenetic inheritance occurs on top of the genome with no change to nucleotide DNA. It is vital to developing tissues. Epigenetic refers to covalent modifications of DNA and histones that help guide processes like forming heterochromatin and euchromatin. Modifications can be perpetuated across cell division.
Histone coding
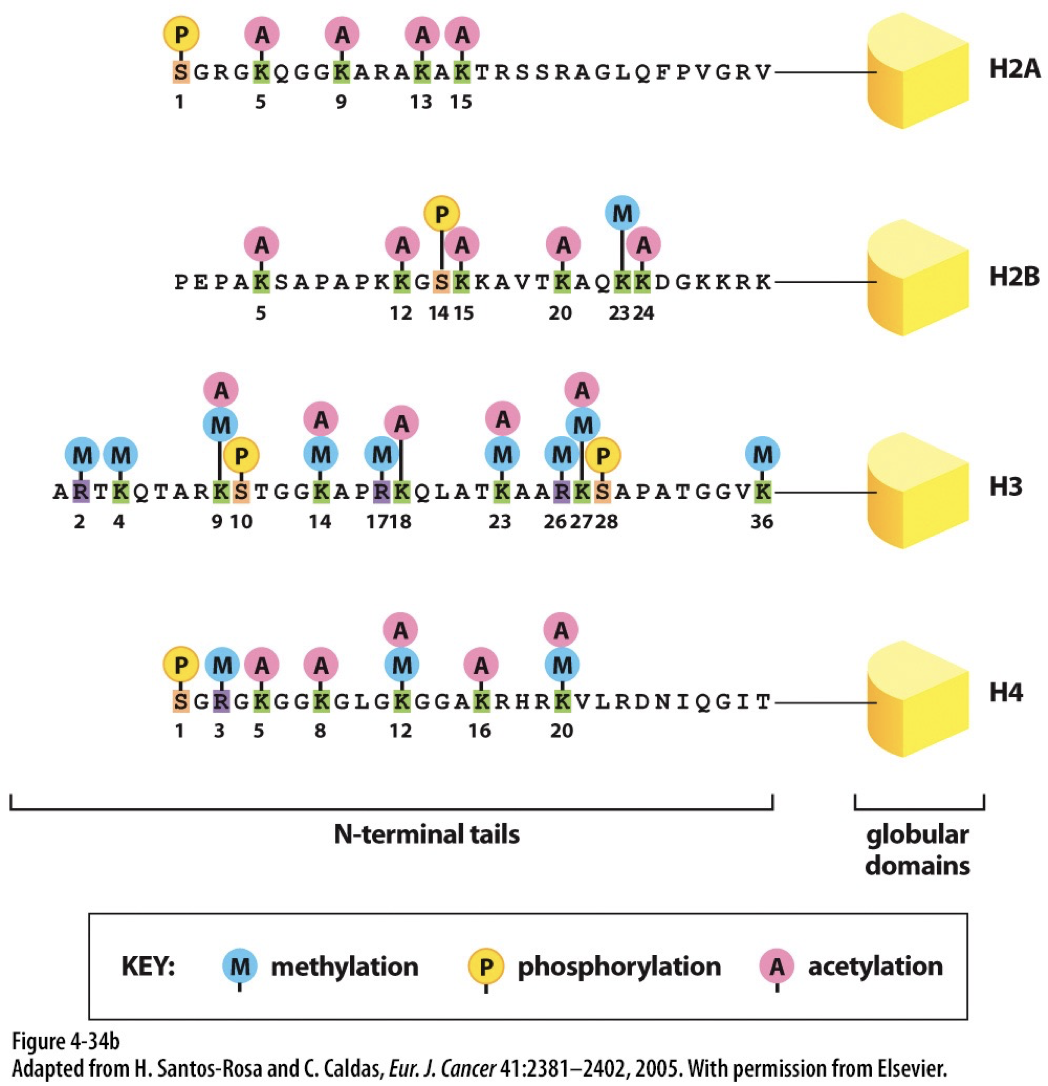
Modifications of histone tails can disrupt or stabilize nucleosome assemblages. This known to regulate chromatin structure.
Acetylation: leads to more open structure
Becomes uncondensed when H2, H3, and H4 are acetylated
Methylation: leads to less transcription
Becomes more condensed when H3 and H4 are methylated
Histone acetyltransferases (HATs): acetylate histone proteins by transferring acetyl group from acetyl-CoA to specific lysine residues. Associated with euchromatin.
Histone deacetylases (HDACs): Removes acetyl group
Histone methyltransferases (HMTs): add methyl groups to lysine or arginine residues
Histone demethylases (HDMs): remove methyl groups
Organization of chromatin in nucleus is determined by this.
Methylation of DNA and histone code work together. The methyl group attaches directly to a cytosine base in DNA. The proteins that bind to methylated DNA can recruit enzymes involved in modifying histone tails. Methylation of cytosine stabilizes nucleosomes and can prevent proteins from binding to DNA sequences (prevent initiation of transcription).
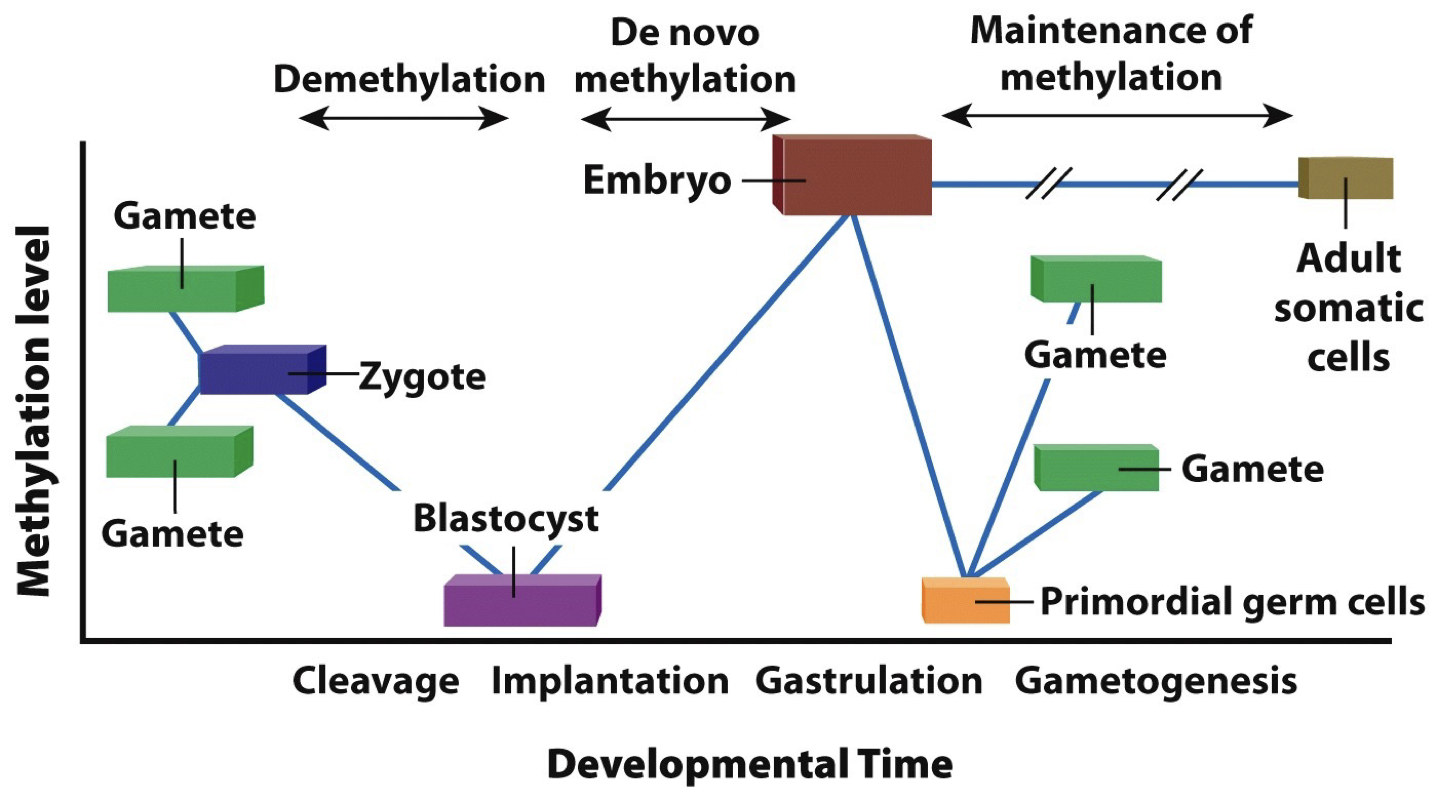
Epigenetic memory of DNA methylation patterns
DNA methyltransferase can add the methyle group to DNA at sites where C is followed by G (5’ to 3’ CpG)
DNA is methylated as it is replicated so that methylation patterns can be passed on to daughter cells (symmetry is critical)
Silencing genes by heterochromatin formation occurs in regions (“position effect”) and is maintained in replicated DNA.
Epigenetic modification is inherited during cell division and some but very little is passed onto offspring. Propagating heterochromatin after DNA replication: epigenetic signals that regulate chromatin are propagated through space and time (as they are changing while the grow, they need to be maintained).
Reading histone code
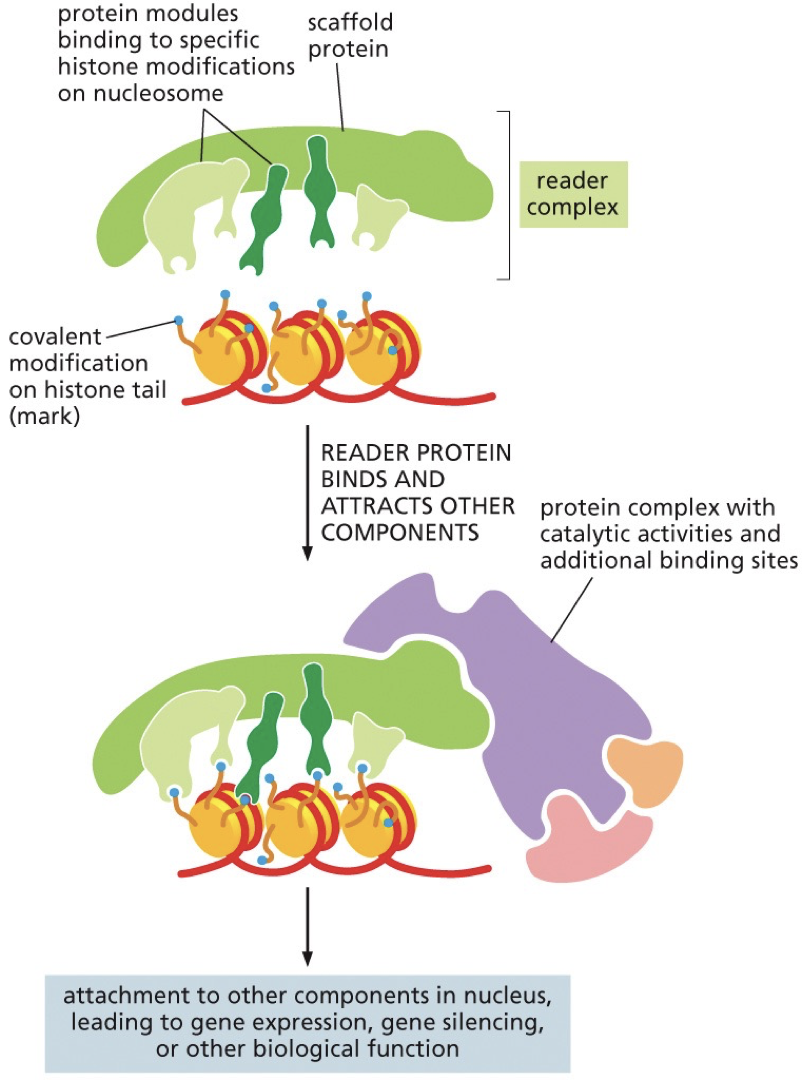 Histone modifications are recognized by specific proteins that are often part of multi-protein complexes.
Histone modifications are recognized by specific proteins that are often part of multi-protein complexes.
Reader complex “reads” the histone code and positions and activates “writer” enzymes that can act on adjacent DNA/histone
Chromatin remodelling enzymes
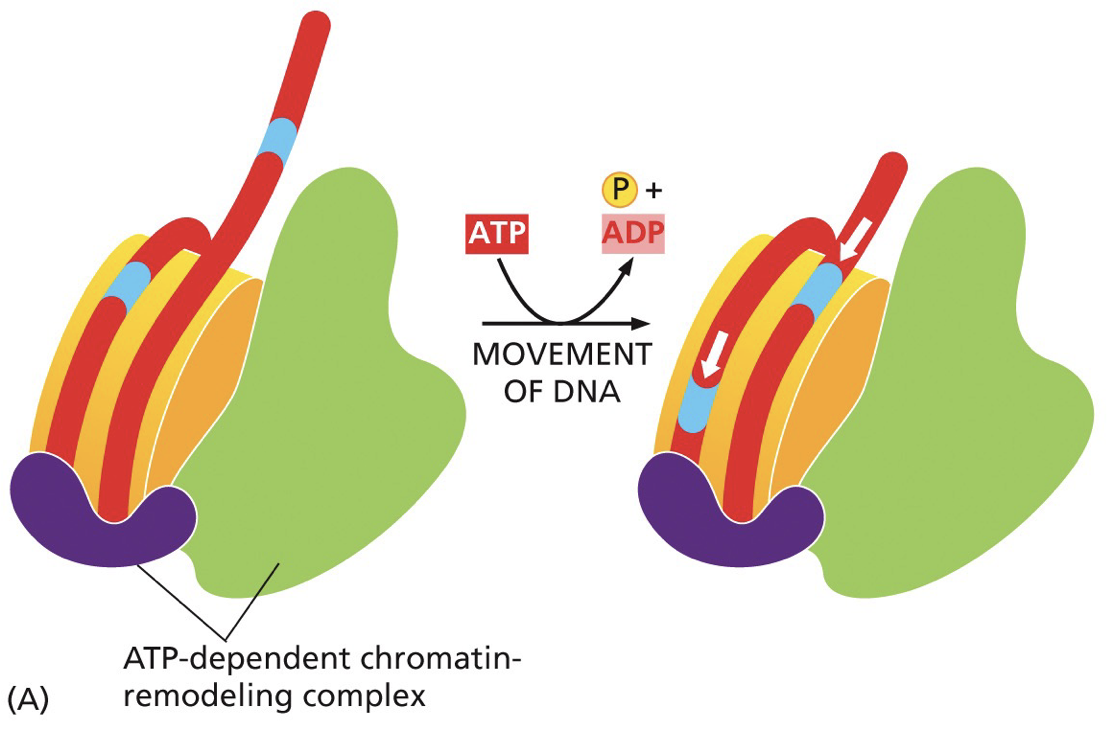
Readers/writers of the histone code work together with chromatin remodelling enzymes. Perform functions like altering position of nucleosomes on DNA, removing histones and switching in histone variants associated with particular functions.
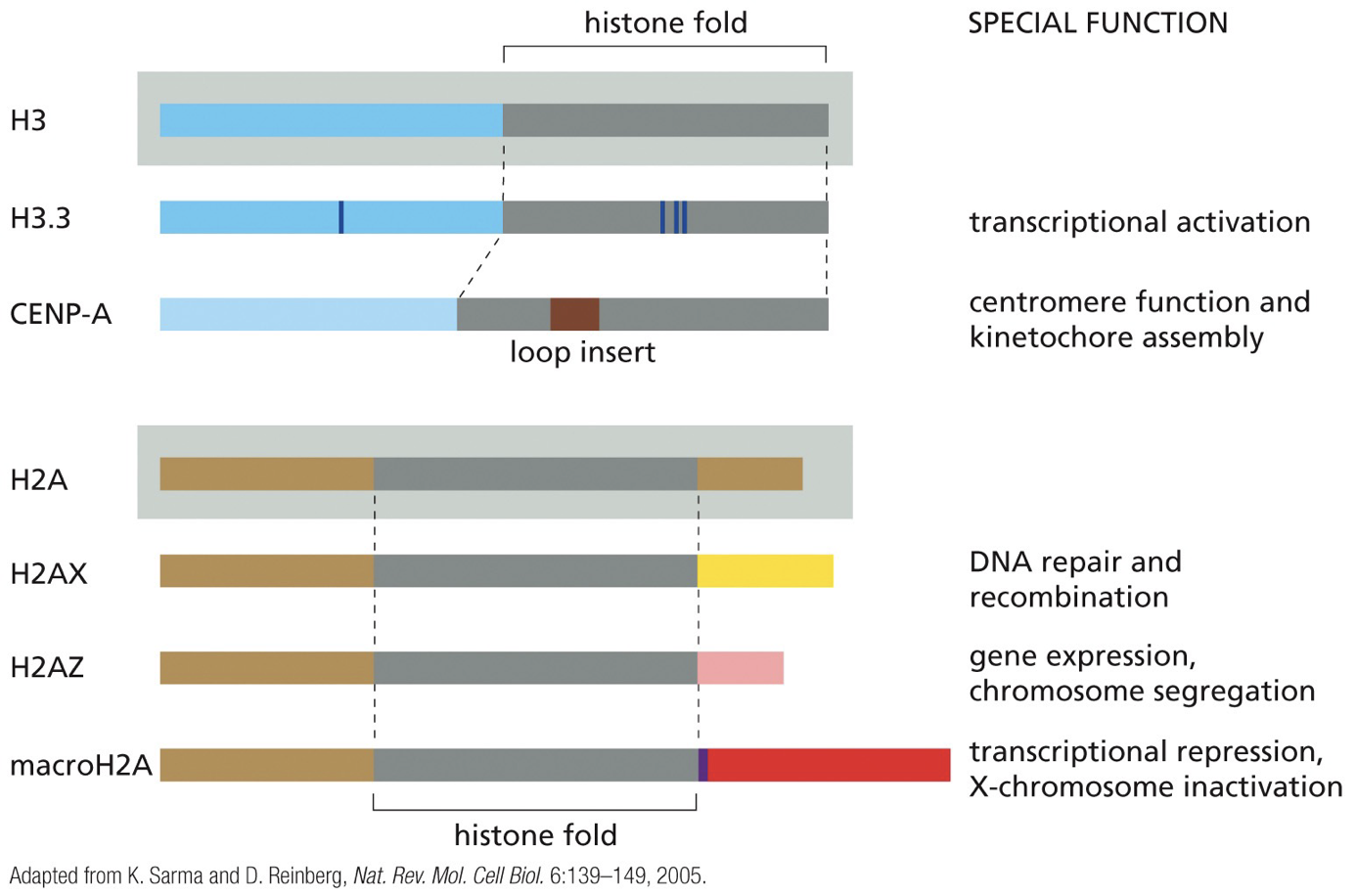
Histone variants
Barrier DNA sequences
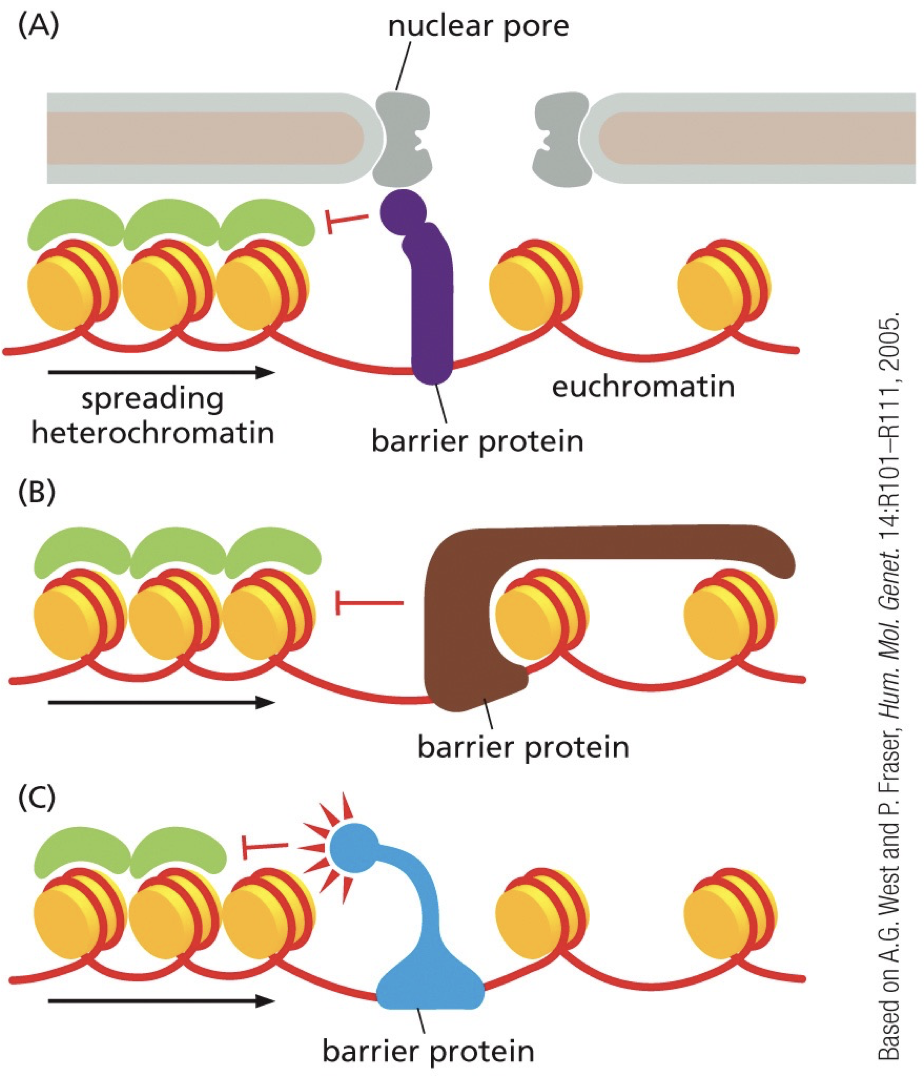
They recruit protein complexes that block that spread of reader-writer complexes and separate chromatin into domains. Insulators. Bind to barrier sequences and create physical barriers or recruit opposing chromatin modifying enzymes. Chromatin becomes separated into domains with different transcriptional activation regulation.
-
Divergence between species is reflected in amount of change in genome. Some closely related eukaryotes have large differences in genome size that reflect differences in loss/addition of non-coding DNA.
****
Genome Maintenance
Exons are highly important, and are something that need to be contained for the organism. Rate of change works in millions of years. Rate of change is in millions of years.
Not all genes and non-coding portions of DNA accumulate at the same speed. There is no selective pressure to keep certain parts the same. Change from bacteria to eukaryotes can be seen by tracking a gene that does not matter. All regions accumulate change, just at different rates. Between and within genes may have different levels of importance controlling their rate of change.
Multispecies comparison
Repetitive DNA correlates with more genes. There are regulatory sequences (5%) that are conserved.
Protein coding sequences: more highly conserved than genome size and organization
Exons: more highly conserved than introns
Humans v mouse: certain areas outcompeted

Mechanisms of change in DNA sequences
Permanent changes are in sequences of nucleotides.
Changes occur by damaging agents, and despite repair mechanisms, some mutations squeak through.
Bacteria error rate is 3 mistakes for 10^10 nucleotides copied
Human error rate is 1 mistake for 10^10 nucleotides copied
But given a million years and a population size of 10 000 diploid individuals, this means every nucleotide substitution would be tried out 20 time.
Mutation: Change in DNA sequence
Point mutation: switching one nucleotide for another nucleotide
Large-scale rearrangements: include deletions, duplications, inversions, and translocations.
Mutations, point-mutations and large scale rearrangements all come in and influence genes that exist.
Only 5% of genes show conservation, and coding genes only make up 1/3 of that.
Repetitive DNA
Portion can be classified as highly repetitive DNA, also known as tandem repeats.
Tandem repeats: To be highly repetitive you would need 10^5 copies of sequence repeated over and over without interruption.
Satellite DNA: 5-500 base pairs in tandem
Minisatellite DNA: 10-100 base pairs with up to 3000 repeats. Highly variable. Differ between individuals and generation. Gets into DNA fingerprinting. Change quickly enough to tell a difference between humans. Criminal and paternity tests.
Microsatellite DNA: 1-5 base pairs in clusters, 10-40 base pairs scattered quite evenly. Highly variable mutations, can be used to compare closely related populations. Good for phylogenetic trees.
Portions can be classified as moderately repetitive DNA. about 20-80% of genome depending on the organism. Repeats a few times to tens of thousands of times. Can include genes or non-coding DNA.
Ex. Many copies of rRNA and histone. They are sequences are usually identical and present in tandem.
Repetitive DNA sequences are unstable. Easily expanding and shrinking due to slippage, where you hydrogen bond with the wrong base pair. Misalignement occurs and leads to deletions or expansions.
Microsatellite instability contributes to progression of some diseases.
Large-scale chromosomal rearrangements
Each chromosome in a mouse genome was coloured and then sought out in human genomes. The genes coding for things in a mouse’s 7th chromosome is not the same as what is happening in a human’s chromosome. This occurs by events that move blocks of DNA. Same genes in same order in different sections.
Regions of synteny: a block that carry genes in a conserved order
X-chromosomes in mice and humans similar with x-inactivation. If the genes are brought elsewhere they may end up being inactivated.
Changes within and between chromosomes
Intrachromosomal rearrangement: usually in euchromatin, predisposed to large deletions, inversions or further duplications. Within 1 chromosomes.
Interchromosomal rearrangement: common in pericentromeric or subtelomeric regions.
DNA moves itself: Jumping genes
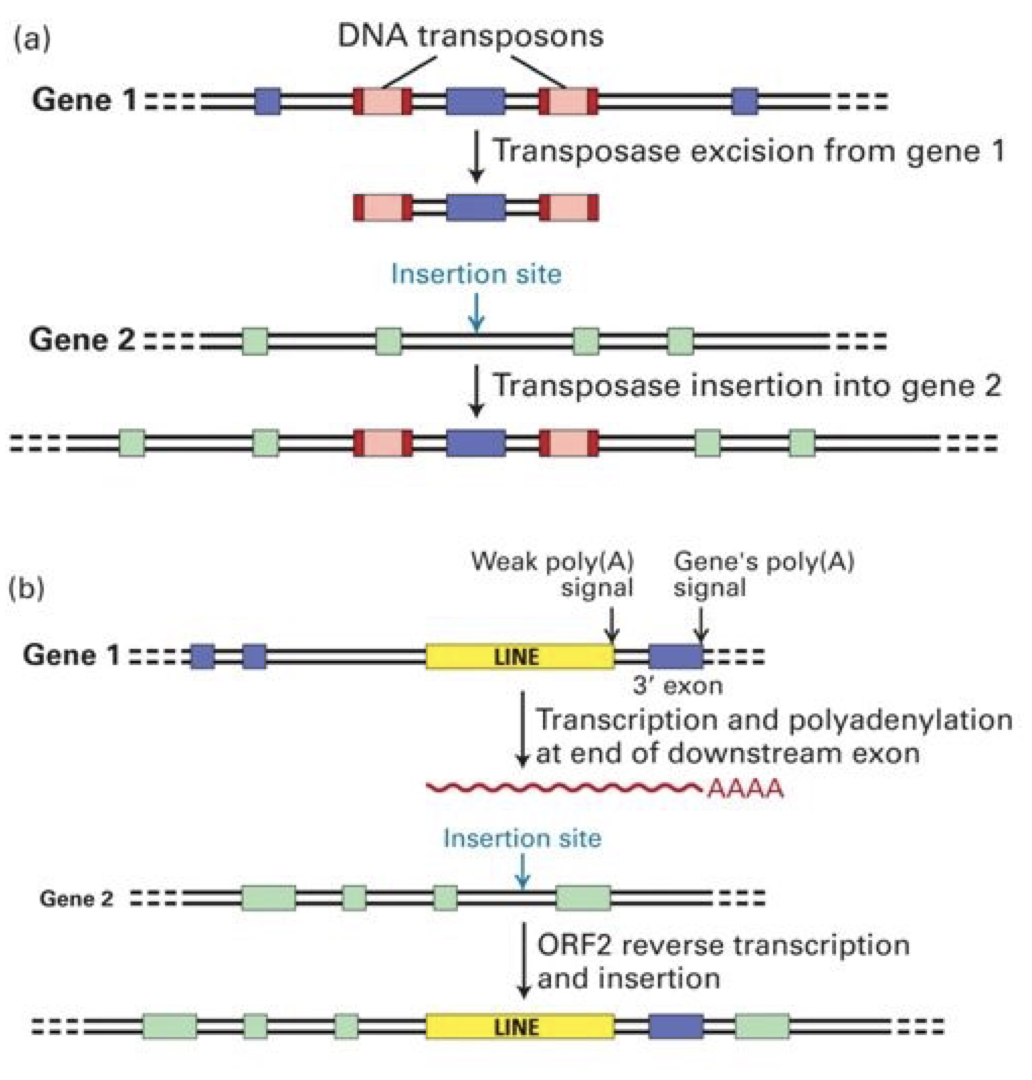
Mobile DNA: DNA that moves from one place to another in the genome. This genetic rearrangement is called transposition and moves mobile genetic transposable elements. Transposition contributes to repetitive DNA sequences.
Transposons: Jumping genes
DNA transposons: Cut and Paste.
Catalyzed by transposase enzyme. Creates repetition.
Inverterted repeats on the end of the transposon are required for recognition by transposase and excision from the donor DNA.
The direct repeat is generated in the recipient DNA.
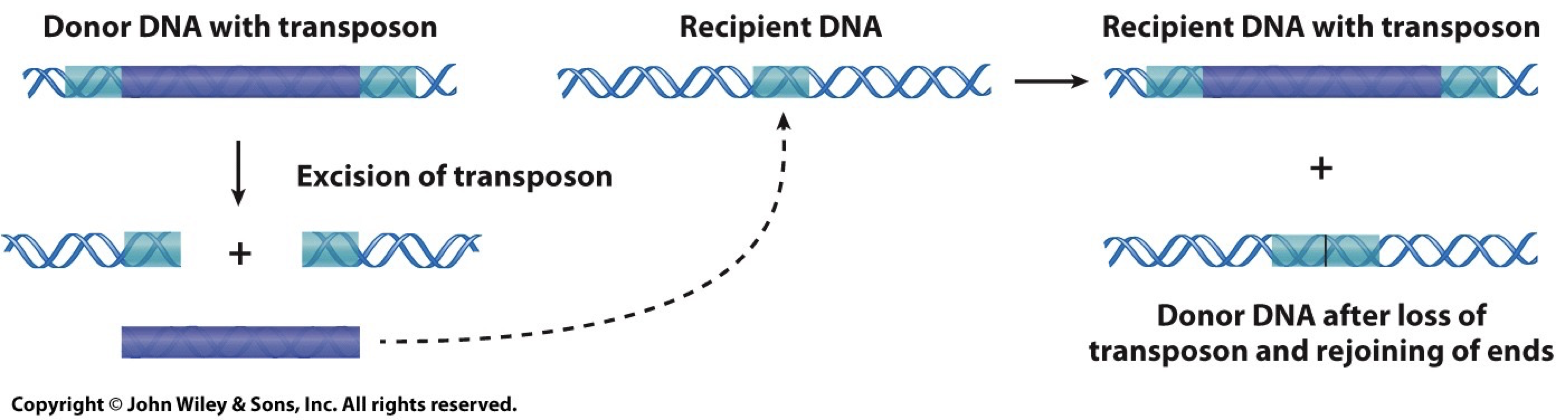
Retrotransposons: Copy and Paste.
Involves RNA intermediate
LINEs and SINEs
Can encode reverse transcriptase enzyme to catalyze production of DNA and RNA
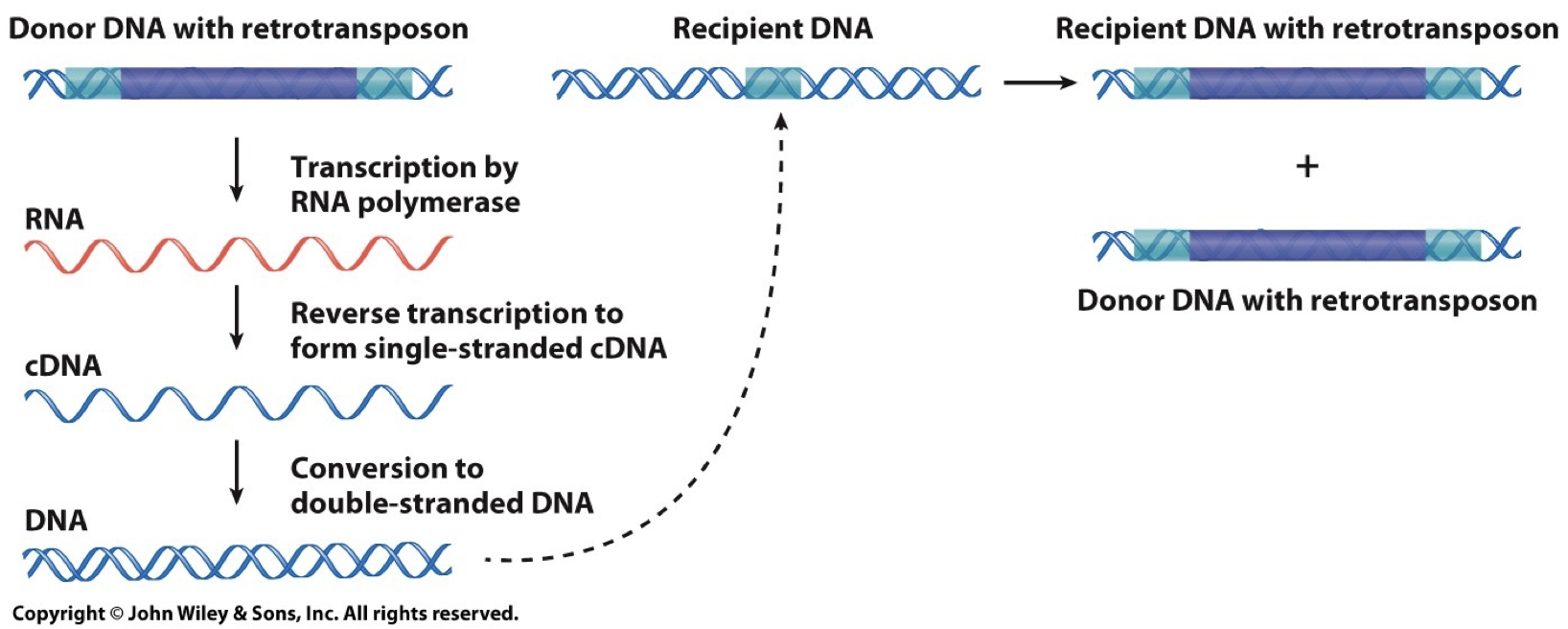
Transposon derived repeats are the major source of interspersed repetitive DNA. Less than 40% of the genome derived from this, though only a minority are actively transposing.
Transposition can cause mutation and play an important role in evolution. Some viruses use transposition to move into host genomes and source genetic change.
Animals and plants high higher frequency transposition (10-100 transposition per offspring) opposed to bacteria (one per every 100 000 divisions). This occurs because there is more non-coding DNA.
Transposons cause genetic change
Exon shuffling: transposons can pick up other sequences. If your genes pickup the transposon, it is more likely to land in an intron. Transcribed and then reversed transcribed, and it could also transcribe additional sequences.
 Unequal crossing over
Unequal crossing over
A situation in meiosis where, during crossing over, the genes misalign and one gene gives and another genes gets, but there is no reciprocation. One gene will have extra DNA and the other will be missing DNA.
Successive unequal crossing over leads to tandem arrays in DNA.
Gene duplication
Point mutation changes one and that creates a new allele, may function just slightly differently. Genes duplicate because it creates redundancy. If we have two copies and mutate one, the organism will still survive because of the original copy. Gives us an opportunity to experiment.
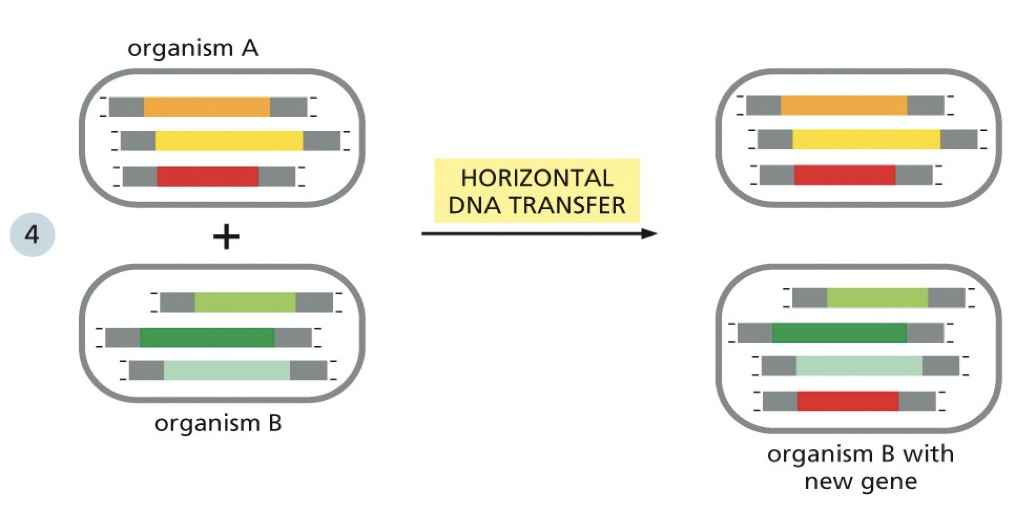
Horizontal gene transfer
Can create genes. Small portions get added in. When a change occurs that is very damaging to the organism, it won’t survive to have offspring or the offspring won’t survive. Only beneficial mutations are passed down.
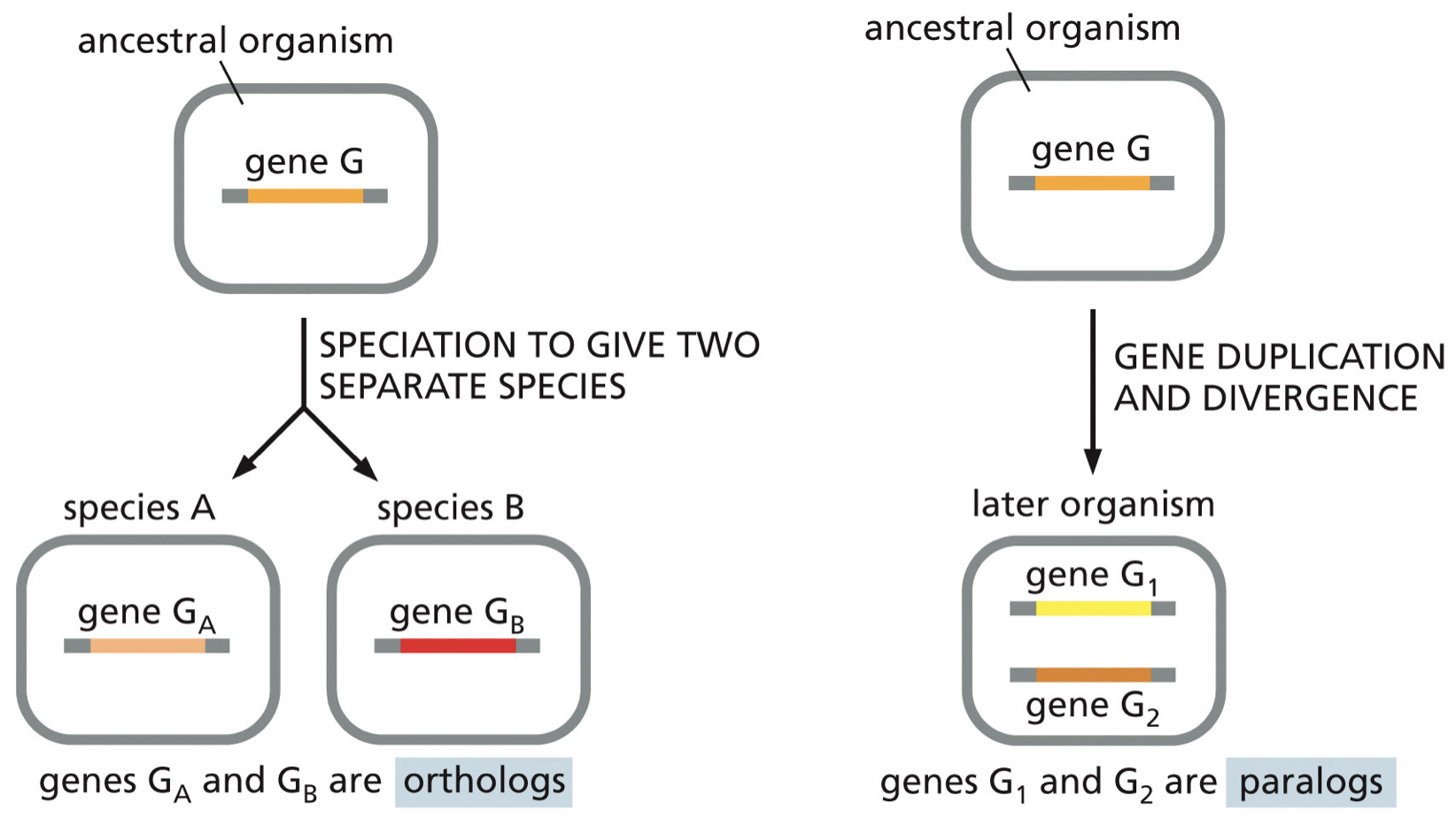
 Duplication can result in two genes
Duplication can result in two genes
Orthologs: speciation given two separate species
Paralogs: gene duplication and divergence
Homologs: any genes that are similar due to common ancestry (include orthologs and paralogs).
 Globin gene family
Globin gene family
Different family with different functions. Similarities in different structure. Lots of mutations. Comparing their DNA sequences will show a lot of similarities but also some divergence.
Pseudogenes: when we have redundant replication and mutations have been accumulated in genes to the point that they’re now non functional.
Globin genes in humans are orthologs (and homologs)
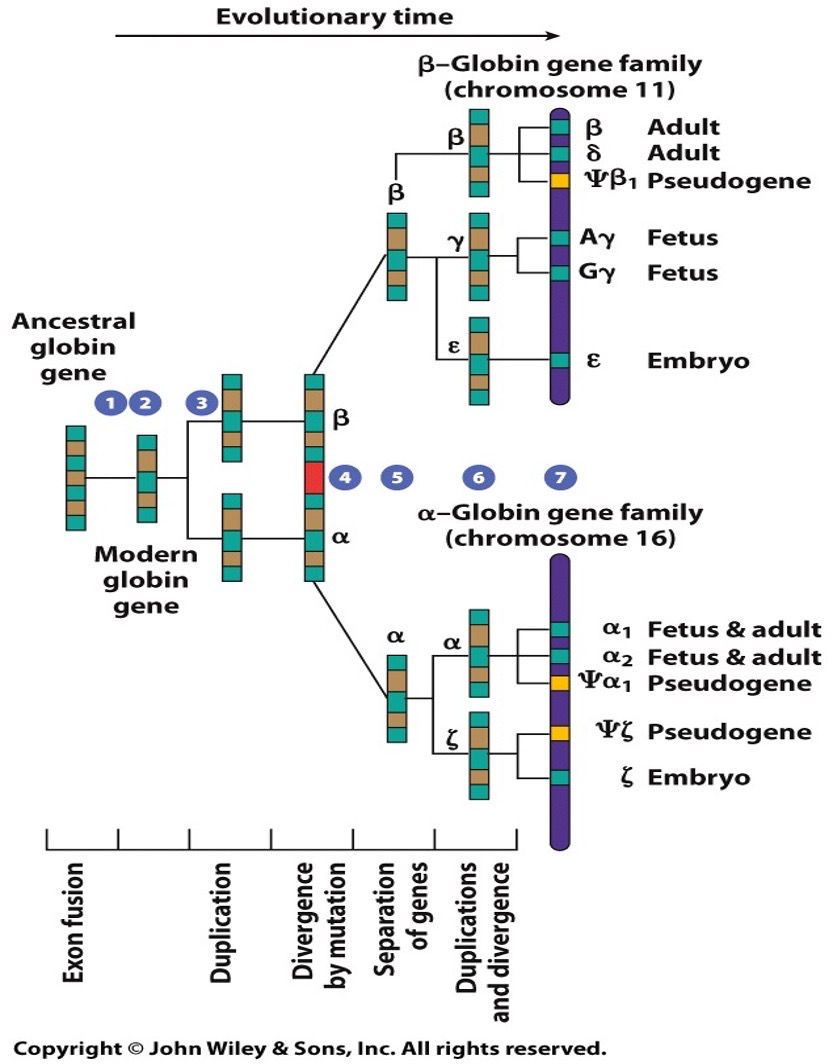
Humans differ from chimpanzees by about 4%. The FOXP2 gene encodes a transcription factor where the protein sequences differs by 2 amino acids. Human sequences of FOXP2 is required for specific lip and tongue movement (oral communication). Mutations in FOXP2 stop oral communication.
Single Nucleotide Polymorphisms (SNPs)
Most common. In protein coding regions, they contribute to phenotypic differences in humans (alleles). Changes on nucleotide and its base pairing.
Set of SNPs we can track (are always inherited together) can help us with understanding genetics. Splitting them up and having Mendelian probability that they’ll end up in the same gamete. Find them in the same place and they never undergo crossing over.
Series of these may be all “AAA” or “AGA”
Haplotypes: a particular combination of alleles on a chromosome that are inherited together
SNPs can be useful in understanding why certain complex diseases tend to run in families. Not all are carried on and given to offspring (meiosis).
Copy Number Polymorphism (CNPs)
Differences in the number of copies of a particular sequence. In protein coding region, extra copies mean more protein, and there is a phenotypic difference.
Structural variation
Large segments of DNA change by duplication, inversion, deletion, etc.
DNA mutations and structures of replication
Lowering error rate
A low error rate is essential to survival. DNA polymerase adds new nucleotides to a growing DNA strand in the 5’ to 3’ direction by adding a new nucleotide onto a correctly positioned 3’OH.
The existing DNA strand serves as a template and if an error disrupts complimentary-base pairing, then it usually detected and corrected
Proofreading function
DNA polymerase has a catalytic site for polymerization and for editing. DNA is pushed into the editing site when a mismatch is detected.
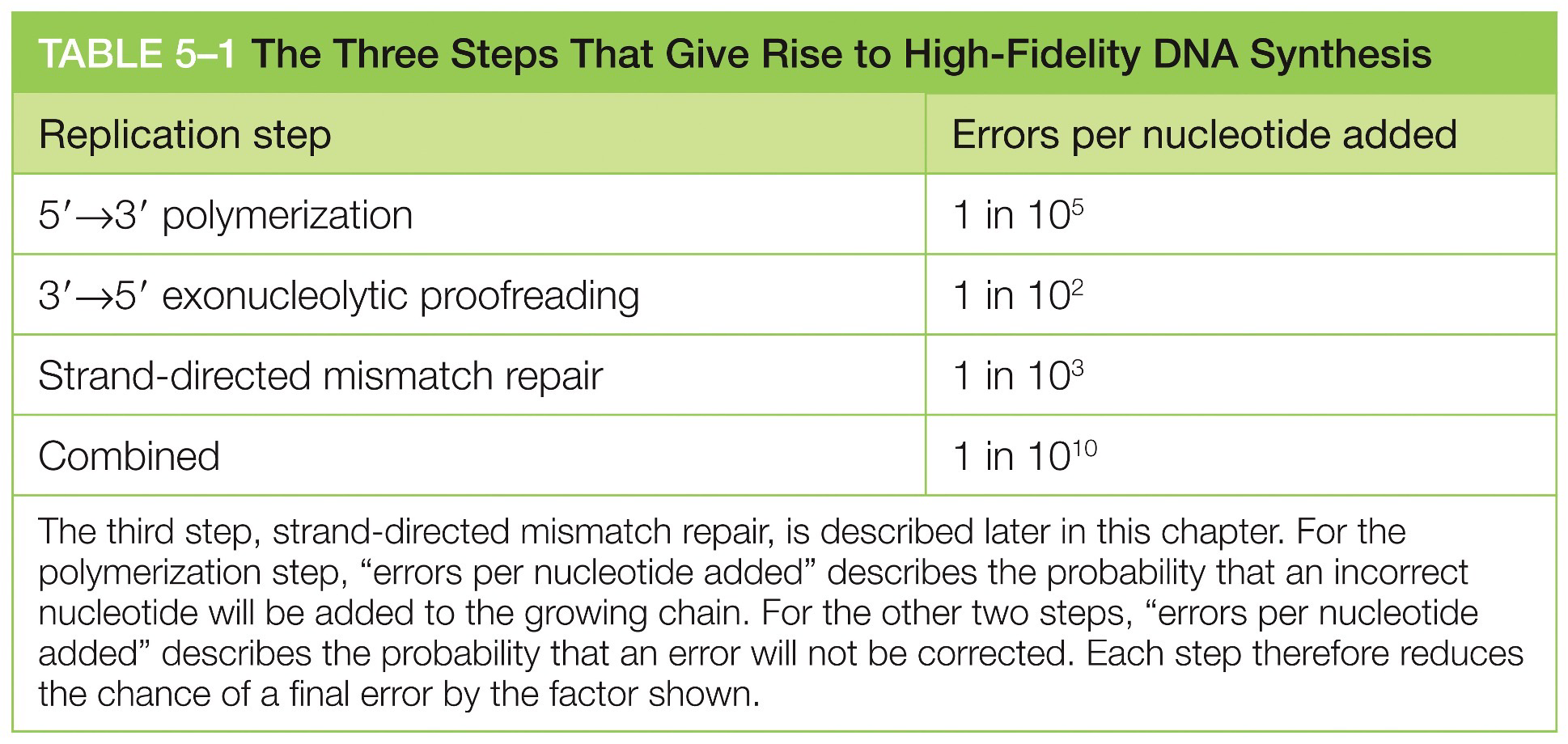
Increased error rate
Cancer would come up in somatic cells. Mutations in germ cells may cause infertility.
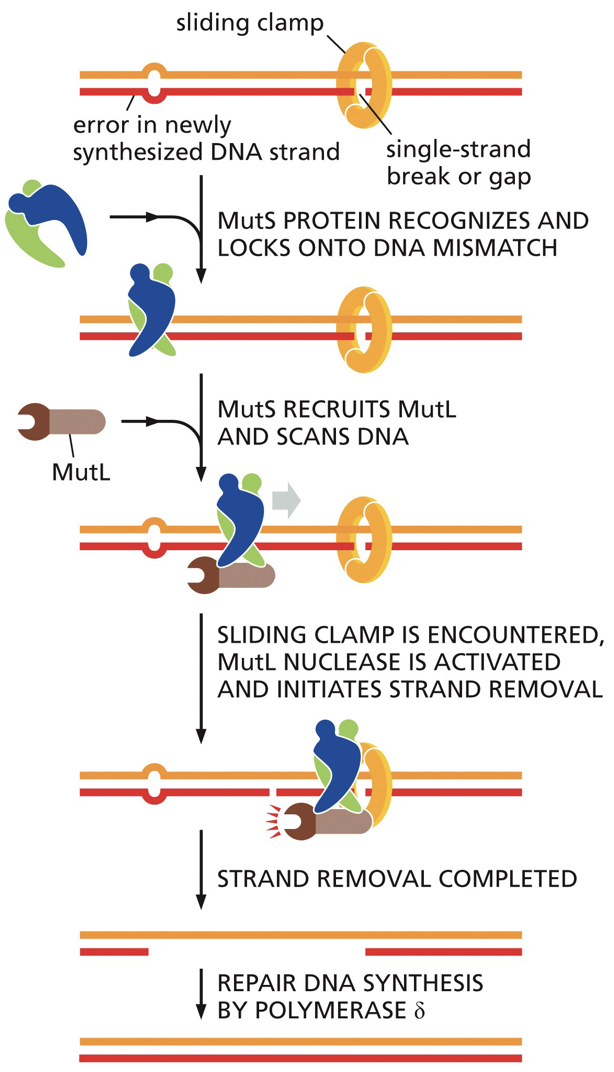
Strand-directed mismatch repair
This involves a recognition of a mismatch, identification of newly synthesized strand, removal of incorrect nucleotides from new strand, resynthesis of excised section, ligation to seal DNA backbone
Distortion of double helix: it is used for recognition of mismatched base pairings.
We can distinguish which strand is newly synthesized and target that strand for correction (using the other strand as the template)
 DNA replication models and structure
DNA replication models and structure
Early proposed models
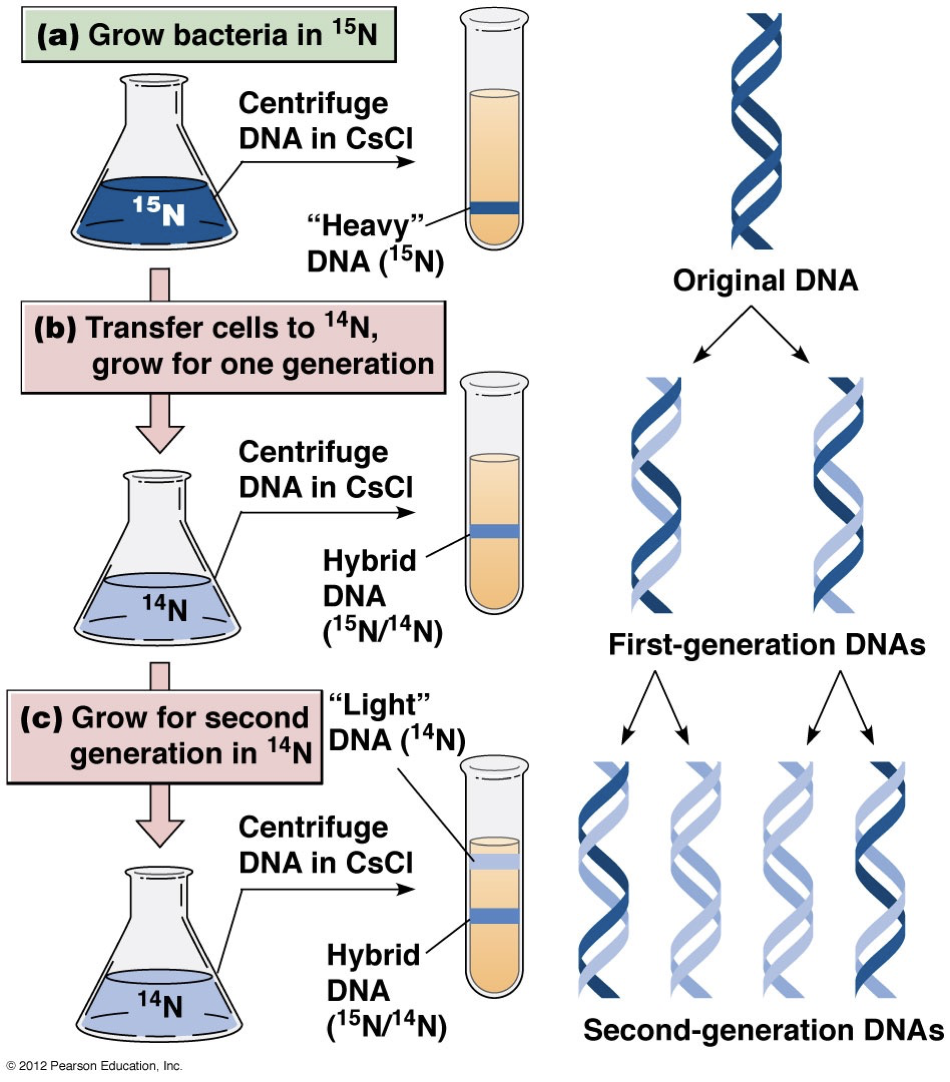
Semiconservative replication: Pull it apart and have each strand duplicate and have a different (anti-parallel identical) strand attached.
Conservative replication: A completely separate new DNA strand duplicates and the strands stay intact.
Dispersive replication: Parent strands are broken into smaller fragments and used for daughter strand synthesis.
Experiment
The semiconservative model was proved correct. One band is labelled and heavier (15N). We check where it is.
The newly synthesized DNA includes a lighter strand (14N), so it won’t sink as much in the CsCl solution gradient.
After the first generation, we see a single band in-between where we expect to see all heavy and all light DNA. This eliminates the conservative model.
In the second generation, there are two bands for the lighter DNA and heavier DNA. This eliminates dispersal method.
 5’ to 3’
5’ to 3’
Incoming nucleoside triphosphate are added to 3’OH as we create the backbone. Energy forming is already contained in the phosphate bonds of incoming nucleoside triphosphate. Catalyze the energy by breaking it.
How structure of DNA makes replication possible
Hydrogen bonds are pulled apart through a process called denaturation, and re-associated by renaturation.
Renaturation is critical for RNA transcription and DNA replication.
Denaturing works by heating up so you get enough heat energy to break and pull bonds apart. There is a small temperature range. Thermal DNA denaturing is called DNA melting.
We can monitor via UV light absorption. We get no change for awhile, and then after a specific temperature, there is a big jump in absorption of UV light by the DNA/cell, and this occurs until DNA is completely denatured. This helps us learn the temperature of melting DNA sequences.
Tm = the temperature at which the shift in absorption is half completed.
Thermal denaturation of DNA is effected by base composition
G/C is held together by 3 hydrogen bonds so it has a higher melting point, while A/T have 2 hydrogen bonds so they have a lower melting point.
Similarly, the rate-limiting step of renaturation is complementary base pairings. Once they find each other they zip up quickly. Base pairings have to find each other.
Small genome with fewer strands had less wrong answers to sift through and will go faster.
DNA renaturing graph:
Fragment DNA molecules into small pieces → raise temp to denature DNA → slowly cool to see how long it takes to renature
X-axis: Initial DNA concentration (C0) by time of reaction (t)
Y-axis: fraction of original DNA concentration that is renatured (dsDNA)
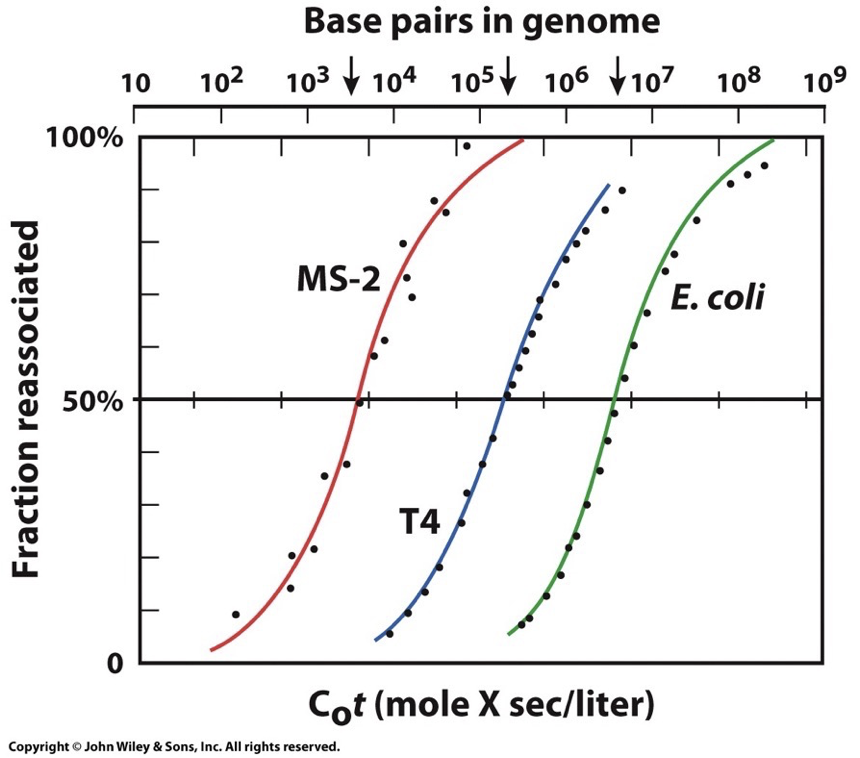
Repetitive sequences will be faster because they have more options. Unique base pairings take the longest.
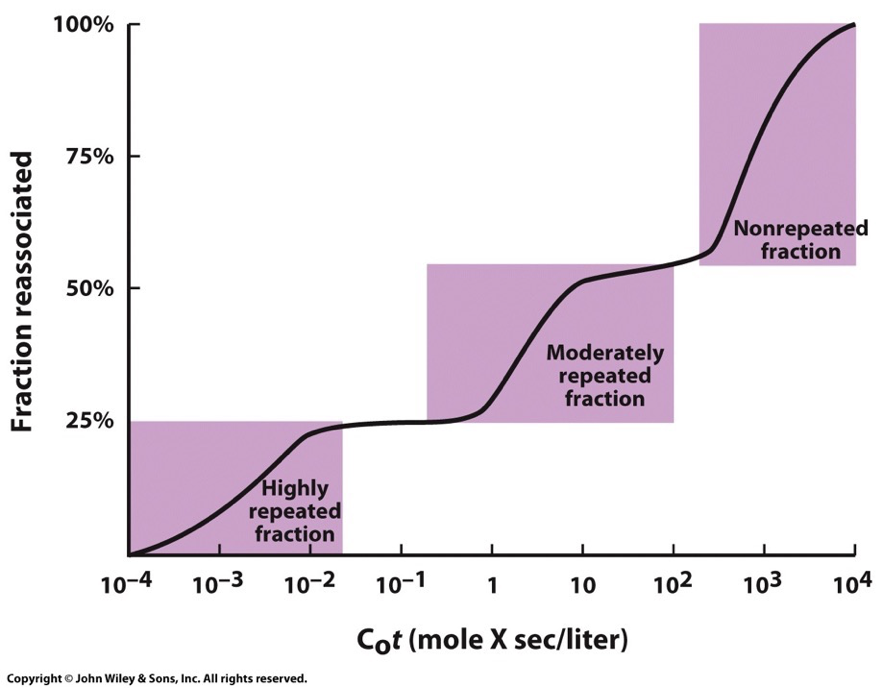
This plot for eukaryotic genomes shows that fractions of DNA reanneal at very different rates. Usually three distinct steps that represent these broad classes of DNA sequences which reanneal at different rates due to number of times the nucleotide sequences are repeated within the population of the fragments.
****
DNA Replication
Prokaryotes
Many proteins are required for DNA replication in prokaryotes. Initiation of this process occurs at the origin of replication by sequence recognition. To be able to begin, we have to open up the double helix and make the “bubble”.
Replication will proceed outwards in both directions (replication forks). Initiation sequences are rich in A-T base pairs because they have less hydrogen bonds and so they require less energy to pull apart.
Replication forks: points where a pair of replicating segments come together and join the non-replicated segments. They move in opposite directions at 1000 nucleotides per second. They eventually move around the circular genome and meet back together, once entirely around it separates.
1 origin of replication, 2 replication forks, both forks move away from each other, DNA polymerase moves 3’ to 5’ on the template strand and 5’ to 3’ on the new strand.
Systems
The systems required in DNA replication consist of Helicases, single stranded DNA-binding proteins (SSBs), topoisomerase, primase, DNA polymerase(s), and DNA ligase.
Helicase: DNA is pretty stable, the double helix has to be pulled apart. A-T rich sequences and their hydrogen bonds interact with helicase and ATP hydrolysis. The helicase opens up the DNA double helix ahead of the replication fork, making the template strand accessible.
Single-stranded binding proteins (SSBs): Single stranded DNA has a tendency to fold on itself to make hairpins, and the SSBs can help to stop the DNA from refolding. They bind and stabilize single stranded DNA after the helicase unwinds the double helix.
DNA Primase: DNA polymerase can’t start from nothing, and it needs 3’OH to build from. Polymerase can’t do something from nothing. DNA primase starts a short segment of RNA to then build off of, they will be removed, but they allow DNA polymerase to start (about 10 nucleotides).
Topoisomerase: When double helix is opened up there is overwinding ahead of the fork and this aids it. Replication requires unwinding of DNA which causes torsional stress (positive supercoiling), which can stall replication. Topoisomerase relieves the stress and can be ahead of helix.
Topoisomerase I: catalyzes to breaking/nicking of one DNA strand to allow for rotation (relieve strain).
Topoisomerase II: catalyzes a double strand break, detangling the DNA.
Sliding clamp: DNA polymerase has a tendency to fall off the DNA template and this prevents that. It is associated with polymerase to keep it in association with the DNA molecule. If it is bounded to clamp, it won’t fall of. A clamp loader will open up the ring and then add the sliding clamp to the DNA.
Okazaki fragments: DNA polymerase CAN’T synthesize 3’ to 5’ and these aid in that. Synthesis of each fragment waits for parental strand to separate and expose additional templates (lags behind leading strand synthesis). Primase adds RNA primer to each segment, and is later removed and the gap is filled in. DNA ligase covalently connects fragments into a continuous strand.
Leading strand: synthesis continuously in direction of replication fork
Lagging strand: synthesis discontinuously in direction of replication fork. The first one made is farthest from replication fork.
DNA polymerase: Synthesis of new DNA has to be carried out by family of DNA polymerase (different kinds that all require DNA template, primer for 3’OH, and all four deoxynucleotides available). New DNA strand is complementary and anti-parallel to the template strand.
Proofreading: If DNA polymerase adds the wrong nucleotide, it can remove it by 3’ to 5’ exonuclease activity. It can tell because mismatched nucleotides don’t fit well into the enzyme’s active site.
5’ to 3’ exonuclease can remove RNA primers.
In DNA polymerase I, both 5’-3’ and 3’-5’ exonuclease can degrade DNA or RNA.
Replisome: We have to keep everything coordinated and going at the same time. The replisome is a multiprotein complex with contact to both leading and lagging strand to keep their replication coordinated.
Trombone model is designed to keep the strands in same physical location by looping.
Eukaryotes
Eukaryotic genomes have more issues
Timing with mitosis (requires extra coordination)
Nucleosome (are going to be in the way, need a remodelling enzyme)
Large genome size (cannot replicate as fast)
Chromosome ends (linear and have to deal differently)
Mitosis
Bacteria can initiate DNA replication continuously, however eukaryotes are more complex and both DNA replication and progression through the cell cycle must line up. Nucleus and cell divides in M phase. S phase is when DNA synthesis occurs. S phase lasts 40 minutes to 8 hours
In S phase, histone mRNA production increases a lot.
Nucleosome
Chromatin remodelling complexes: destabilize nucleosomes before fork gets there. Histones are displaced and re-established behind the replication fork. Position of nucleosomes determine length of Okazaki fragments.
Each nucleosome takes up half old and half new histones. This happens because each strand carries half of the previous histone modifications to pass on epigenetic regulation for each region of DNA. We want our epigenetics to be stable as we make the new cells. Keep the liver cells as liver cells.
Large genome size
There are multiple origins of replication in eukaryotic cells. Eukaryotic cells replicate their DNA in small portions called replicons. Each replicon has an origin of replication, we have 10k to 100k different origins in a human cell.
Initiation of DNA synthesis in replicon is highly regulated and requires assembly of an ORC (origin of replication complex).
Chromosome ends
Telomeres are sequences at the end of the chromosomes and are composed of tandem repeats. The sequences at the end are going to shorten with each replication which is why we need telomeres to protect the end. Our leading strand will be easy, but lagging will be an issue without a 3’OH to complete lagging strand. Telomeres work to stop this shortening.
RNA template serves to create DNA. Reverse transcriptase is capable of making DNA from and RNA template. Telomerase contains its own RNA template. Telomerase extends template strand. Primase + DNA polymerase action then fills in the end of the lagging strand.
Expression of telomerase is regulated (some cells with more or less). This is because some cells divide faster than others. If we had high levels of telomerase activity, cancer would happen. Cancer cells activate telomerase so they can continuously divide.
Stress shuts down telomerase activity.
Fidelity of DNA replication
Low mutation rate is a result of accurate selection of nucleotides, immediate proofreading, post-replicative repair, DNA damage from sources unrelated to replication.
More damage slows down replication and will cause apoptosis.
DNA damage and repair mechanisms
Issues can come from ionizing radiation, reactive chemicals, UV radiation, and thermal energy.
DNA repair mechanisms
Damage | Repair | Key protein |
|---|---|---|
Double strand break in non-dividing cell | Non-homologous recombination (NHEJ) | Ku |
Pyrimidine dimer | Nucleotide excision repair | Excision nuclease |
Double strand break at replication fork | Homologous recombination | Rad51 |
Uracil | Base excision repair | DNA glycosylase |
Point mutation with incorrect base-pairing | Mismatch repair | MutS |
Depurination: very common, 18k events to repair everyday. Removes the base from DNA.
8-oxo-guanine: oxidative damage is also common. It is a tell for amount of oxidative damage in a cell.
Deamination: most commonly converts cytosine to uracil. This results in the cause of a mutation, and we get a point-mutation.
May end up in an A-T base pairing and cannot tell if it was a mutation
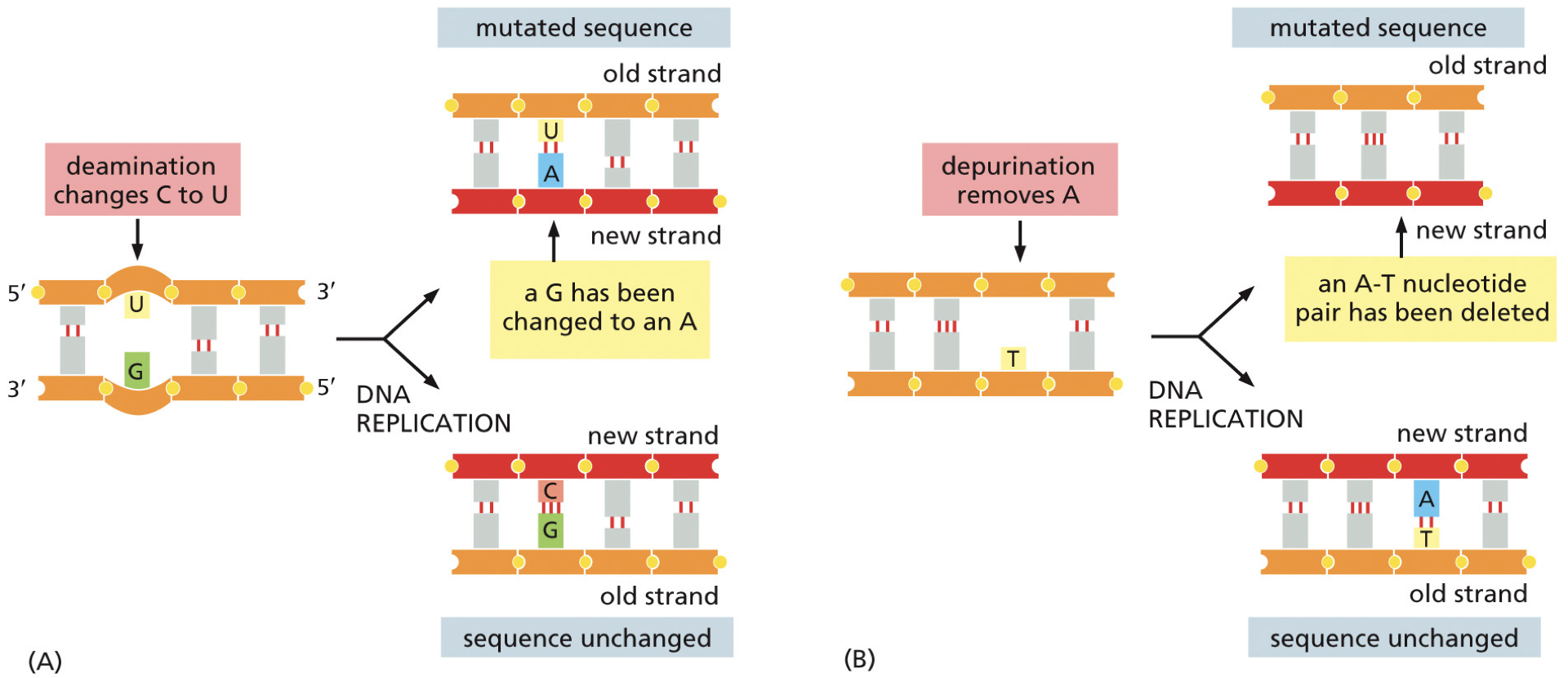
5-methylcytosine is about 3% of C bases, results in thymine. Because thymine is a natural base we cannot recognize with most, can only use mismatch repair
Nucleotide excision repair
Nucleotides are removed; removing bulky lesions (pyrimidine dimers, chemical alterations)
There are two distinct pathways
Transcription coupled pathway: repairs actively being transcribed DNA. High priority of the genes that are of great importance to the cell.
Global genomic pathway: slower, less efficient. Repairs DNA in the remainder of the genome (less important).
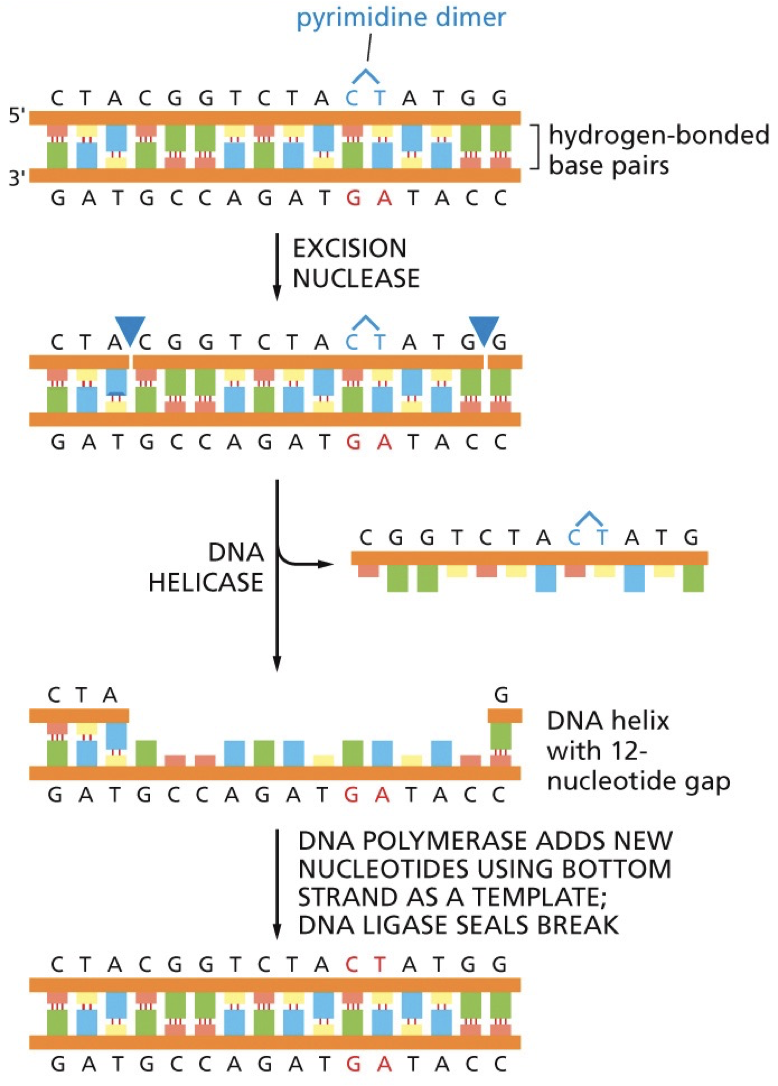
Xeroderma pigmentosum: patients are unable to use NER to repair damage from the effects of UV rays and so they cannot go in the sun.
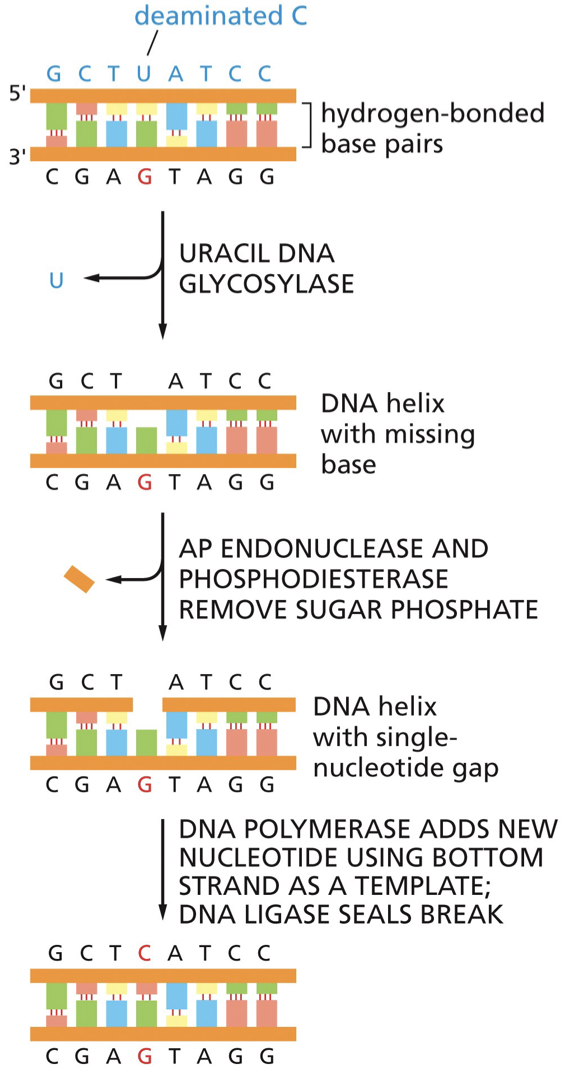
Base excision repair (BER)
BER repairs altered bases. It is initiated by DNA glycosylase enzymes that recognize specific alterations.
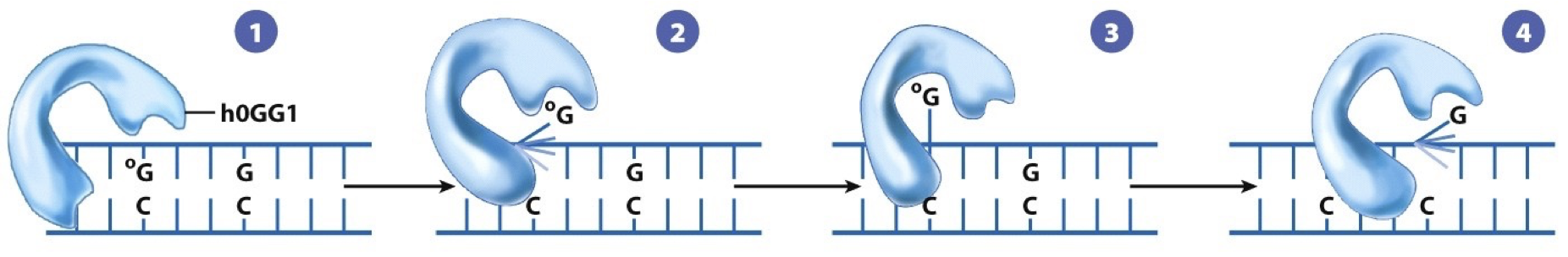
Remove altered bases by cleaving the glycosidic bond attaching the base to deoxyribose sugar. May have distinct transcription-coupled or global repair pathways. DNA glycosylase recognizes an oxidized guanine (8-oxoguanine). The nucleotide is flipped out by the DNA glycosylase, and the incorrect nucleotide fits into the active site and is cleaved from the deoxyribose sugar. Sugar phosphate is removed and replaced by DNA polymerase. DNA ligase seals. G is put back into regular position.
Double stranded breaks
Ionizing radiation causes breaks in both strands, called a double strand break (DSB). Can be repaired through non-homologous end joining or homologous recombination
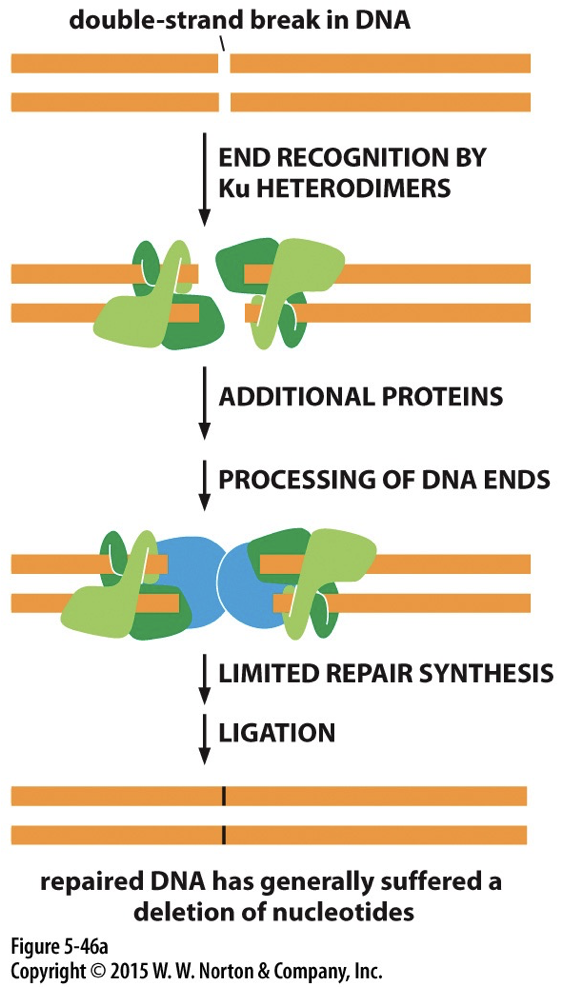
Non-Homologous End Joining (NHEJ)
A highly active pathway. Ku detects the DSB and binds to both ends. The ends are processed and brought together. DNA ligase IV seals the gap.
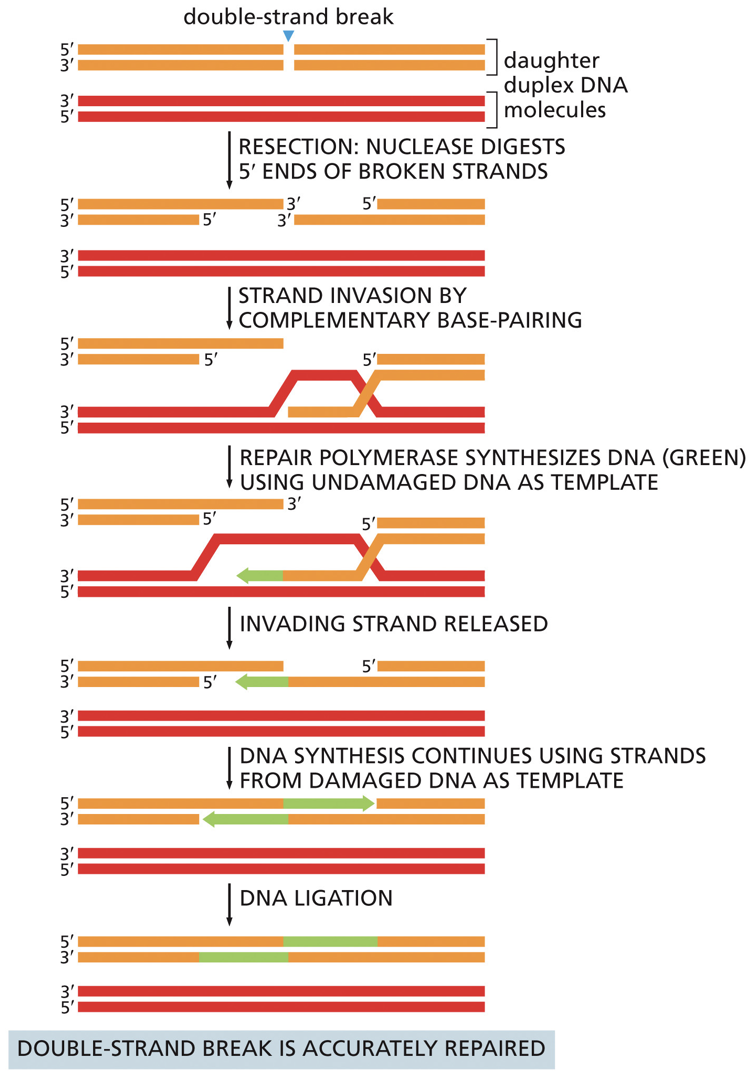
Homologous Recombination (HR)
Nuclease digests a 5’ end. RecA/Rad51 catalyzes strand invasion into the other DNA duplex (must have homology). Strand invasion is similar to crossing over. The idea of joining two molecules together. Undamaged DNA is used as a template and DNA ligase seals backbone.
HR v. NHEJ
HR uses a template and is thus more accurate than NHEJ. NHEJ doesn’t recognize anything and will often lose things. HR is possible because of the source of homologous material because we have homologous chromosomes. NHEJ is more likely to cause translocations. NHEJ is more damaging because at least you have all your DNA in HR.
What could go wrong?
HR is more likely to cause loss of heterozygosity. This can affect cell function because if there is a genetic disease, and you’ve copied it over you can have bad things happen. This is called loss of heterozygosity.
Crossing over is different from HR that occurs because you will still have all the DNA sequences and in turn won’t lose heterozygosity.
****
Gene Expression
Variations in gene expressions occurs with different phenotypes and DNA diverging to create new phenotypes. In our body we have a lot of different cell types which have different morphology, and this is because of differentiation through euchromatin and heterochromatin.
Differential gene expression: rather than changes in DNA, we get changes in what RNA is represented. Not all genes have to be expressed the same. Gene A may produce many copies of RNA, while B doesn’t produce as much. Gene A is more actively transcribed. Because of amplification, you can get more than one protein out of an mRNA.
When staining DNA, darker bands are higher density DNA. The lighter areas are less condensed areas that are being transcribed. We get to see that different regions of the DNA are being expressed differently. This differs in different cell types.
Messenger RNA (mRNA) is translated into protein.
Structure and Function of RNA
Similarities to DNA:
Single strand structure is similar.
Sugar backbone, 5’ to 3’ directionality, complementary base pairings (RNA can fold on itself)
Overall structure of a nucleotide has same rules apply.
Differences from DNA
RNA has ribose and uracil
It is single stranded
RNA folds differently. It does not stay the same, it folds and makes complex structures. RNA can have functional catalytic activities due to forming complex base pairings.
Non-conventional base pairings: not repaired as often, the backbones flip around and bound A-G, U-C. DNA would be more active in their repair mechanisms than RNA.
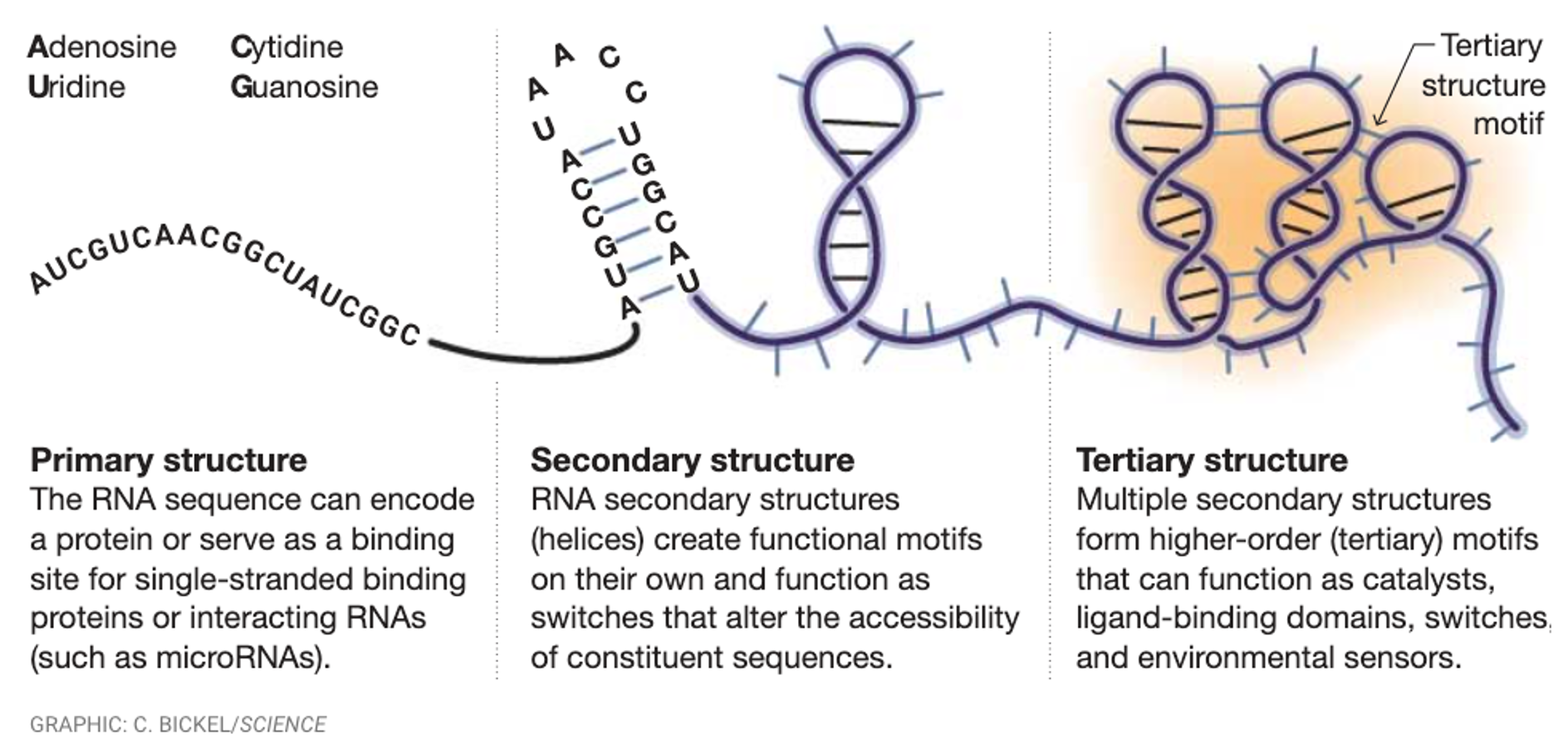
Types of RNA
mRNA - intermediate between DNA and proteins. Template for protein synthesis.
rRNA - doesn’t have informational purposes, doesn’t code, but it provides structural support and catalytic activity.
tRNA - not translated themselves, they work as RNA molecules and bring correct amino acids into translate
rRNA and tRNA stay for awhile and mRNA has a shorter lifespan because there is not higher higher order repair mechanism. This is because if you want to do differentiation you need old RNA gone, as well as overturning RNA quickly gives it more regulation.
Transcription
RNA created by RNA polymerase with one DNA serving as a template strand. Some will be translated into proteins some won’t. Information storage to information usage.
Promoter sequence tells where transcription will begin and on what strand.
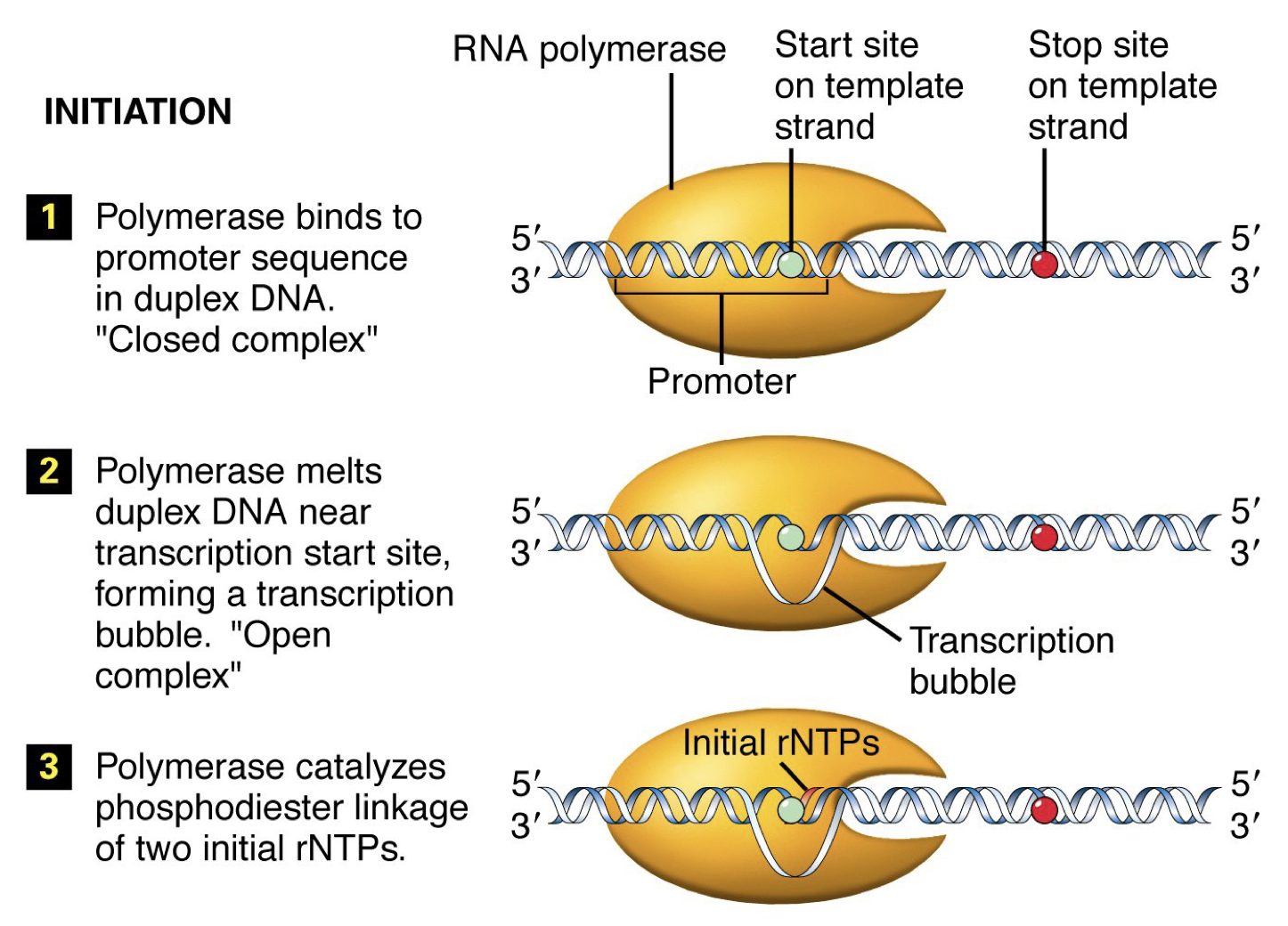
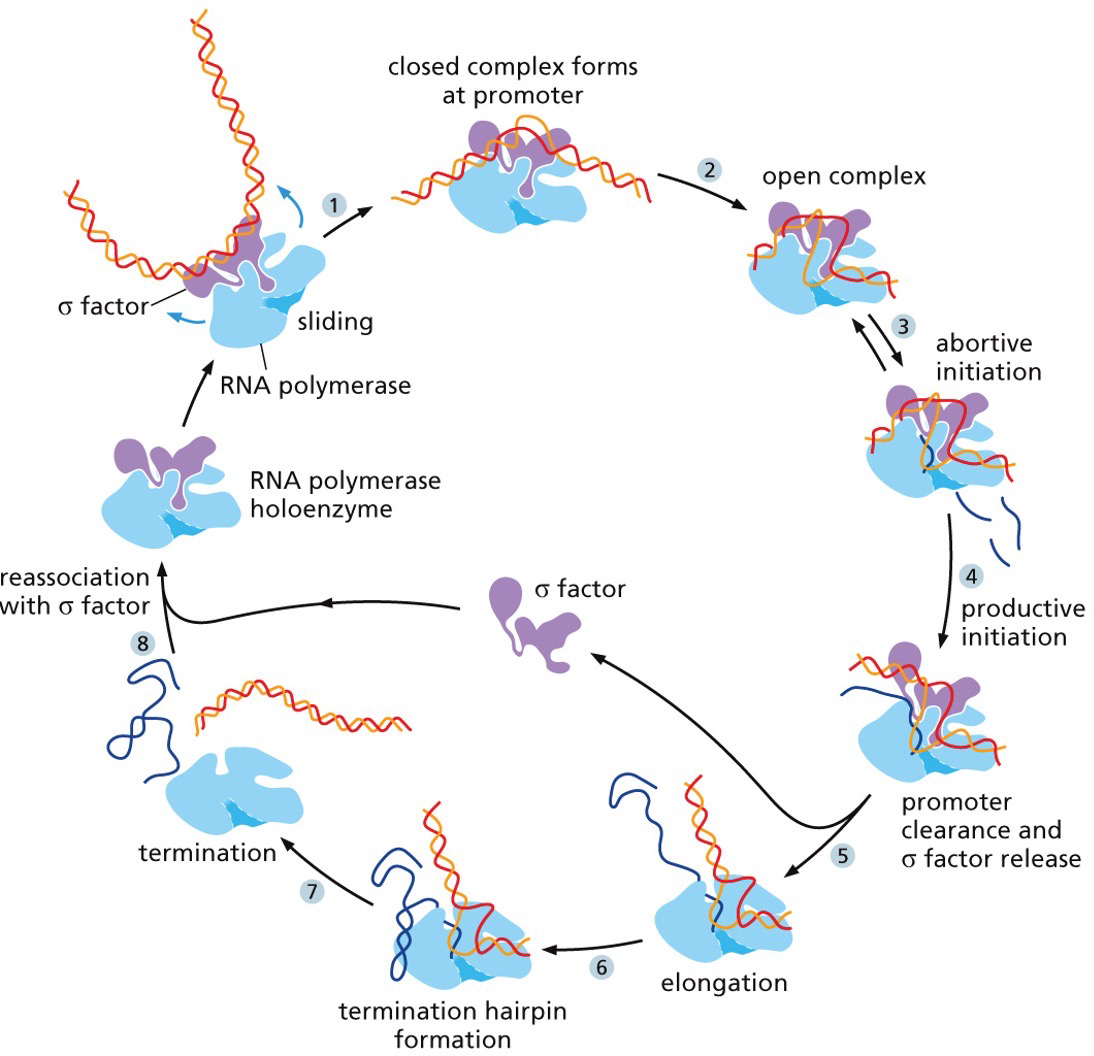
Initiation: RNA polymerase binds to the promoter sequence and helps move transcription factors.
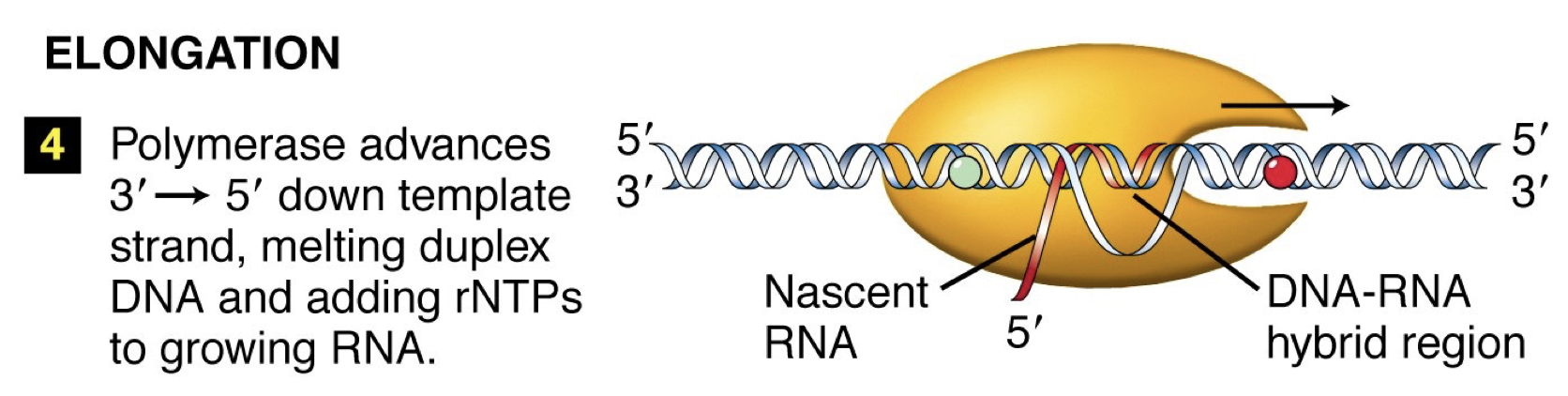
Elongation: Conformation change occurs in RNA polymerase. Move down, unwind, synthesize complementary DNA. Linking by phosphodiester bonds in a 5’ to 3’ direction like DNA replication.
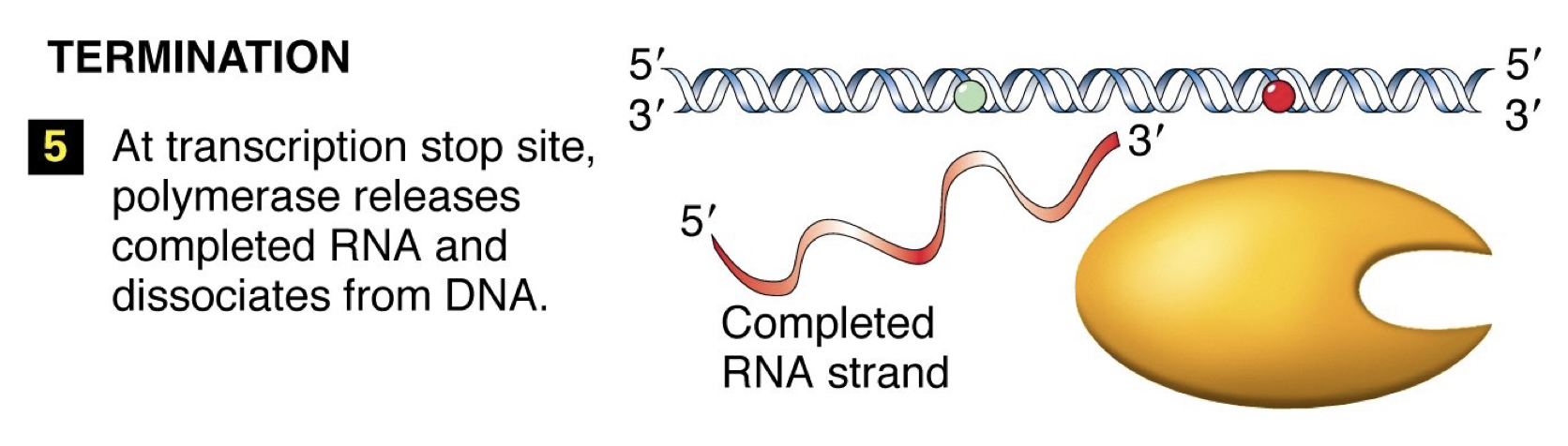
Termination: RNA polymerase reaches signal sequence to cause extended pause of synthesis. Transcript is released from the enzyme. Polymerase floats off to find another promoter.
 Template strand: Antisense or noncoding stand (3’ to 5’)
Template strand: Antisense or noncoding stand (3’ to 5’)
Opposite strand: Sense or coding strand (5’ to 3’)
RNA Polymerase
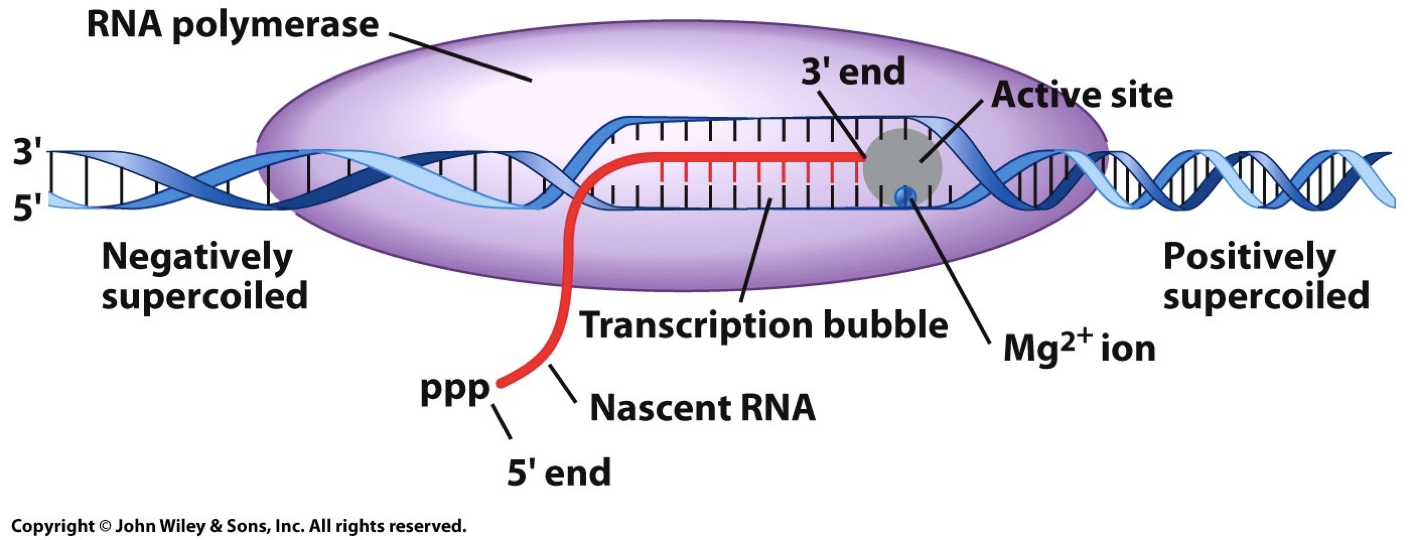
Promoter sequence
In prokaryotes (and eukaryotes), transcription cannot get started on its own. RNA polymerase on its own is no good, and thus there is a sigma factor there to increase affinity for promoter sequence. RNA polymerase holoenzyme is five subunits and the sigma factor.
 The promoter sequence has a +1 sequence where transcription begins, and then a -35 and -10 elements
The promoter sequence has a +1 sequence where transcription begins, and then a -35 and -10 elements
Consensus sequence: most common sequence for a particular task, and they may vary a little, but there’s a similar set. In some sites T is most common and others have more wiggle room. See how proteins interact.
Termination sequence
When RNA transcription is terminated there are 2 mechanisms
Intrinsic mechanism: a hairpin structure stops the sequence
Protein mediated system: a particular sequence is transcribed and have a consensus sequence. The protein will recognize it and help separate the transcript.
Transcription in eukaryotes vs. prokaryotes
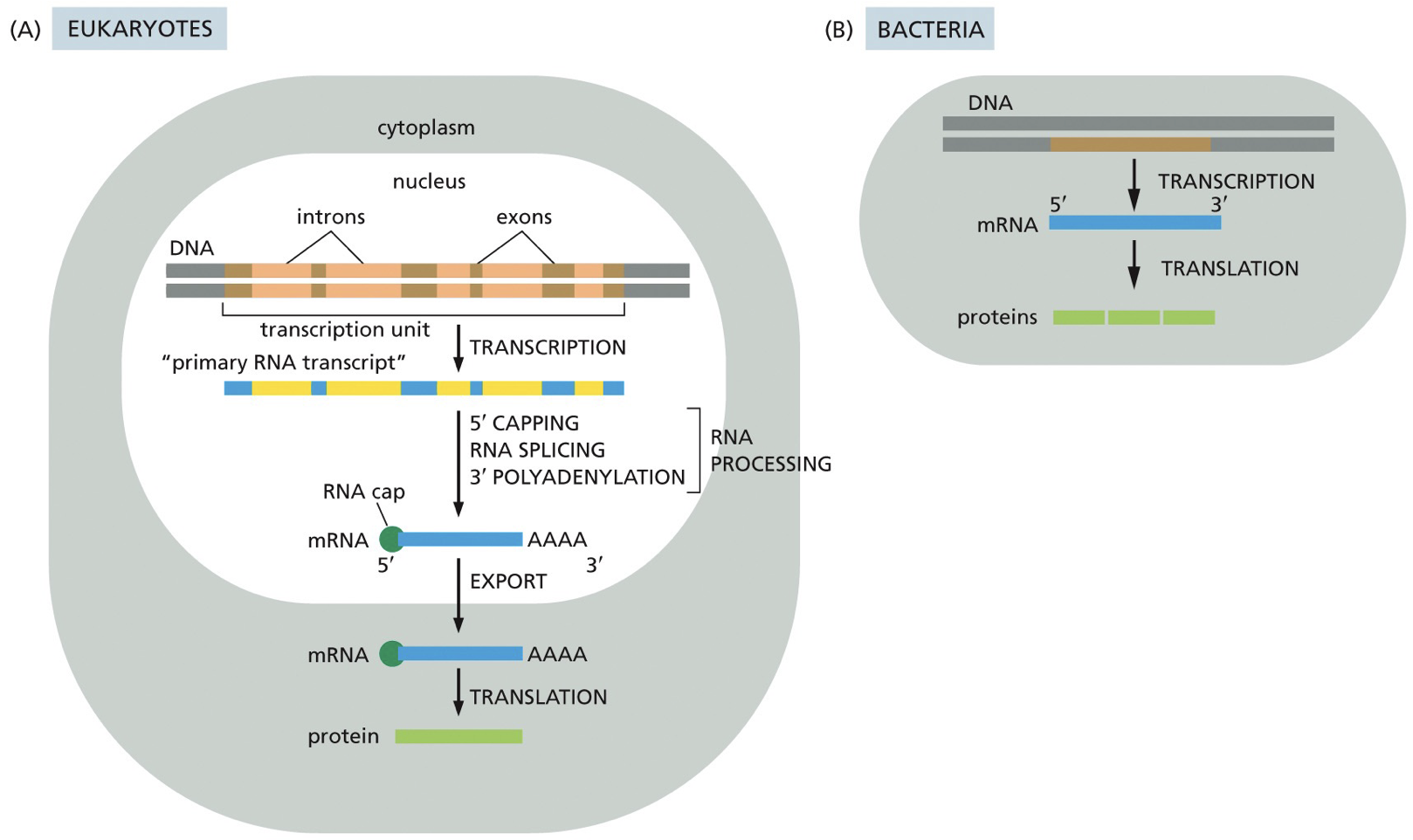
Eukaryotes have introns and exons, ends are going to be translated, and have a nucleus. RNA is going to be exported from nucleus to cytoplasm.
A fair bit of bacterial RNA is polycistronic mRNA; meaning they have multiple genes and proteins coding sequences of same RNA molecules, while eukaryotes are monocistronic; each gene. has its own promoter.
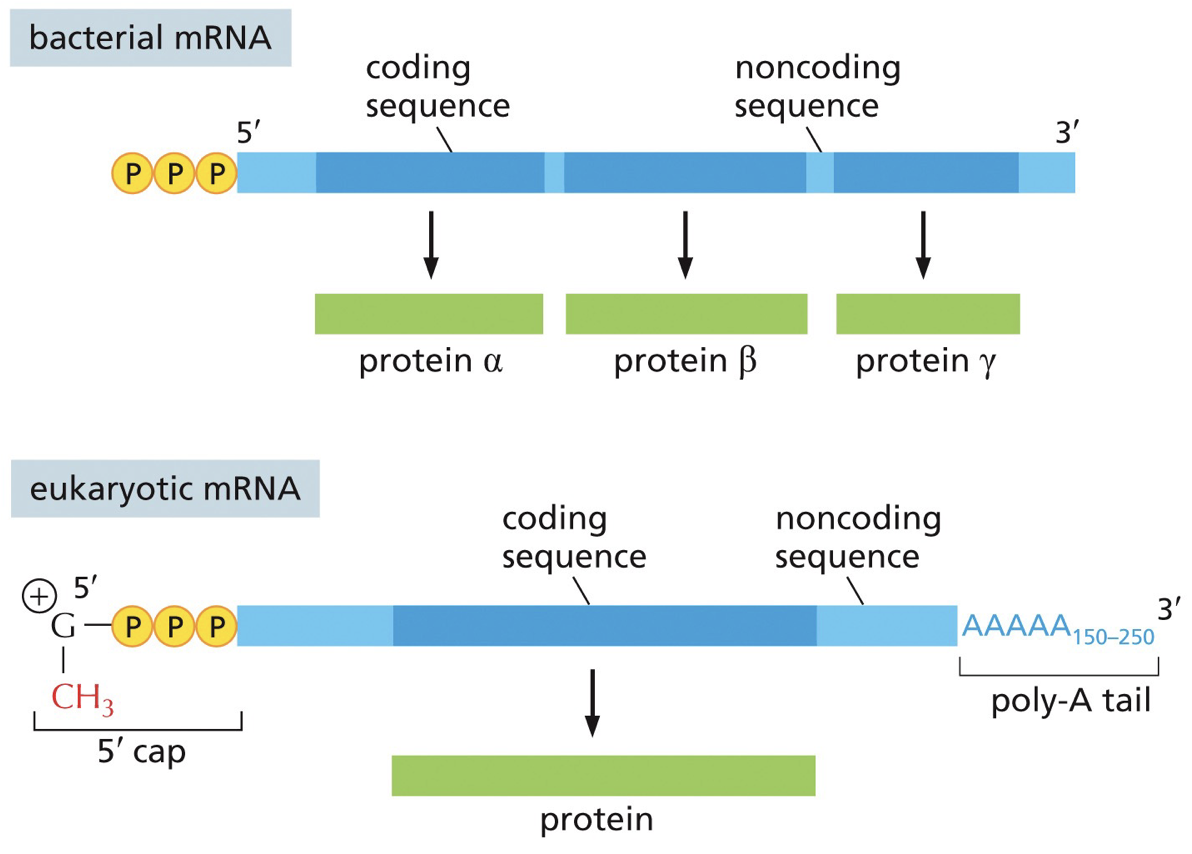 In monocistronic (tryptophan synthesis), each section’s (1-5) wil be separate production of different genes. In polycistronic, tryptophan, translation and transcription are coupled. I would benefit to be polycistronic because we save space and don’t need multiple mRNA. Polycistronic is also regulating because for tryptophan there are regulatory and you need A-E and polycistronic makes them all at once.
In monocistronic (tryptophan synthesis), each section’s (1-5) wil be separate production of different genes. In polycistronic, tryptophan, translation and transcription are coupled. I would benefit to be polycistronic because we save space and don’t need multiple mRNA. Polycistronic is also regulating because for tryptophan there are regulatory and you need A-E and polycistronic makes them all at once.
Eukaryotes (+ exons)
Different exons are picked over others
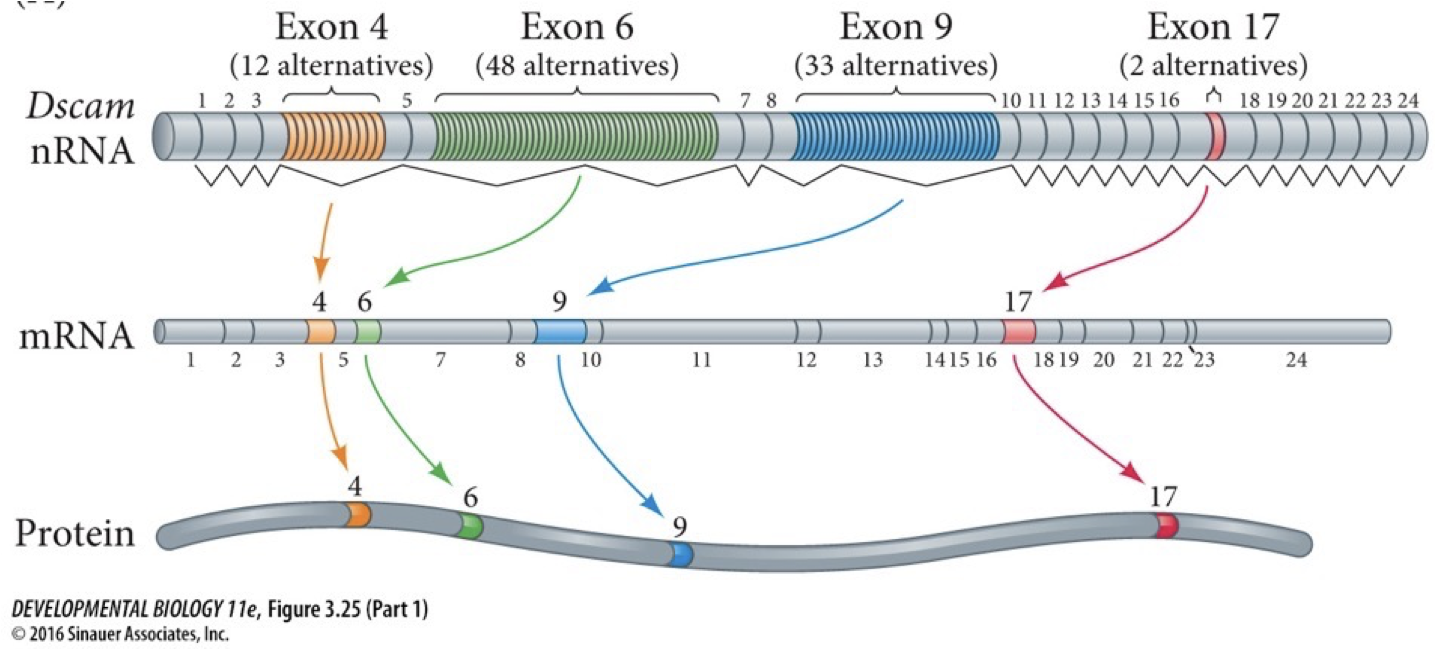
Alternative splicing: allows one gene to produce multiple sequences of proteins.
Splice isoforms are the different sequences of proteins. Both proteins and sequences in the RNA regulate this process.
Advantages: adapting and regulating; save space compared to polycistronic, making different proteins for different functions in response to different stimuli instead of having to code different things for each.
We have many exons positioned and can have many different effects in different cells. Each exon has a bunch of different alternatives. Each time we get a different one selected we can make different compilations for many different versions of a protein. If each had to be encoded separately, we would need more genes. Splicing helps us not have to triple or quadruple genome size.
 All the major RNAs are derived from precursors the are longer RNA molecules that are shortened.
All the major RNAs are derived from precursors the are longer RNA molecules that are shortened.
Transcriptional unit: encoded in the DNA includes promoter, coding sequence and termination stop sequence.
Primary transcript: pre-RNA, the initial RNA molecules from template DNA. Proteins with smRNA.
RNA polymerase
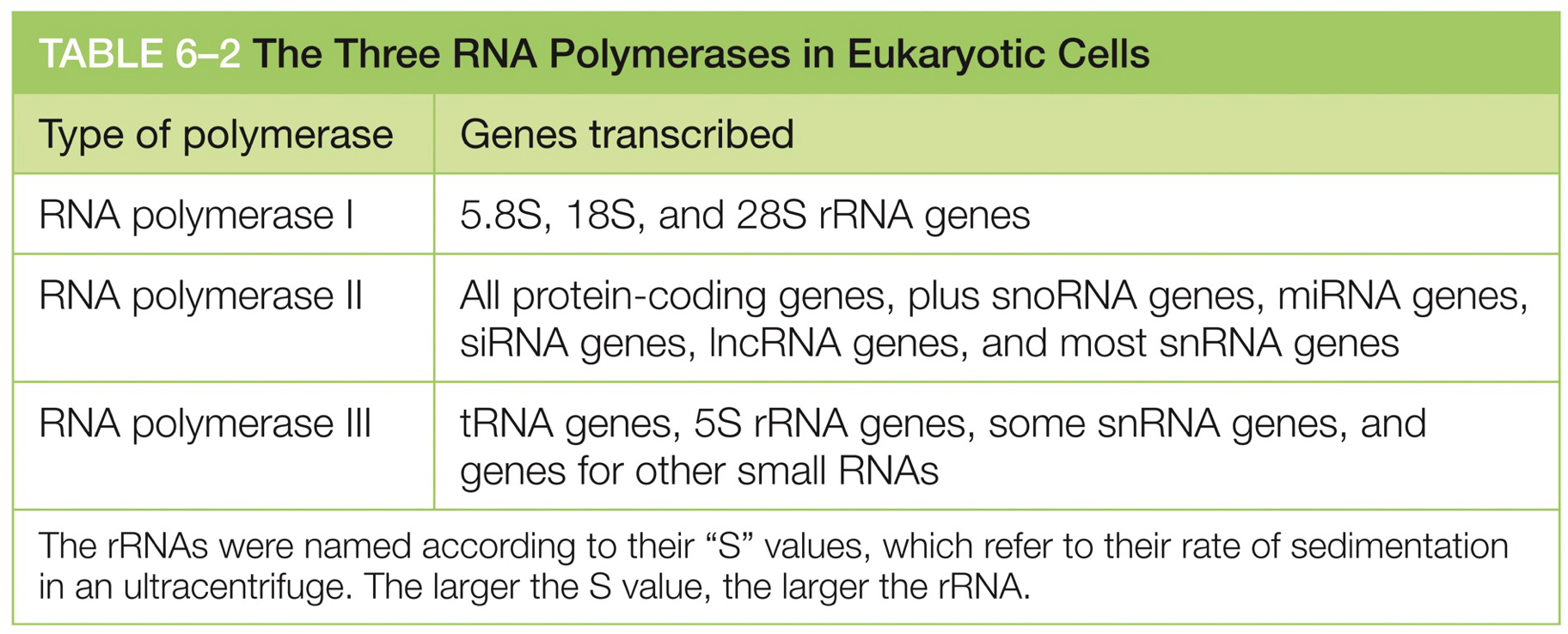
Each RNA polymerase is responsible for different things.
TATA Box: Key consequences, before transcription start site. TBP is what is going to bind to TATA box.
It is the site of assembly of preinitiation complex.
TBP binding, GTFs, RNA polymerase
GTFs: general transcription factors, what pattern and amount of transcription is influenced by.
TBP is a subunit of TFIID.
All of these factors and enzymes form the preinitiation complex (PIC)
TFIID recognizes and binds to the TATA box; occurs through TBP subunit, binding to minor groove of DNA, which also bends DNA.
Transcription in Eukaryotes steps
TFIID recognizes and binds to TATA box, occurs through subunit TBP binding minor groove of DNA, which also bends the DNA.
Enables TFIIA and TFIIB to also bind
RNA polymerase and TFIIF are recruited to the promoter, followed by TFIIE and TFIIF
TFIIH - kinase (phosphorylate) and helicase, change how things interact. It phosphorylates the RNA polymerase II tails (called the C-terminus), and begins elongation (by being kicked off it trigger elongation). It starts by opening up the DNA double helix through ATP hydrolysis, and then phosphorylates the C-terminus.
TFIID - usually stays bound at the promoter while other GTFs are released once elongation begins.
RNA polymerase → TFIIF → TFIIH → TFIID
As polymerase moves along, it recruits a bunch of proteins to help it (the first being the elongation factors).
TFIIH - Unwinds DNA at the transcription start point, phosphorylates Ser5 of the RNA polymerase. C-terminal domain (CTD); release the RNA polymerase from the promoter.
TFIID - Recognizes TATA box and other DNA sequences near the transcription start point.
Once the PIC assembles, transcription can start. TFIID is left behind and it’ll start another process of transcription again.
Elongation factors (Spt4, Spt5, Elfl) are important for it to keep transcribing through. These factors can help pry away histones or directly destabilize histone, and can also include help from chromatin remodelling complexes and histone chaperones.
C-terminal domain
RNA pol II transcribes DNA to RNA. The processing proteins are carried on its tail.
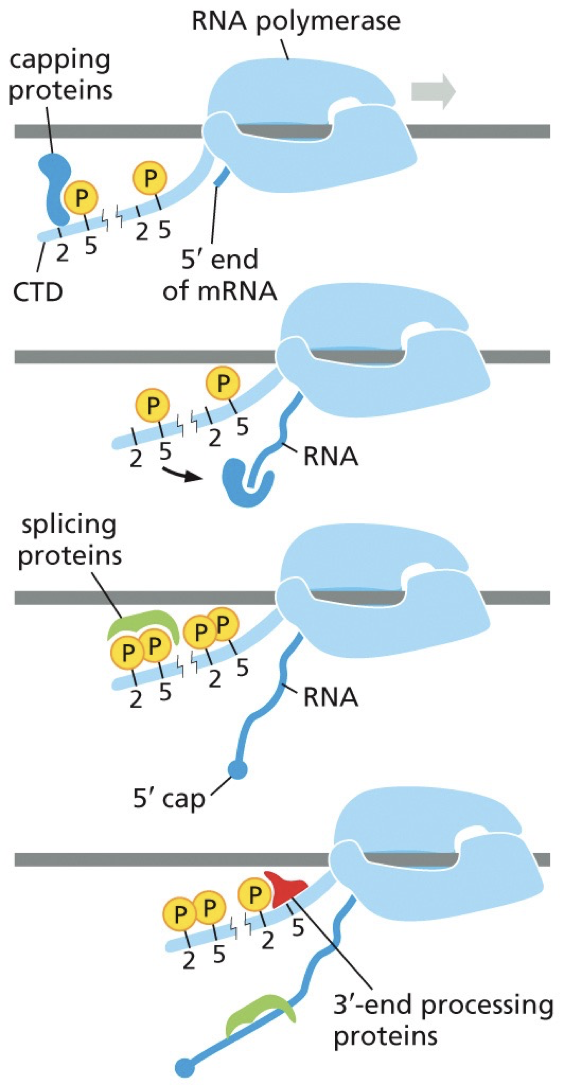
Proteins are recruited to it. The tail is there, and it’s a good plot to recruit complexes needed. 5’ end is coming out, and the first protein recruited is phosphorylates and bound to capping proteins. This makes a 5’ cap. Capping proteins can jump to 5’ end and begin cap process. Process occurs at the same time as transcription.
The 5’ cap has methylguanosine as the first step, before introns are dealt with, which prevents 5’ end from exonuclease.
Plays a role in transport out of nuclease, and initiation of mRNA translation.
Distinguishes mRNA from other RNA
Capping enzyme: two active site for two different enzymes (RNA triphosphatase and guanylyl transferase)
Steps:
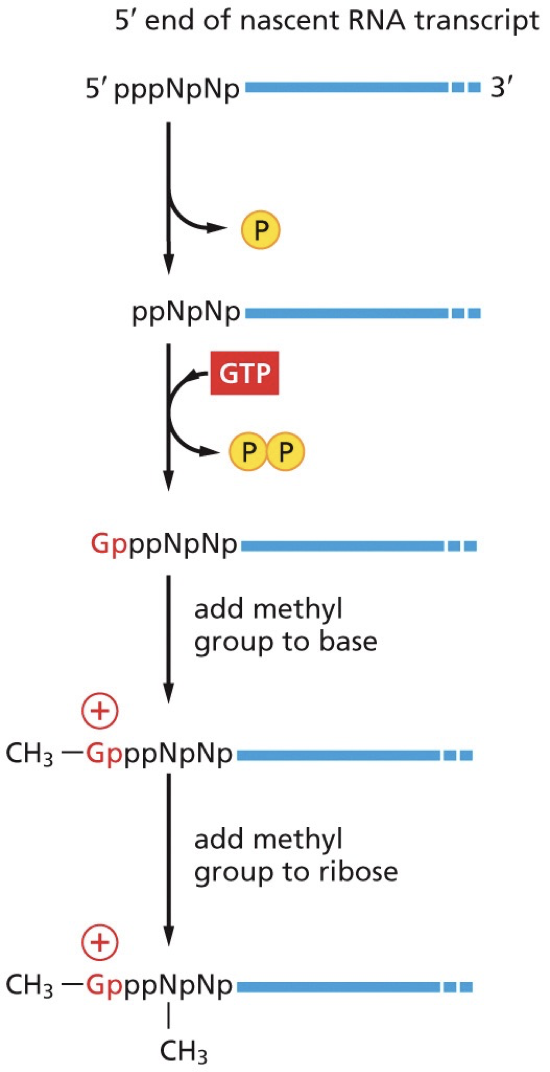
RNA triphosphatase removes the terminal phosphate group leaving a diphosphate
Guanyl transferase adds GMP in an inverted (5’-5’) orientation, forming a triphosphate bridge.
Guanine base is methylated by RNA methyltransferase
The next set of proteins aid in splicing. As polymerase processes, tail changes where splicing proteins can get on. Multiple introns are removed. There is an intron to splice and it occurs until en of RNA molecule.
Finishes off with the 3’ end and its Poly(A) tail. Untranslated regions between tails and coding regions play a regulatory role.
Poly(A) tail - series of adenine. 50-250 adenosine.
Polyadenylation: addition of A to 3’ end
20 nucleotides downstream of a special cleave and poly(A) signal sequence
Sequence site is a site of for assembly of protein complex for polyadenylation
Poly(A) tail varies in length
protects from exonuclease
Isolate and stabilize mRNA from other RNA using oligo dT column (base pair with a bunch of A’s to pull out all the good RNA).
Consensus sequence is responsible for recognition, and the preferred sequence leads to cleavage at a particular site. Extra RNA that gets degraded, and then the addition of the Poly(A) tail.
CstF & CPSF are going to hang on C-terminal domain until RNA polymerase moves past. Gets onto site. Poly(A) polymerase will get on and make all the As without complimentary base pairings