BIOC15: Genetics Midterm
Lectures 1-5, labs, textbook readings
Table of Contents
Introduction
Mendelian Genetics
Chromosomes
Linkage and Recombination
Lab Stuff
Introduction
Genetics: study of genes and heredity, including the inheritance of traits from one generation to the next. Biological function of genes. Investigates genetic code and its role in determining an organism’s characteristics. Genetic variation, mutations, and their impact on an organism’s development and functioning.
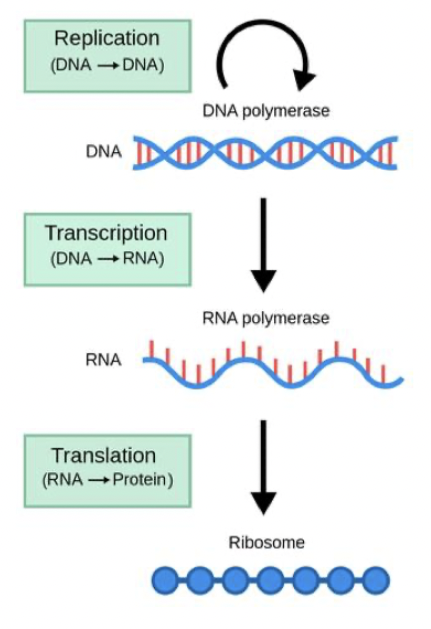
DNA: Hereditary material that encodes all information from an organism, composed of 4 nucleotides (A, C, T, G).
Genes: segments of DNA that are transcribed to RNA to make proteins (mRNA) or functional RNA molecules (rRNA, tRNA, regulatory RNA).
Mendelian genetics still hold true today. All our cells have nucleus with DNA organize into chromosomes.
Beadle and Tatum’s ‘One gene, one enzyme’ hypothesis: each gene encodes for a single enzyme. Incomplete picture. A linear reaction of DNA to RNA to Protein.
DNA may encode transposable elements, which have no function. Not a lot of actual coding RNA. We don’t call it junk DNA anymore. “Non-coding DNA” is 98% of DNA.
A gene can encode for several enzymes with different functions (pleiotropy), or for other molecules that are not enzyme.
Genomic region encoding the foraging gene in D. melanogaster. Shows protein coding genes with several alternative gene products. Non-coding genes (long noncoding RNAs, microRNAs). Mobile genetic elements.
A gene can encode for several enzymes with different functions (pleiotropy). All the different products affect a bunch of different phenotypes. Fat stores, feeding, gut and brain.
Alleles: two versions of the same gene, that differ in DNA sequence and/or function.
Foraging gene has ‘rover’ and ‘sitter’ allele. Mendelian genetics is based on multi-allelic traits.
Genetic variation gave us alleles. Alleles have different types. We are diploid. Almost, all animals are - means to have 2 versions of every gene, one from father, one from mother.
Polymorphic: gene has multiple wild-type alleles. multi-allelic.
Wild-type: over 1% of the population has it, otherwise it is considered a mutation
Homozygous: individual (diploid), has two of the same allele.
Heterozygous: individual (diploid), has two different alleles.
Hemizygous: individual has one copy of a gene.
Phenotypes: Measurable characteristic as the manifestation of the genotype of an organism (morphological, developmental, physiological, behavioural), depending in part on which combination of alleles are present. ex. a given allele can make more proteins than another allele. A given allele might not make any protein.
Genotype: genetic makeup of individuals, given alleles for a particular gene.
In mendelian genetics, we have either dominant or recessive.
History of genetics
1800s: genes had not been discovered yet. Humans had been selectively breeding plants and animals for generations. Better wheat
Mendel: Artificial selection was the first applied genetic technique. Purposeful control of mating by choice of parents for the next generation. Domestication of plants and animals was a key transition in human civilization. Important crops, for example, rice, wheat, corn, and herds of pigs, sheep, and cattle are the results of selective breeding over many generations.
Mendel process: prevent selfing (removed stamen) → observed discrete traits → pure-breeding (true breeding lines, inbred) → reciprocal crosses (reversed whether trait was coming from male or female to see egg and sperm importance).
The results of selective breeding were regularly unpredictable. Plants mating provides distinct characteristics.
Humans select for desired traits when breeding animals. Offspring don’t always resemble their parents. Despite artificial selection being widespread, it can still be shocking and unpredictable. Come up in the progeny, was not expected before.
Mendelian genetics
Four general themes:
Variation is widespread in nature and provides for continuously evolving diversity
Observable variation is essential for following genes from one generation to another
Variation is inherited by genetic laws, which can explain why like begets like and unlike
Mendel’s laws apply to all sexually reproducing organisms.
Mendel and Darwin were contemporaries, but did not work together. Darwin was widely accepted post-mortem, because he did not have the key information on inheritance at the time that Mendel did. Darwin believed in pangenesis (blending inheritance).
Darwin: Organisms and species are changing over time by the process of natural selection, but he didn’t have a hereditary mechanisms
Mendel: Genetic model of inheritance, but he couldn’t explain the implications of allele frequencies changing over time. He inferred laws of genetics that allowed predictions about which traits would appear, disappear, and then reappear.
Pangenesis: if you mix different things, you get a mix of the two.
Male-dominated inheritance: an issue with understanding inheritance in Darwin and Mendel’s theories.
Mendel proved Darwin’s theories.
“Experiments in plant hybrids” by Gregor Mendel, 1866. Became cornerstone of modern genetics. Wasn’t discovered until 1900s. Wrote it in a monastery in Austria, selectively breeded plants. Studied in the garden. Experiments in plant hybrids.
Success - a near perfect study system. Self fertilizing male and female parts in the same flower. Observable phenotypic variation.
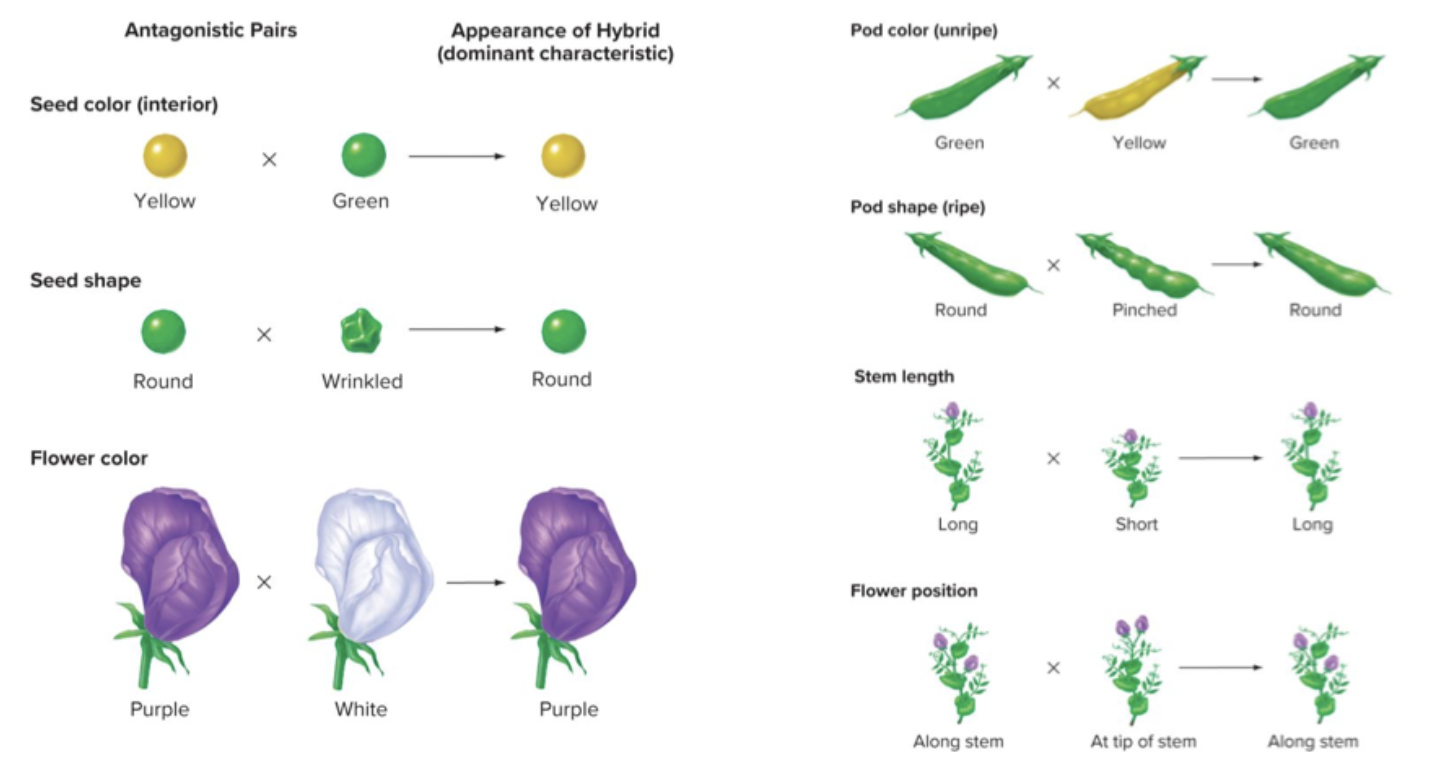
Traits:
Colour of peas (yellow or green)
Colour of pod (yellow or green)
Colour of flowers (purple or white)
Shape of peas (round or wrinkled)
Shape of pods (smooth or indented)
Position of flowers (along stem or at tip)
Plant height (tall or dwarfed)
Multiple bi-allelic and discrete traits. lots of offspring. short generation time. controlled crossings (self-fertilization).
True breeding lines: homozygous breeding lines.
Crossing two different true breeding lines, all offspring would show phenotype of one parent. Dominant x recessive = dominant. F1 is hybrid, heterozygous.
Selfing F1 - You see some recessive traits again. Stipulated blended inheritance. Saw both parental forms again.
F2 progeny of two true breeding lines = 3:1
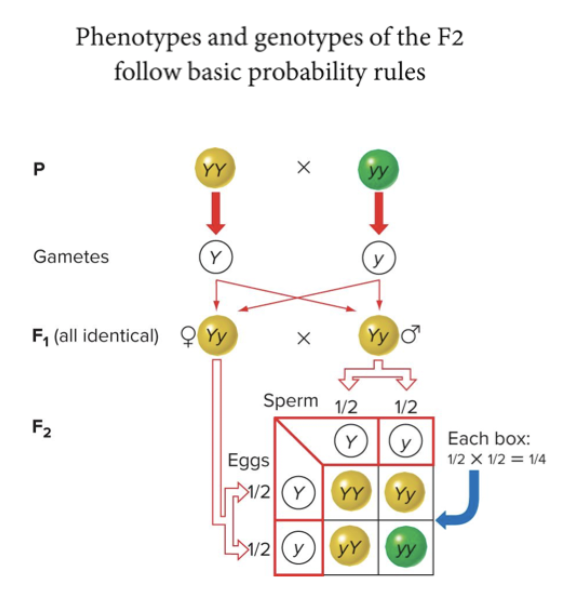
Product rule: probability of two independent events occurring together is the product of their individual probabilities P(1 and 2) = P1 X P2
Sum rule: probability of either of two mutually exclusive events occuring is the sum of their individual probabilities P(1 or 2) = P1 + P2
Testcross can reveal an unknown genotype
Testcross: cross individuals with dominant phenotype to a true breeding recessive individual (yy) and score progeny. If you cross with a recessive phenotype, it can only give yy, while dominant phenotype could give Yy or YY
Monohybrid: Individuals have two different alleles of a single gene
Mendel followed a mathematical approach to inheritance. Simple antagonistic traits, controlled breeding via cross-fertilization or selfing. Large numbers of progeny produced within a short time. Reciprocal crosses. Traits remained constant in crosses within a line. Many replications. Mendel’s monohybrid crosses disproved blending and male-dominated inheritance, and revealed units of inheritance. Mendel basically looked at big numbers to see general ratios. It’s actually closer to 1:2 or 1:4.
Single
Monohybrid cross: 1 gene is crossed, 2 possible alleles. genotypic ratio of 2 heterozygotes is 1:2:1.
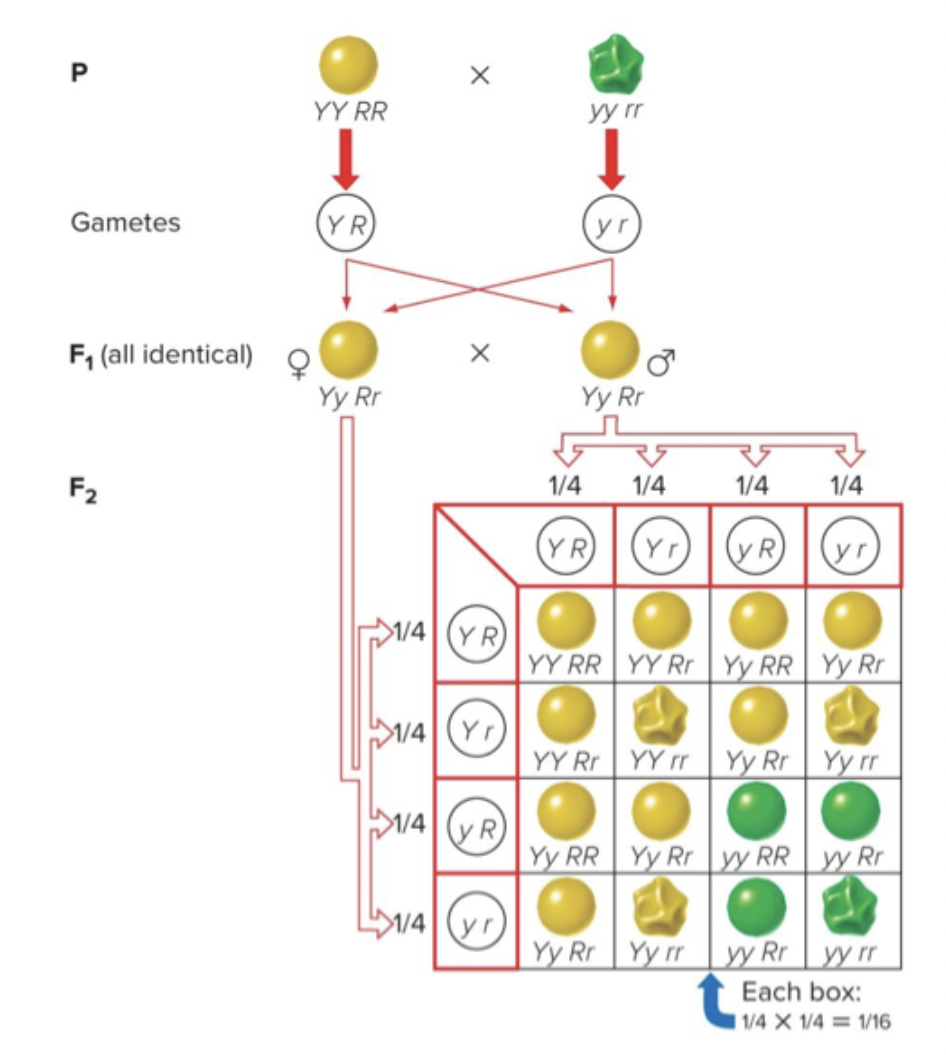
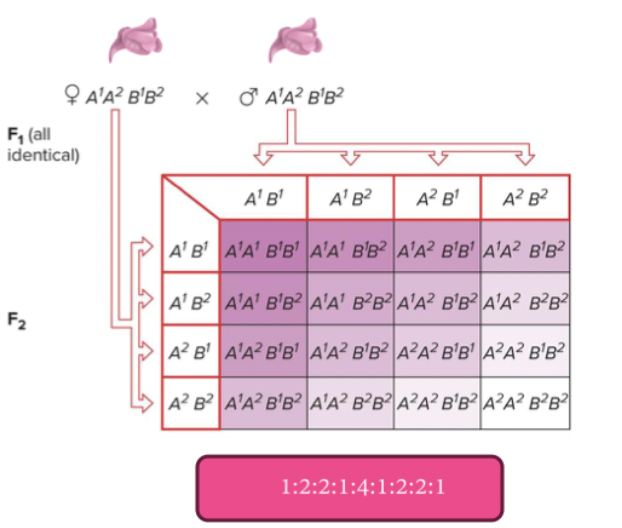
Inheritance with two genes: mendel investigated what would happen when he tracked the inheritance of phenotypes by two genes. Peas have 8 antagonistic traits he could track. Yellow genes break down chlorophyll. Round peas make branch sugars that makes branch sugars that makes round shape.
YYRR x yyrr = YyRr, you will only see one allele combination/phenotype in the offspring.
Self F1 → YyRr x YyRr = 9:3:3:1 (yellow round, green round, yellow wrinkled, green wrinkled) (2 homo dom, 1 homo dom 1 recessive, 1 homo dom 1 recessive, 2 recessive). You see all possible combinations of alleles.
Two phenotypes are like parental phenotypes, and two are a mix of parental combinations (recombinant phenotypes).
Recessive traits occur at a lower proportion.
Different pairs of alleles segregate independently of each other during gamete formation. Y is just as likely to assort with R as it is with r. y is just as likely to assort with R as it is with r.
Law of segregation: alleles of the same gene segregate during gametogenesis with equal probability (proven with monohybrid crosses)
Law of independent assortment: pairs of alleles segregate independently of each other during gamete formation, results in parental and recombinant genotypes in the offspring (proven with dihybrid crosses)
Alleles in parental cell → gamete formation → possible allele combinations in gametes.
Dihybrid test crosses: you cross an organism that is recessive for two genes, with an organism that is dominant for both genes, and their possible outcomes will determine the genotype of the dominant plant.
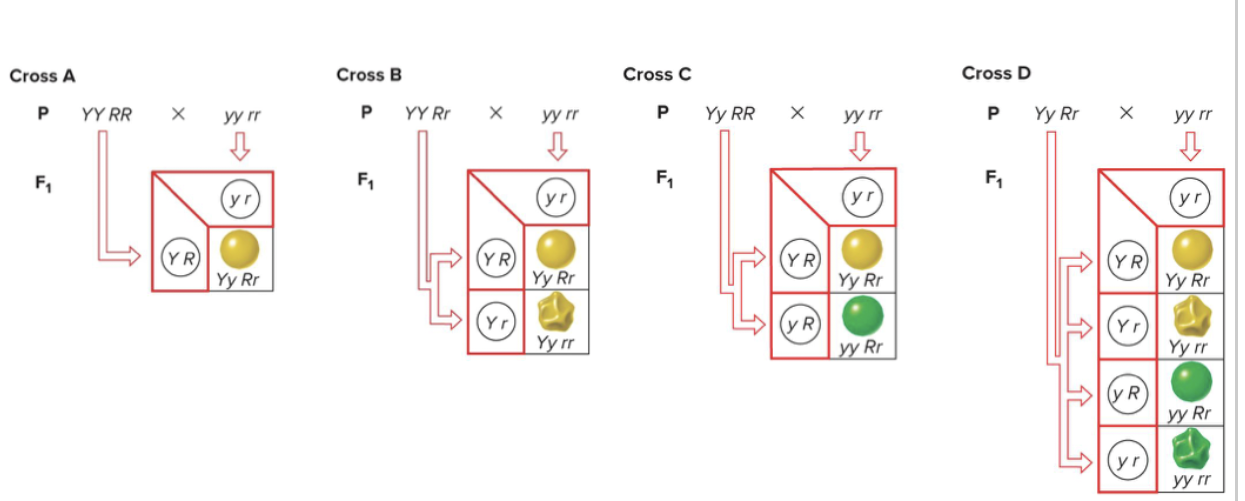
Punnet square gets progressively more complicated as you go on. n is the number of traits that are heterozygous:
2n = the number of different gametes → Aa Bb Cc Dd = 24 = 16 kinds of gametes
You can break this down into 4 monohybrid crosses instead of 1 with 16 different rows.
RR Tt Bb yy = 4 gametes possible - this is in terms of selfing. 12 × 22 = 4
If you make like 4 different monohybrid crosses, in order to find the probability of a genotype, you can use the product rule based on the chances in the given crosses.

you can do this with genotypes or phenotypes.
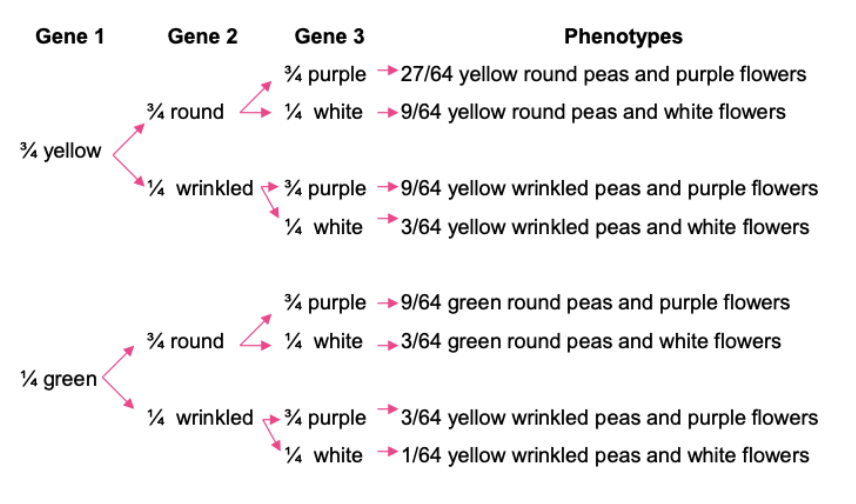
Branched line diagram: predict proportions of progeny from multi-hybrid crosses
Pedigrees: visual representation of the inheritance of a phenotype within a family. As many generations as possible. Ideally at least up to grandparents. Can use Mendel’s Laws to analyze patterns of inheritance. Dominant vs. Recessive traits.
Rare traits may not be seen in family: we assume it’s not running in the family.
We use consistent symbols across diagrams. Single line - mating. Double line - mating with cousins/siblings line. Birth order read left to right.
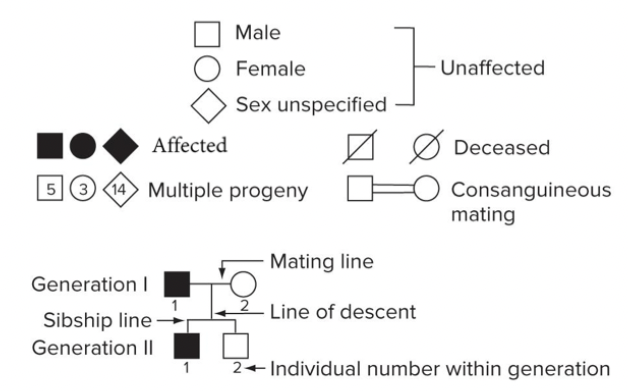
Pattern of inheritance: rare dominant traits. Every affected person has at least one affected parent. Vertical inheritance (very generation affected). Two affected parents can produce unaffected children, if both parents are heterozygous.
Vertical pattern of transmission: a trait that appears in an affected individual also appears in at least one parent, one of the affected parent’s parents, and so on. If a trait is rare, a pedigree with a vertical pattern usually indicates that the disease-causing allele is dominant. Huntingtons.
Horizontal pattern of transmission: a trait that appears in an affected individual may not appear in any ancestors, but it may appear in some of the person’s siblings. A pedigree with a horizontal pattern usually indicates a rare recessive disease-causing allele. Affected individuals are often products of consanguineous mating. Cystic Fibrosis.
Affected children can have unaffected parents. Horizontal inheritance (skips generation).
Probability that they’re a carrier if they don’t have it - you would only include the versions that have a dominant allele in it. Use context clues, look at siblings for information about dominant parents.
P(affected child) = P(of parent 1 being heterozygous) x P(of parent 2 being heterozygous) x P(offspring of heterozygous parents being aa)
Extensions of Mendel’s Laws - One Gene
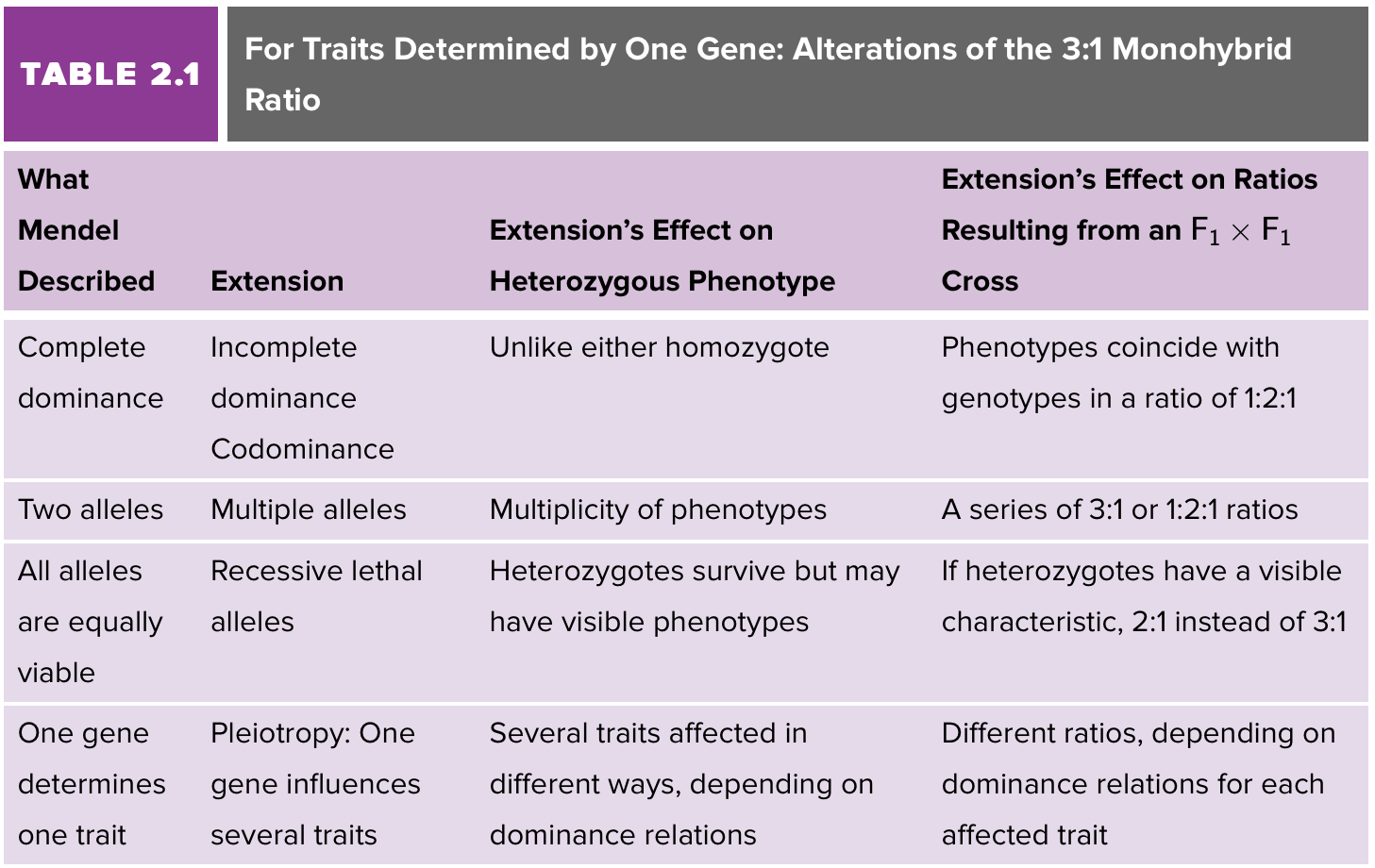
Phenotypes in pure-breeding crosses don’t always follow Mendel’s rules. Why? For a single gene - Dominance relationships of alleles, genes with more than two alleles, recessive lethal alleles, genes/alleles that affect more than one phenotype (pleiotropy). Phenotypes often don’t follow bimodal results.
Dominance
The phenotype of heterozygote determines allele dominance. It’s not intrinsic property of alleles.
Pea shape is determined by an enzyme that breaks amylose down into amylopectin. Double recessive makes it a lack of function allele. It is enough to have 1 functional copy to break down sugar. Biochemical pathways determine the effect of each protein on the phenotype.
Complete dominance: hybrid resembles one of the two parents.
Incomplete dominance: Phenotype ratio reflects the genotype ratio. A dominant allele doesn’t necessarily make something yes or no, it makes it more like yes (AA), no (aa), OR in between/some (Aa)
Codominance: Both phenotypes show up equally in the heterozygote. often occurs because both alleles code for a functional protein. Phenotype reflects genotype ratio again. Gives small or big blotch? Heterozygote gives both.
More than two alleles
5 different alleles determine the colour, and they all have different dominance relationships with each other. Dominance relationships are only meaningful when examining pairs of alleles.
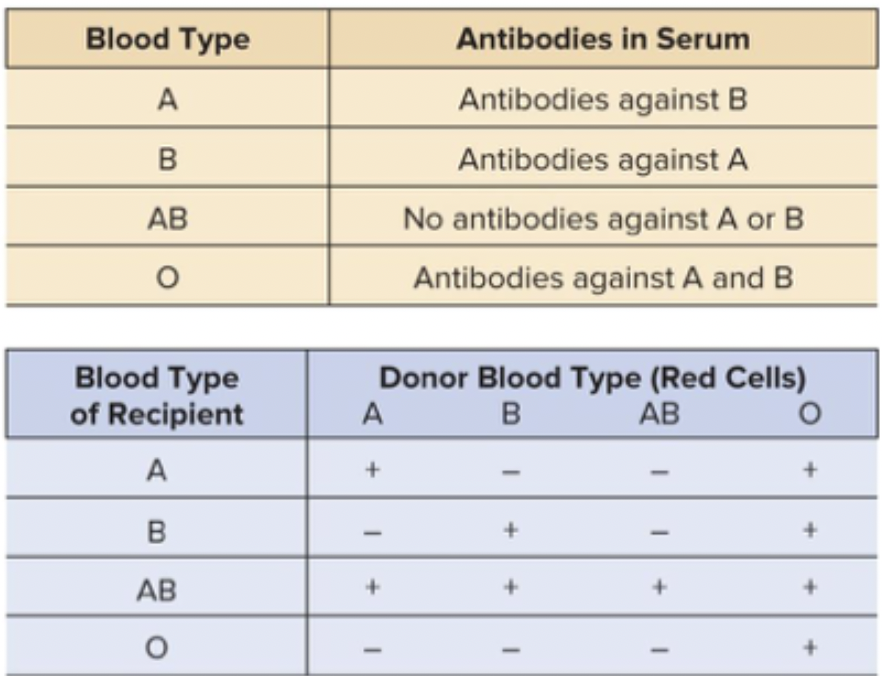
ABO gene determines blood type. Each individual carries two or three possible alleles. Sugar determines it. A and B are codominant. Gene involved in catalyzing the sugars, recognized by our body’s antibodies. O doesn’t deposit any sugar, which is why it’s recessive. The other two do their own things.
Antigens in blood are more complicated because they are also determined by histone compatibility antigens. You have millions of combinations of antigens. This is the reason why it is so hard to find an organ donor match. Need to be on immunosuppressants. 400-1200 version of the 3 genes.
Origin of new alleles: mutations in germ-line cells (gametes).
Allele frequency is the percentage of total number of gene copies for an allele in a population.
Monomorphic genes: only one wild-type allele
Polymorphic genes: more than one wild-type allele. Called common variants.
Recessive lethal alleles
CyO mutation in drosophila. CC is lethal. C is dominant for wing shape, but recessive for lethality.
Double dominance can be lethal - it’s not the curly wings, but the gene has 2 functions. Important for liability.
Genes/Alleles affect more than one phenotype (pleiotropy)
Like curled wing and lethality. This can often result in syndromes.
Ciliary Dyskinesia: Genes that encode proteins necessary for the function of cilia and flagella. A non-functional recessive allele results in both respiratory issues and sterility.
Frizzle chickens: mutation on the a-Keratin (KRT75) gene. Effects feather shape and egg laying ability. They wanted the get these cool feathers and bred for them, but there were side effects.
Phenotypic ratios depend on the individual dominance relationships for each phenotype affected. 60% of genes have multiple effects. Side effects you didn’t account for.
Extensions to Mendel’s Laws - Two Genes
Calleds extensions: don’t change the laws, just change the phenotypic outcome. You can have interactions between genes, you can have all this stuff stopping Mendel’s laws.
Interactions between genes
You get colour because you have enzymes making precursors into pigments. You need a dominant of at least one colour.
Additive gene interactions can lead to new phenotypes popping up on the progeny, based on how much you have.
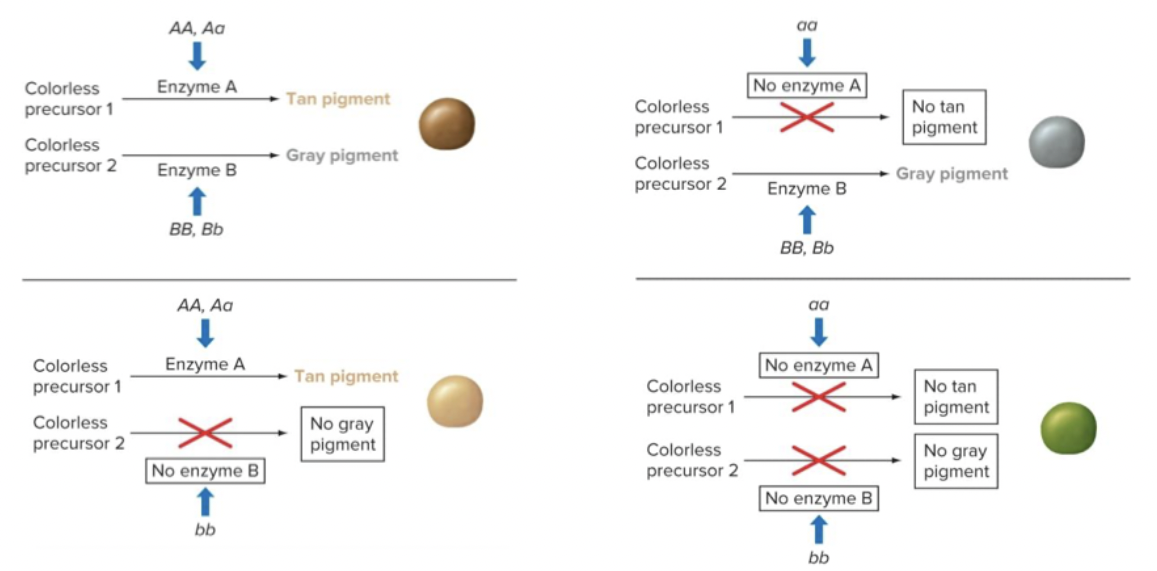
Pretty much has the same ratio as dihybrid cross - but it’s only looking at a single trait.
Incomplete dominance of two genes: white all the way to deep purple. All these different gene combinations affect the phenotype. Not mutually exclusive extensions to Mendel’s laws, you can have multiple things going on.
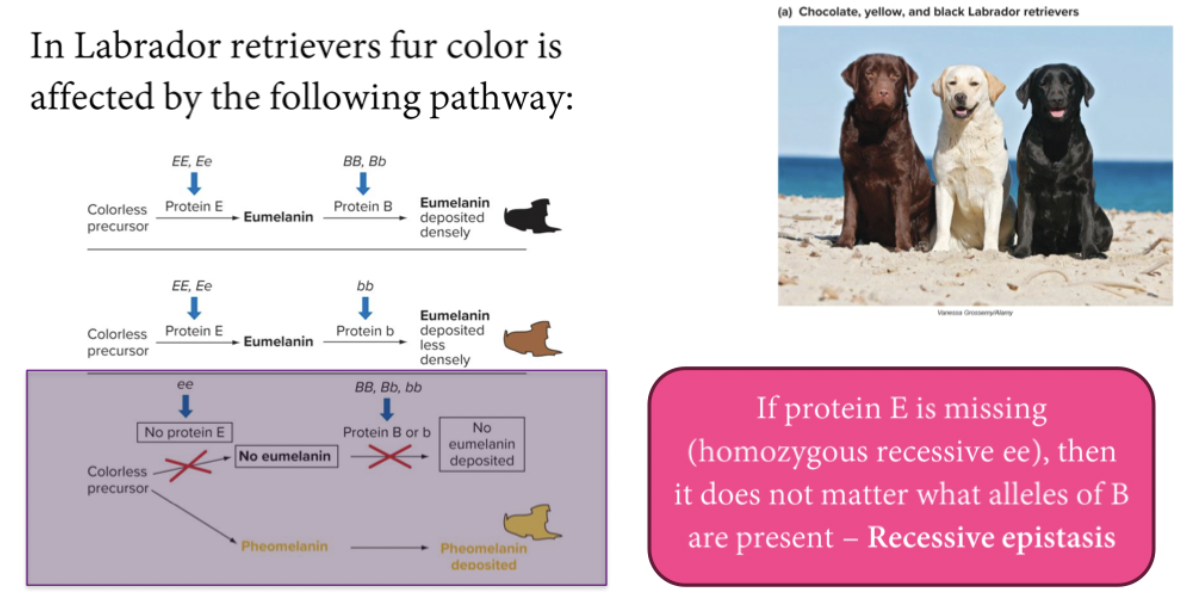
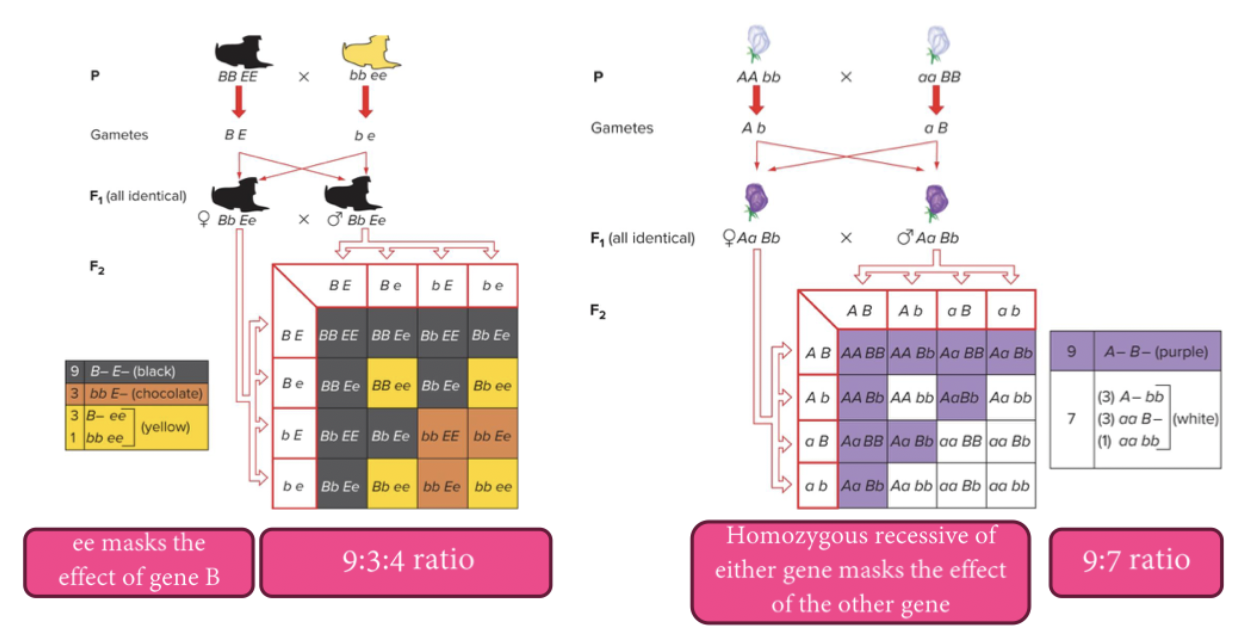
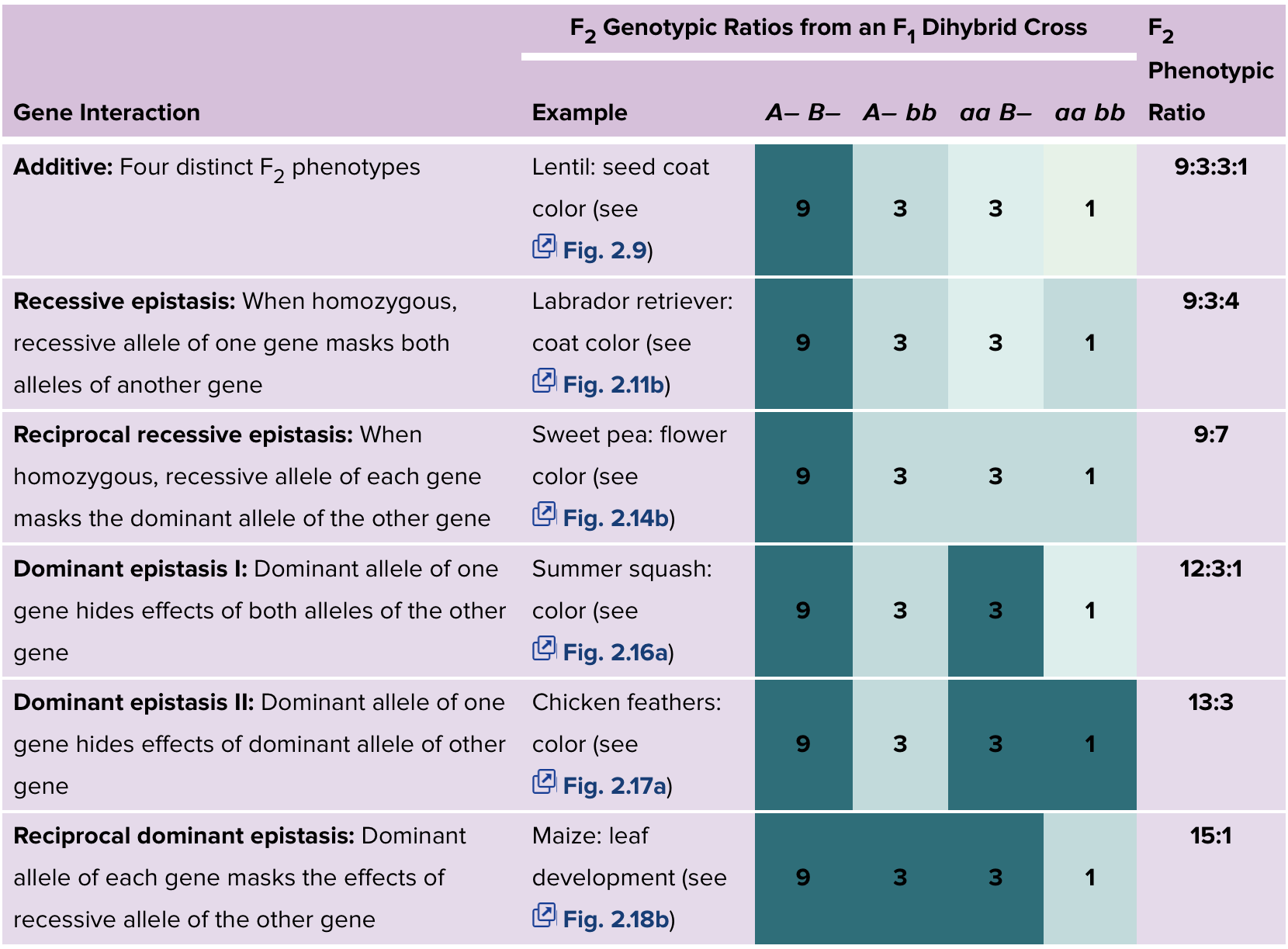
Epistasis: when one genes’ allele masks the effect of another genes’ allele. Can reduce the number of phenotypes in typical Mendelian ratios. Both recessive and dominant epistasis can occur.
Recessive epistasis: There will be a functional pathway made. Then dominance and colour are determined further. No functional pathway will produce a separate phenotype. You need to create the pathway, for the other genotypes to even have an effect. Recessive gene does the blocking. The dominant alleles of both genes function in a common pathway to affect a common outcome.
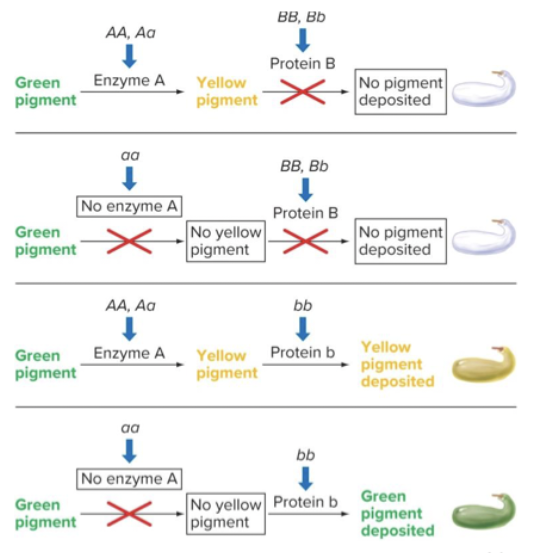
Dominant epistasis: the recessive allele may allow for colour, but the dominant does not. If the dominant dysfunctional allele of gene B is present, then it does not matter what allele A has. Dominant allele does the blocking. The dominant alleles have antagonistic functions.
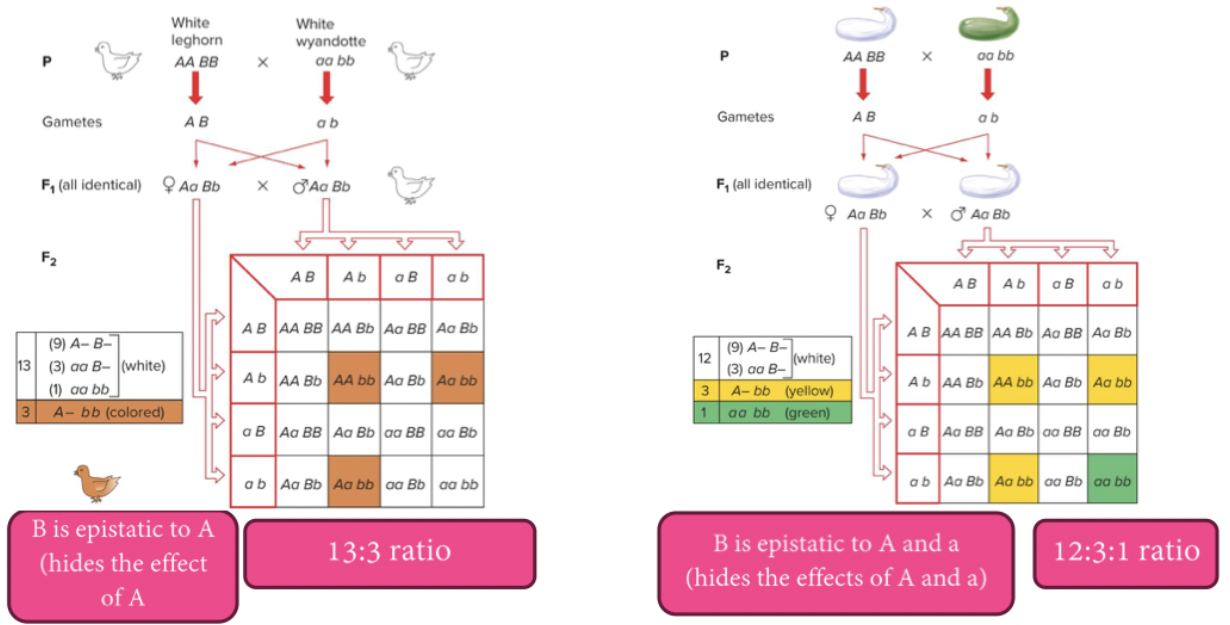
Epistasis in which the dominant allele of one gene hides the effects of another gene is called dominant epistasis (12:3:1 ratio).
Redundancy
One or more of the genes affecting a phenotype are superfluous. Occurs when genes function in parallel. Two genes that are redundant to each other, two or more effecting. Still get broad leaf, you just need one of the genes to have a dominant allele. Two genes affect the same phenotype, and having a functional allele of either gene is enough. 15:1 ratio for Aa Bb X Aa Bb.
Locus heterogeneity
Mutations in any of a number of genes give rise to the same phenotype. Flower colour in peas is a heterogeneous trait, because two genes act together to produce purple flowers, and mutations in either result in a white flower.
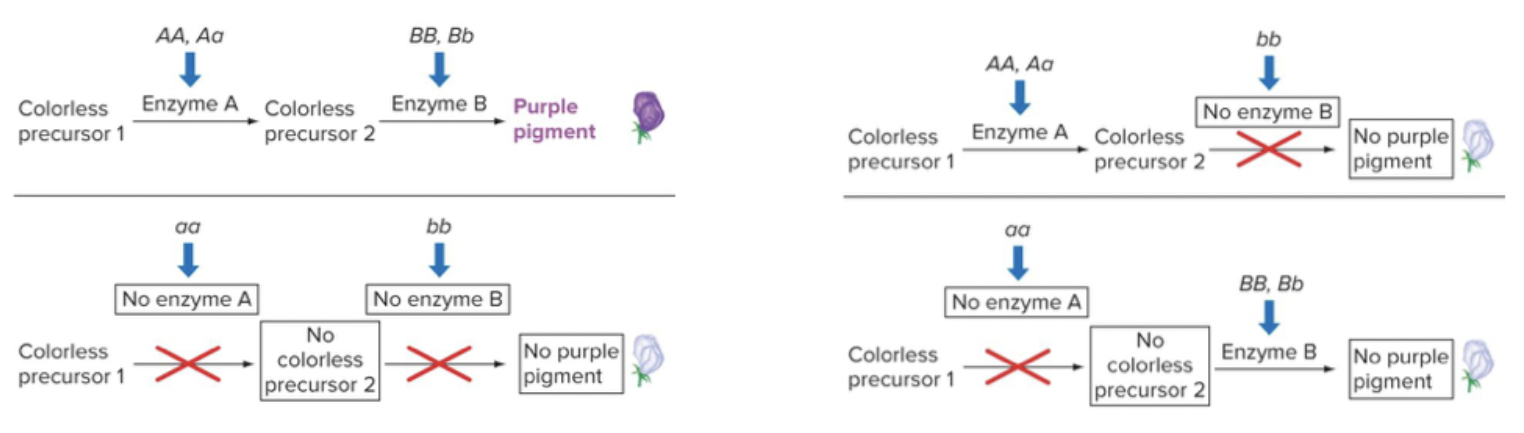
reciprocal recessive epistasis: the dominant alleles of two genes acting together (A– B–) produce a characteristic, while the other three genotypic classes (A– bb, aa B–, and aa bb) do not
Complementation
Recessive mutations in multiple genes have the same phenotype, dominant phenotypes can originate from two recessive parents, if their recessive mutations were in different genes.
Complementation testing: allows to determine the number of genes that result in a given recessive phenotype (eg. the number of genes functionally involved in a phenotype).
Dominant from any two recessive parents if they get recessive from different genes.
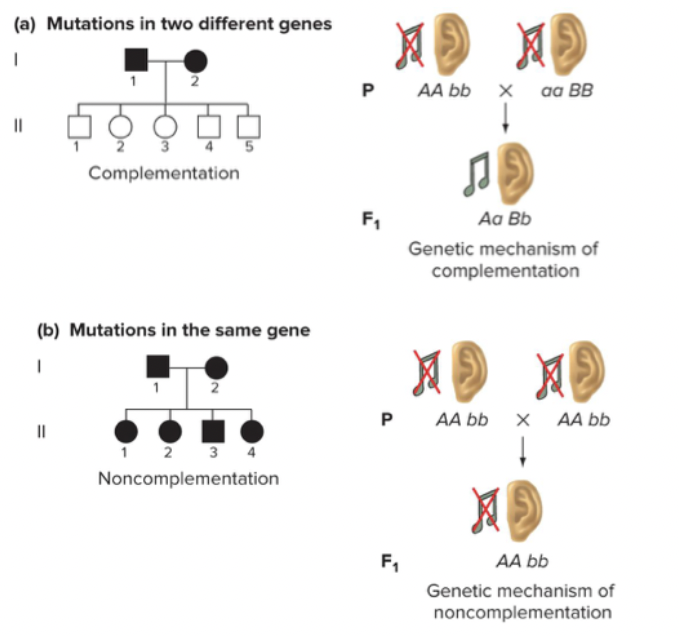
Can get recessives genes from mutations in one genes, and the other is from mutation in another gene.
Recessive epistasis usually indicates that the dominant alleles of the two genes function in the same pathway to achieve a common outcome. In the Labrador retriever example, B and E both function to generate black hairs.
Dominant epistasis usually indicates that the dominant alleles of the two genes have antagonistic functions. Both in the cases of squash and chicken color, the dominant allele of gene B prevents deposition of a pigment whose synthesis depends on the dominant allele of gene A.
Mutations:
Most reduce gene expressions rather than increase
Incomplete dominance loss of alleles
Hypermorphic alleles - more normal protein product or more efficient mutant product
This happens in Dwarfism
Neomorphic alleles - generates novel characteristics
Antimorphic alleles - removes a characteristic
Complexity in traits
Complexity increases with number of genes involved in a trait. More than two alleles, and more than two genes can make genetics very complicated very quickly.
Discrete traits: clear-cut either/or phenotypes. Mendelian inheritance patterns.
Complex traits: quantitative/continuous phenotypes. Controlled by many genes (polygenic). Influence by environment. Most traits are affected by more than one gene.
A given genotype doesn’t reliably produce the same phenotype due to: penetrance & expressivity, modifier genes, environmental effects (epigenetics), random chance. All situations where Mendel’s laws don’t fit.
Penetrance & Expressivity
Penetrance: Does one genotype always equal same phenotype. same genotype doesn’t result in the same phenotype. Can be complete (all the same) or incomplete (not all show same phenotype for same genotype).
Expressivity: levels or shades of a phenotype. Can be variable or unvarying
Both can be affected by the environment, modifier genes, or just chance.
Major genes: have a large influence on the phenotype in question
Modifier genes: have a subtle influence on phenotype. Cause secondary effect that alters the phenotype produced by other genes.
It’s situational on whether something is major or modifier, and it works on a scale.
Environmental effects: Many abiotic factors can affect gene expression and protein function. Ex. temperature → Siamese cats have an allele variant that renders melanin-producing enzyme temperature sensitive, melanin is degraded in cat’s stomach. Previous experience can change gene function long term through epigenetic markers.
Plant height is genetic, but if one gets more sun and water, it’ll likely be taller.
Conditional lethal alleles: cause death under specific conditions. Permissive conditions (wild-type behaviour), restrictive conditions (causes lethality)
Mutant shibire allele in Drosophila, above 29oC → flies with mutant allele become paralyzed and die if no reduction in temperature, while wild-type flies can do it. Outside of those conditions, mutant flies are indistinguishable from wild-type alleles.
The range of temperatures under which the insects remain alive are permissive conditions; the lethal temperatures above that are restrictive conditions
Random chance: influence on traits can be non-heritable, subdivided into whether they are predicted from measurable variable or not. Non-measurable effects result in a certain level of stochasticity for complex phenotypes. We don’t know why something might happen. Mendelian laws still hold true.
Phenocopy: a change in phenotype arising from environmental agents that mimics the effects of a mutation in a gene. Phenocopies are not heritable because they do not result from a change in a gene.
Main Points
Biochemical pathways determine how phenotypes are expressed
Mendelian laws hold true for the inheritance of alleles, but not necessarily for phenotypic ratios
Progeny ratios that diverge from Mendel’s predicted ratios can help us to infer many things about inheritance. Whether a single gene is monomorphic or polymorphic, a single gene or two genes control a trait, alleles are interacting to produce phenotypes.
Monomorphism means having only one form. Dimorphism means having two forms. Polymorphism does not cover characteristics showing continuous variation (such as weight), though this has a heritable component. Polymorphism deals with forms in which the variation is discrete (discontinuous) or strongly bimodal or polymodal
Chromosomes
Chromosomes and meiosis
Chromosomal theory of inheritance: evidence that genes reside in the nucleus and chromosomes. Chromosomes are segregated into different cells. Scientists concluded something is happening biologically.
1854 - 1874: direct observation of fertilization through union of nuclei of eggs and sperm (frog and sea urchin). Something in the nucleus must contain hereditary material.
1880s - Improved microscopy and staining techniques. Long, threadlike bodies in nucleus. Movement of these bodies followed through cell division.
1902 - Sutton and Boveri proposed what we now call the chromosome theory of inheritance.
Chromosomes: packaging of the cell’s DNA. Made of DNA and proteins. DNA wrapped around histones makes a nucleosome, together makes a chromatin fiber.
Diploid (2n) = each chromosome pair has one maternal and one paternal copy of an allele. Meiosis creates haploid gametes from it (n).
Organisms can differ in haploid chromosome number and in ploidy. Human - 23, Fly - 4, Mouse - 20, Polyommatus atlanticus - 224-226. When n gets really high, we can only estimate. Ophioglossum reticulatum - 630-760.
Two types of nuclear division and chromosome sorting.
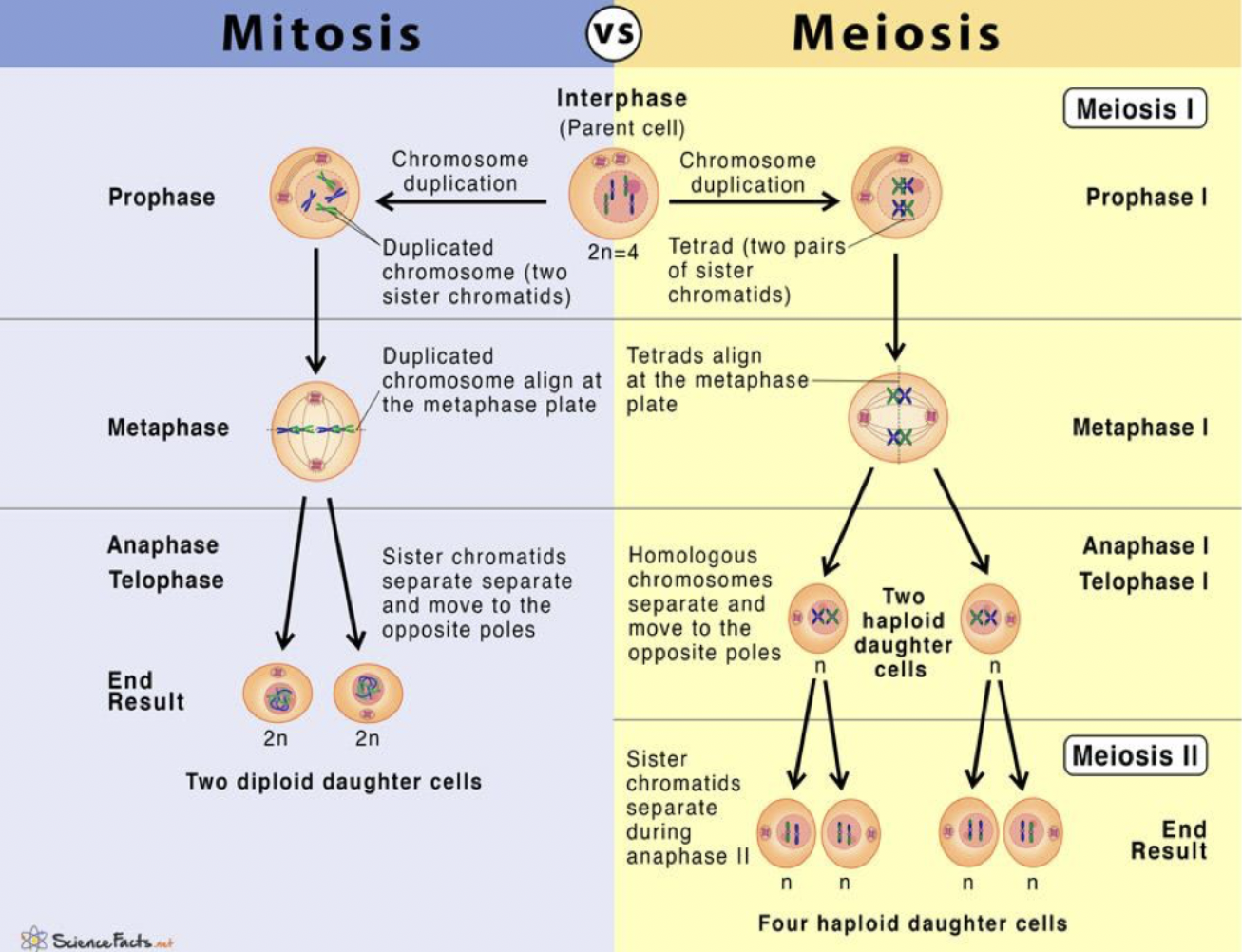
Mitosis - Nuclear division that generates two daughter cells containing same number and type of chromosomes as parent cell
Meiosis - Nuclear division that generates gametes (egg and sperm), containing half the number of chromosomes found in other cells.
Haploid gametes carry only a single set of chromosomes. Two gametes fuse during fertilization to form a zygote. Zygotes are diploid (2n) carry two matching sets of chromosomes, one each from egg and sperm.
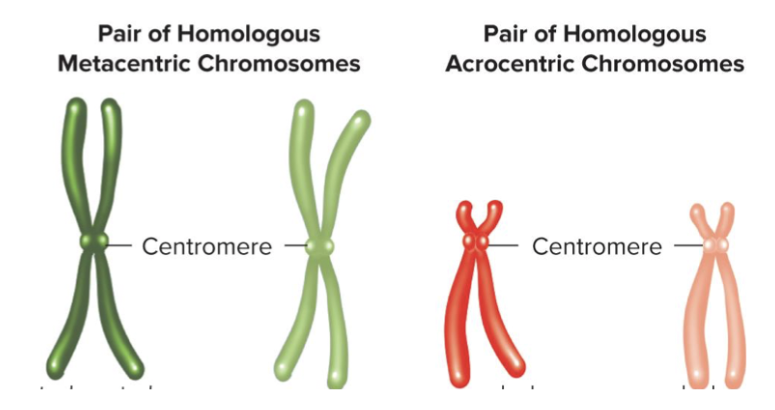
Karyotype - Micrograph of joined chromosomes arranged in homologous pairs. Denser chromosome is darker and length determines where it ends up.
Homologous chromosomes: same size, shape, and banding
Autosomes - all chromosomes except X and Y
Sex chromosomes - unpaired X and Y.
Genetic disorder
DNA test on fetus can check on chromosomal disease. Look at chromosome number, is there abnormality. A lot of genetic syndromes we know.
Chromosome number abnormalities lead to genetic syndromes. Down syndrome was the first human genetic disorder attributable to an abnormal number of chromosomes rather than a gene mutation. The type and amount of genetic material is important for development. Chromosomes transmit genetic information, might transmit too much or conflicting. There are a sensitive amount of chromosomes.
Non lethal: Down’s syndrome (extra 21), Edward’s syndrome (extra 18), Patau’s syndrome (extra 13). Trisomys
Related to sex chromosomes: Turner’s (single X), Klinefelter’s (XXY male), Triple X syndrome (XXX female), XYY syndrome (XYY males)
Chromosome structure in the nucleus. Chromosomes usually exist in the nucleus as chromatin. Only in preparation for cellular division (mitosis or meiosis) does chromatin duplicate and condense into the visible chromosomes of a karyotype. Duplicated chromosomes (chromatids) are joined at the centromere.
Sister chromatid, self preparing for mitosis
Mitosis
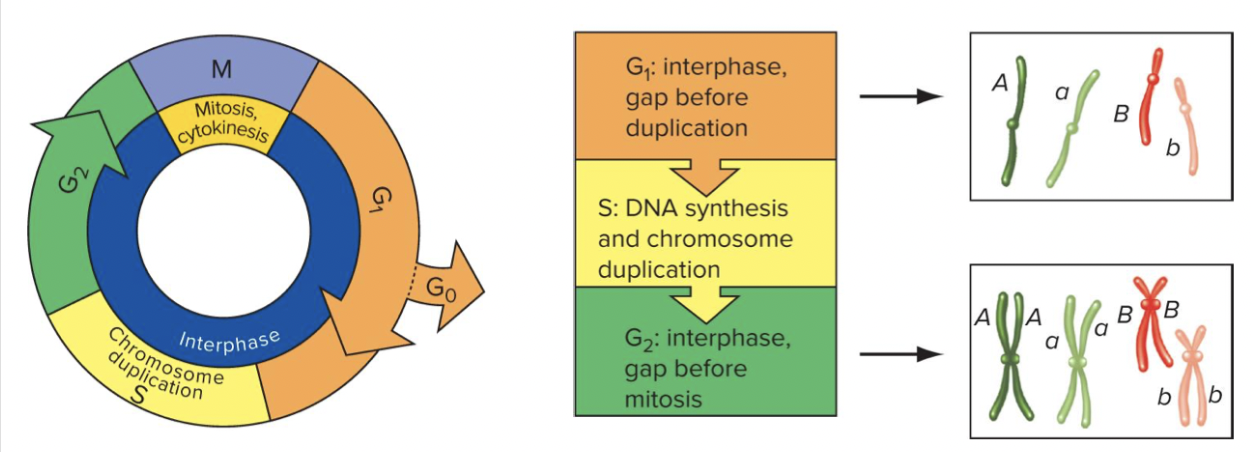
Every cell goes through a cell cycle. interphase and mitosis. Growing and preparing for duplication, the gap phases. Sister chromatid always going to be the same
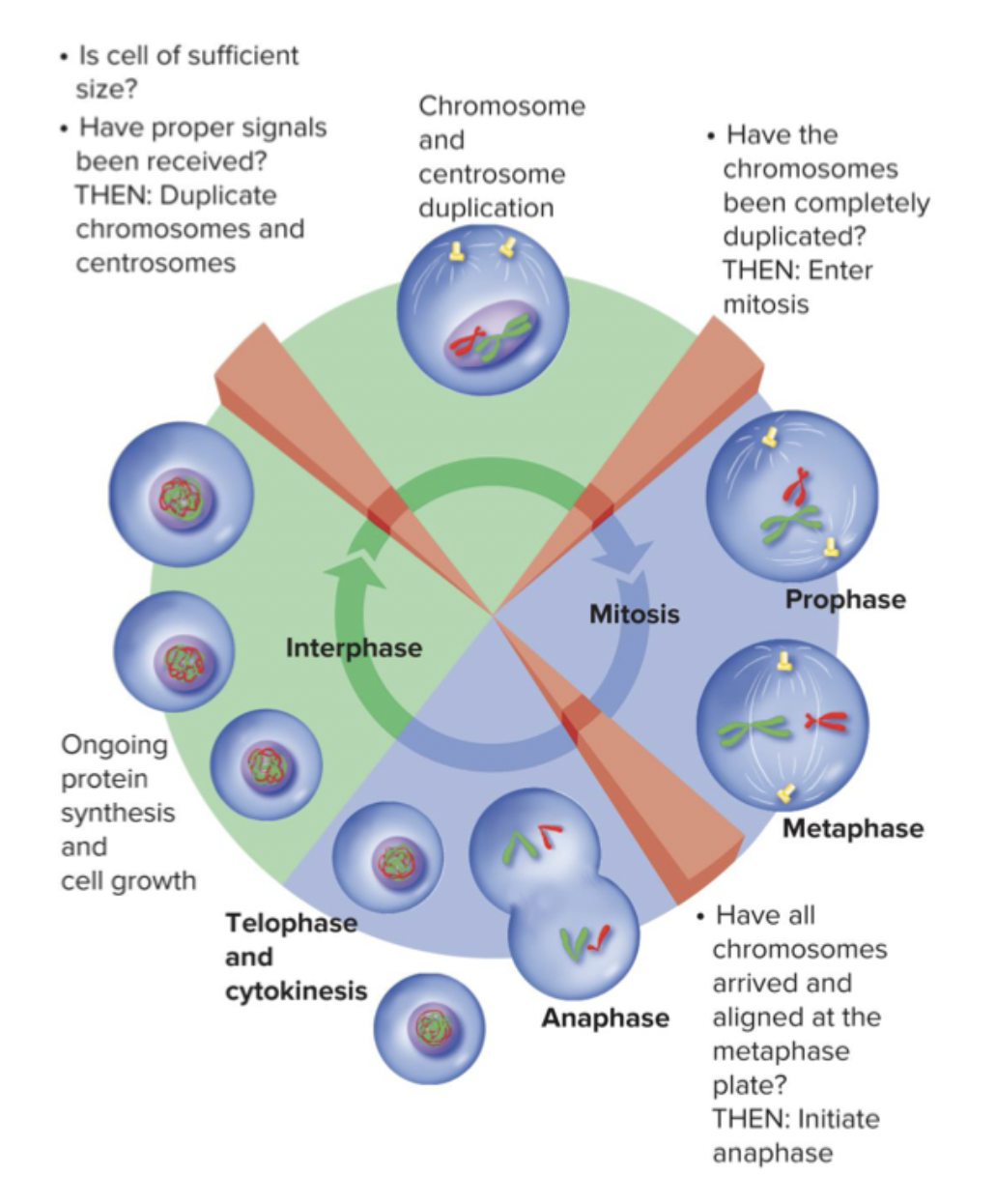
Prophase → prometaphase → metaphase → anaphase → telophase → cytokinesis
Prophase: Chromosomes condense and become visible. Centrosomes move apart toward opposite poles. Nucleoli begin to disappear. Microtubules are created.
Prometaphase: Nuclear envelope breaks down. Microtubules from centrosomes invade the nucleus and connect to kinetochores in the centromere of each chromatid. Mitotic spindle forms from three kinds of microtubules (astral, kinetochore, and polar), first two connect
Metaphase: Chromosomes align on the metaphase plate with sister chromatids facing opposite poles. Forces pushing and pulling chromosomes to or from each pole are in balanced in equilibrium keeping chromosomes in place. Everything is aligned at center of the cell.
Anaphase: Centromere of all chromosomes split simultaneously. Connection between centromeres of the sister chromatids is severed. Kinetochore microtubules shorten and pull separated sister chromatids to opposite poles (characteristic V shape)
Telophase: Nuclear envelope forms around each group. Nucleoli re-form. Spindle fibers disappear. Chromosomes uncoil and reform as a chromatin.
Cytokinesis: Begins during anaphase but not completed until after telophase. Cytoplasm of parent cells split into two daughter cells with identical nuclei. Mitosis finishes.
Produces two daughter cells.
Animal cells: Contractile ring contracts to form cleavage furrow
Plant cells: Cell plate forms near equator of cell
Both: organelles are distributed to each daughter cell (ribosomes, mitochondria, Golgi bodies).
Cytokinesis does not always occur after mitosis. Syncytial embryo. In fertilized drosophila eggs, 13 rounds of mitosis occur without cytokinesis. thousands of nuclei in a single cell.
There are regulation checkpoints at basically every phase. Really complex, more cell bio. Helps prevents errors in cell cycle.
Meiosis
Cell divisions that halve chromosome number.
Somatic cells - make up vast majority of cells in the organism, most often in G0 or are actively going through mitosis.
Germ cells - precursors to gametes. Set aside during embryogenesis. Become incorporated into reproductive organs. Only cells that undergo meiosis to produce haploid gametes.
Meiosis 1 - Reduces the chromosome from 2n to n. Meiosis 2 - produces four haploid nuclei.
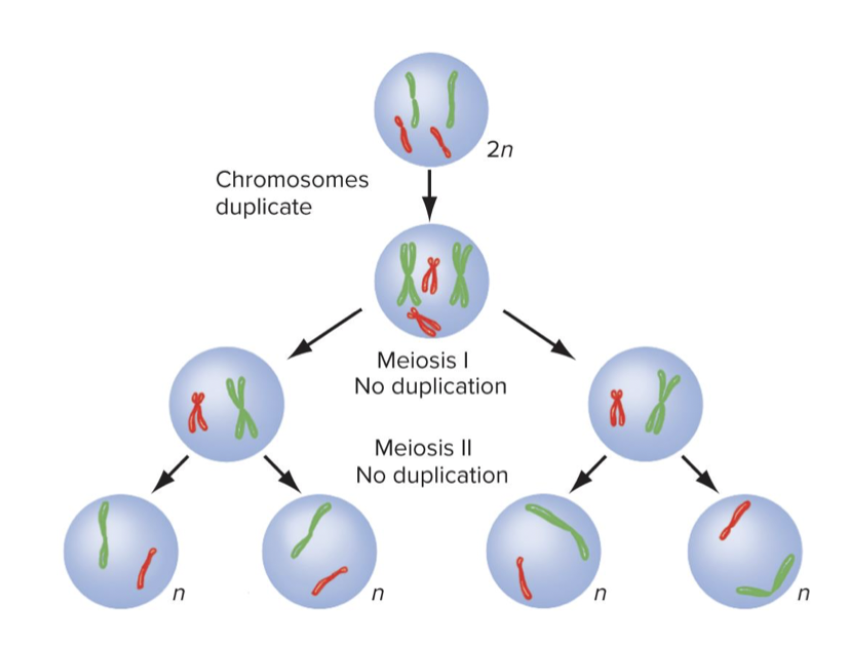
They do crossing over for genetic variability. Somatic cells.
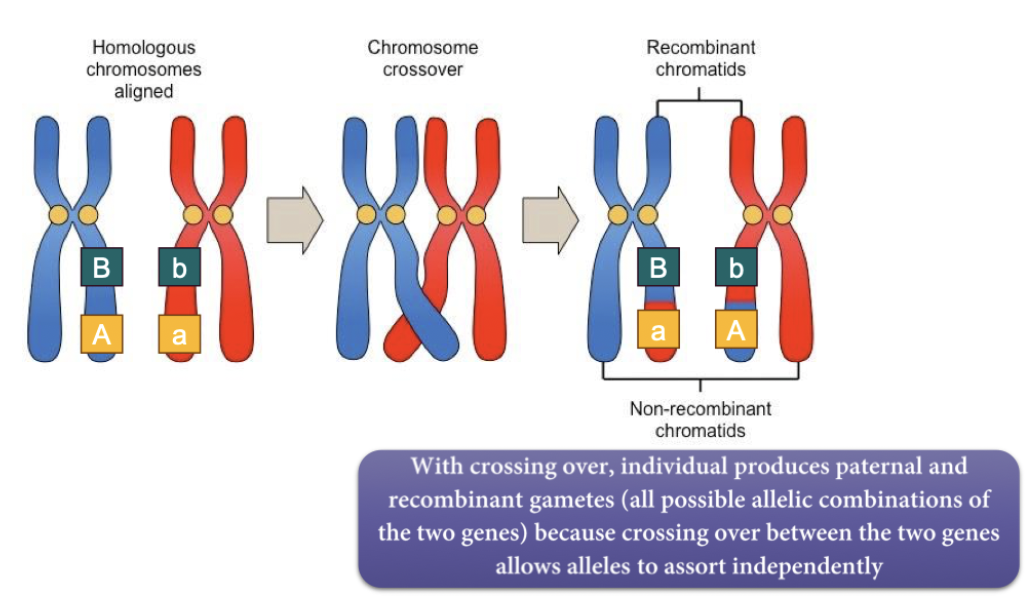
Recombinant chromatid - same combo/same side of chromosomes
Non-recombinant chromatid - different combo. opposite side of chromosomes.
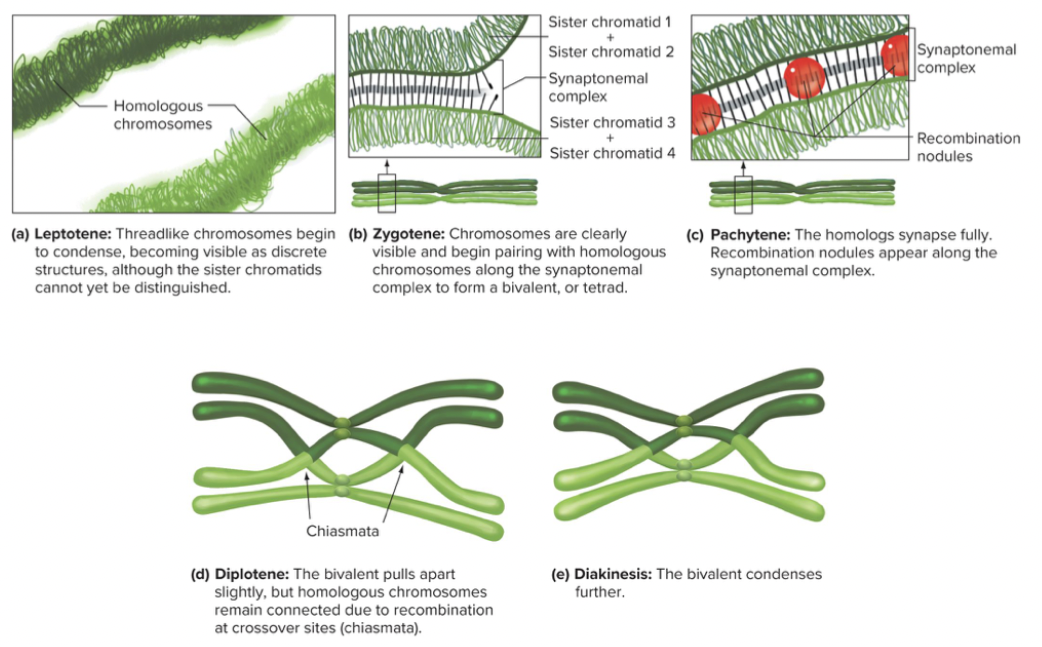
Substages of prophase: Diplotene and diakinesis
Diplotene: Synaptonemal complex (protein structure that forms between homologous chromosomes during meiosis) dissolves. A tetrad of four chromatids is visible. Crossovers points appear as chiasmata holding nonsister chromatids together.
Diakinesis: Chromatic thicken and shorten. At the end of prophase 1, the nuclear membrane breaks down and the spindle begin to form.
Homolog pairing during meiosis 1 forms a tetrad. Crossing over exchanges genetic information. Chiasmata play a role in proper segregation of homologous chromosomes.
Synaptonemal complex - forms a zipper between homologous chromosomes, holding them together, when crossing over occurs, points stay there past complex disappears. Play a role in proper segregation of 4 divided.
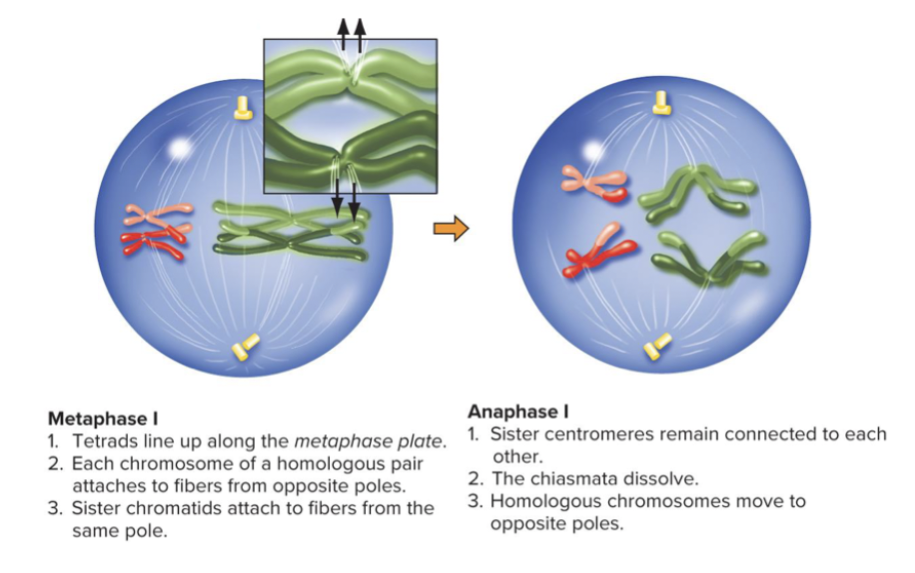
Metaphase 1 and Anaphase 1 = Centromeres do not divide, and sister chromatids do not separate, homologs are separated and move to opposite poles.
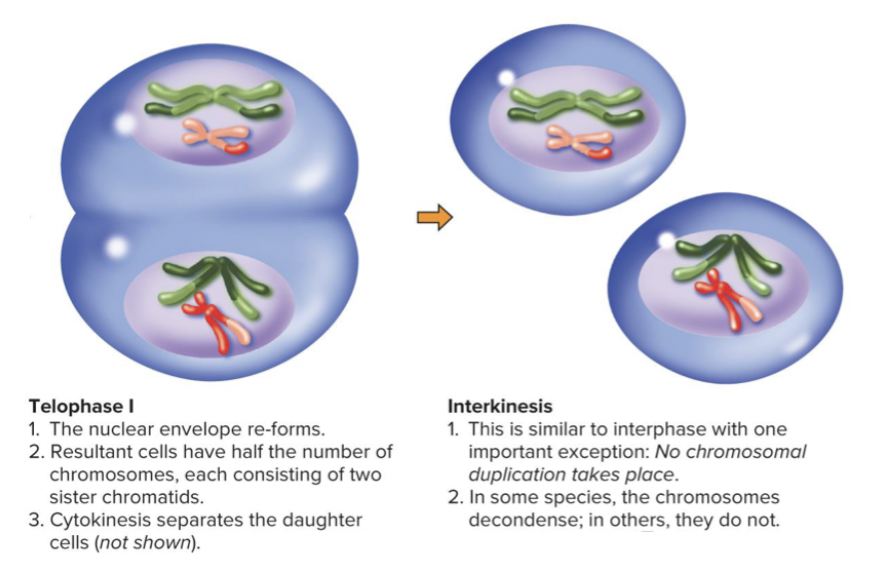
Telophase I and Interkinesis: interkinesis is similar to anaphase, but not chromosomal duplication
Meiosis I is a reductional division: cells contain half the number of chromosomes as parent nucleus each arranged as two sister chromatids joined at centromere.
Haploid nuclei progress through process very similar to mitosis to create haploid daughter cells. Meiosis 2 is an equatorial division. Some cells get stuck in intermediate phase here.
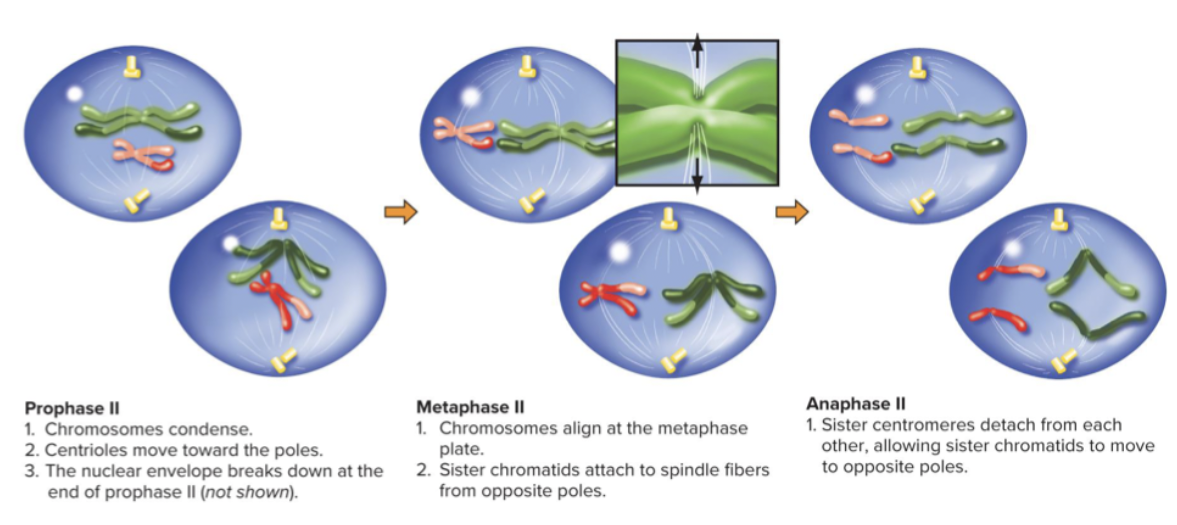
Four haploid daughter cells are result of meiosis 2. Finished by telophase 2, nuclear envelope comes back, and each cell has a copy of 1 chromosomes. 4 daughter cells are not identical to parent cell - most important.
Mistakes in Meiosis
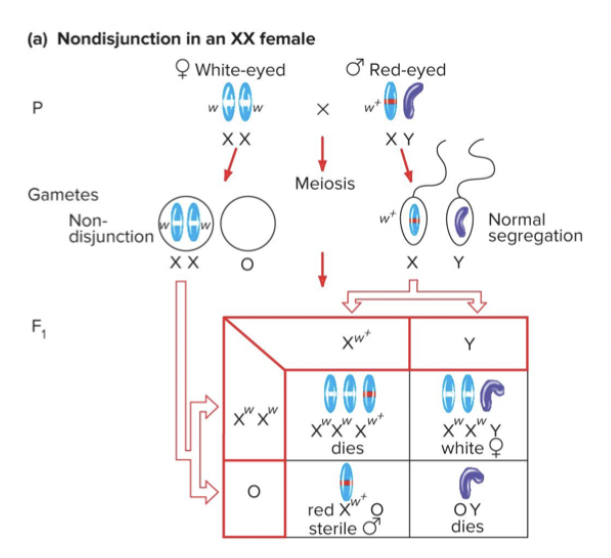
Non-disjunction: homologs of a chromosome pair do not segregate during meiosis 1. May result in inviable gametes or embryos. Can also result in abnormal chromosome numbers in viable individuals (trisomy 21, down syndrome, XXY, Klinefelter syndrome)
Spindle forms and doesn’t attach properly on either side, 2 are pulled to one and 0 pulled to the other.
Meiosis contributes to genetic diversity:
Crossing-over between homologs (Prophase I) creates different combinations of alleles within each chromosomes
Random alignment of homologs at the metaphase plate (Meiosis I) creates equal probability of alleles of different genes assorting together homologs segregating (Mendel’s law of segregation).
Segregation of homologous chromosomes (Meiosis I) and sister chromatids (Meiosis II) ensures each gamete carries one allele of each gene (law of segregation)
Sex chromosomes and sex linkage
W.S. Sutton studied meiosis, testes cells had 24 chromosomes. After meiosis, two types of sperm was formed → ½ had 11 chromosomes and an X, ½ had 11 chromosomes and an Y.
After meiosis, only one type of egg was produced. All had 11 chromosomes with an X.
Fertilization of egg with sperm carrying an X → XX female. with Y → XY male. Sutton concluded that X and Y chromosomes determine sex.
Children receive an X from mother, and X or Y from father. Results in approximately 1:1 ratio of females to males. This is because of the law of segregation.
The primary determinant of maleness is a single gene called SRY (sex determining region of Y). Sex reversal (XX males and XY females) provided evidence implicating SRY. SRY is always present in XX males. SRY is always nonfunctional in XY. Small proportion of Y determining is present in male. Can have 2 X and present as male if you have that small region that determines sex. SRY+ is in males. If you have one, then it makes them male. SRY is always non-functional in XY females. SRY is always functional in XX males.
Human chromosomes contain genes unrelated to sex. Some genes are present on both the X and Y chromosomes of humans, located at the ends of the Y chromosome called pseudoautosomal regions (PARs). Enable sex chromosomes to pair during meiosis. Even though sex chromosomes are different, this all allows them to pair.
Sex determination differs across organisms. In drosophila, the ratio of X chromosomes to autosomes determines sex. In humans, presence or absence of Y chromosome determines sex. Abnormal numbers of X or Y chromosomes have different effects in humans and flies.
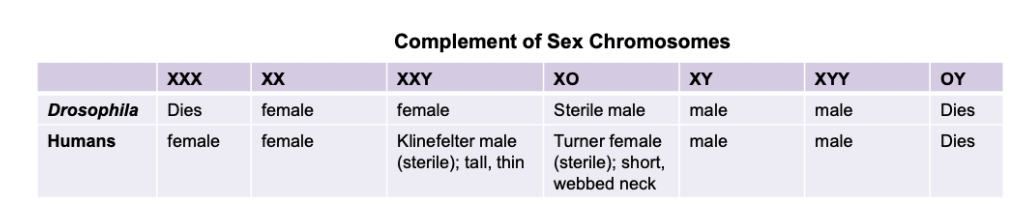
Heterogametic sex - sex with two different kinds of gametes (for example, XY males in humans, ZW females in birds)
Homogametic sex - sex with one type of gamete (for example, XX females in humans, ZZ males in birds)
In some species, sex is determined by environment (for example, temperature).
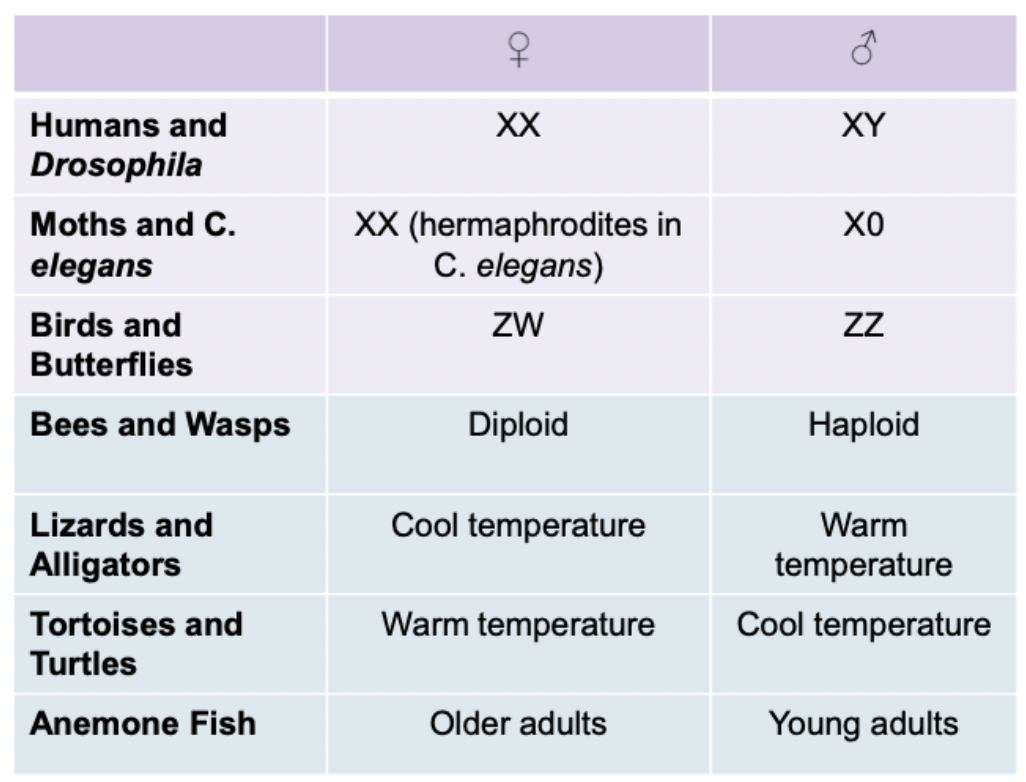
Other species may be completely different. Just the number of sex chromosomes in moths. Anemone fish have sex determined by age. Are male and become female as they age.
Germ-line: specialized diploid cells set aside during embryogenesis
Gametogenesis: gamete formation. Differs across species. Involves meiosis as well as some specialized events after meiosis. Oogenesis produces one ovum from each primary oocyte. Spermatogenesis produces four sperm from each primary spermatocyte.
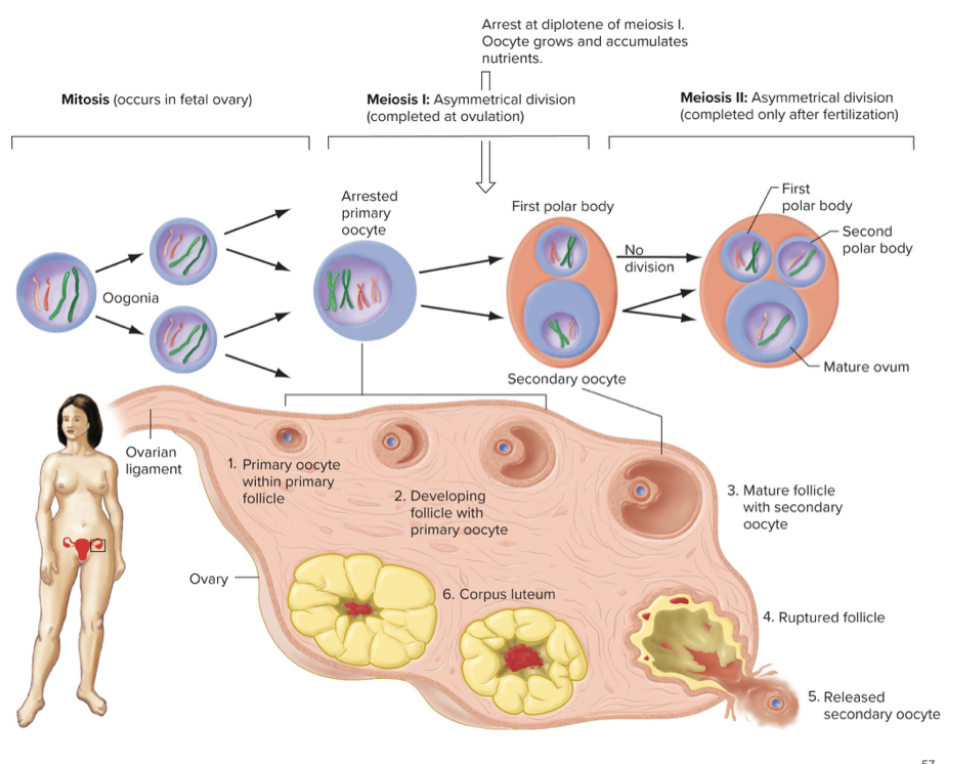
At puberty, one primary oocyte per month resumes meiosis at ovulation. Completion of meiosis I produces a large secondary oocyte and smaller sister cell, the first polar body. Secondary oocyte arrests in metaphase on meiosis II. At fertilization, meiosis II is a completed and produces a mature ovum and second polar body.
You have mitosis of germ cells in the fetal ovary. The cells are oogonia. They go to meiosis 1. when they grow it gets nutrients. Oocyte female is very big. Oocytes get arrested during this phase. Only one of the gametes polar bodies or something with all the cytoplasm. One daughter cell gets all nutrients and matures into a gamete. 2 polar bodies. Division of genetic material is the same but 2 are asymetrical polar bodies.
Long arrest in meiosis 1 where chromosomes are arrest. Longer they're there, they're doing chromosome something.
Likelihood of down syndrome in geriatric patients is higher because they've been in arrest for a very long time.
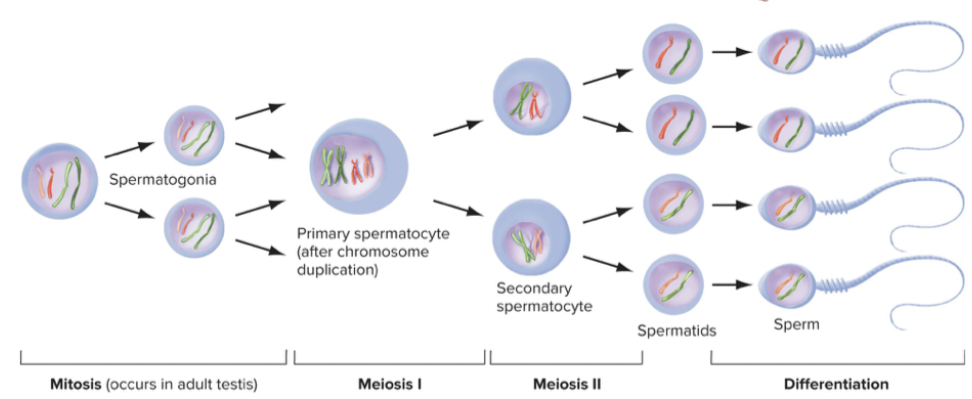
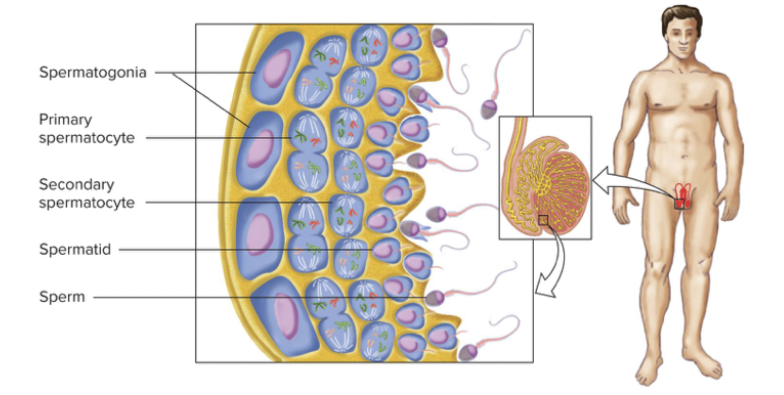
Four haploid sperm produced by symmetrical meiosis of each spermatocyte. Mitosis and meiosis occur throughout adult life. Entire process of spermatogenesis takes approximately 48 to 60 days. Billions of sperm produced over lifetime.
Spermatocyte. Whole process takes two months, no arrest. Produce millions of sperm over lifetime.
2 phases of spermatocyte - spermatid - sperm
Divide many more times than eggs ever do.
Sex linkage
Geneticists of early 20th century tested for transmission of characteristics other than sex via transmission of sex chromosomes. They needed to track chromosomes using microscopy to confirm or refute the correlation of traits with the presence of chromosomes. They needed to track chromosomes using microscopy to confirm or refute the correlation of traits with the presence of chromosomes. T.H. Morgan used Drosophila melanogaster in these experiments based on its short generation time and large numbers of offspring.
Different from another linkage next week. It was coined during 1900s. Even if its not sex itself getting it, correlation of traits help.
Morgan (1910) discovered white-eyes drosophila mutant and did crosses. Eye colour is associated with sex. If he crossed red eye female and white eye male he saw that the offspring all had red eye, because they got red eye from mother.
He found only males that were white eyes. When heterozygous female and red eye male.
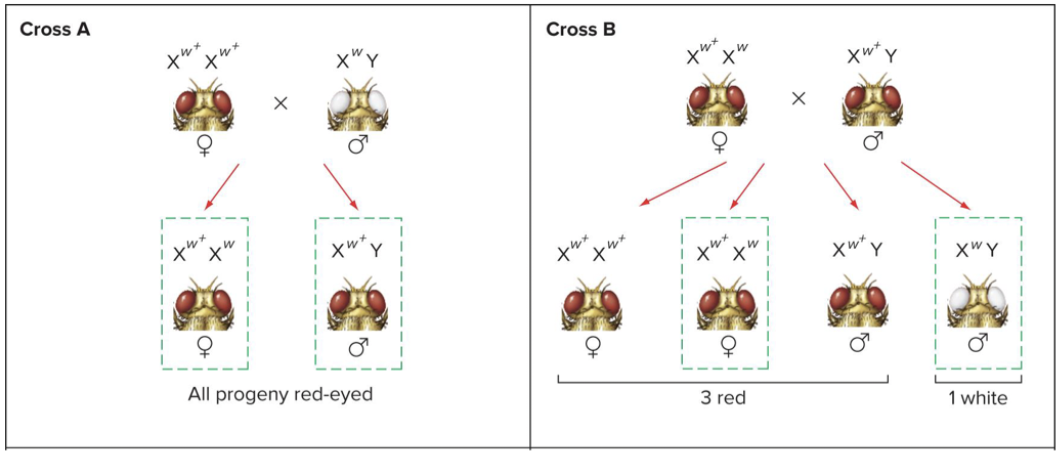
Progeny phenotypes are different depending on whether ww allele is carried by the mother or the father. Sex linked.
Heterozygous female and white eye male. Depending on which direction cross occurred, the proportion of the progeny was different. Not the same in autosomal genes. Depends on parents genotypes.
Crisscross inheritance: daughters inherit the phenotype of their fathers, sons inherit the phenotype of their mothers.
Morgan concluded the white gene was X-linked, carried by the X chromosome.
Hemizygosity: refers to reduced allele number due to sex-linkage and possession of a single sex chromosome (typically X or Y) rather than through heterozygosity of autosomal alleles. Reduced allele number due to sex-linkage.
Rare mistakes in meiosis helped confirm chromosome theory of inheritance. C. Bridges found 1/2000 male progeny of white females have red eyes. Hypothesized that these red-eyed males arose from mistakes in chromosome segregation during meiosis in white-eyed females, called nondisjunction.
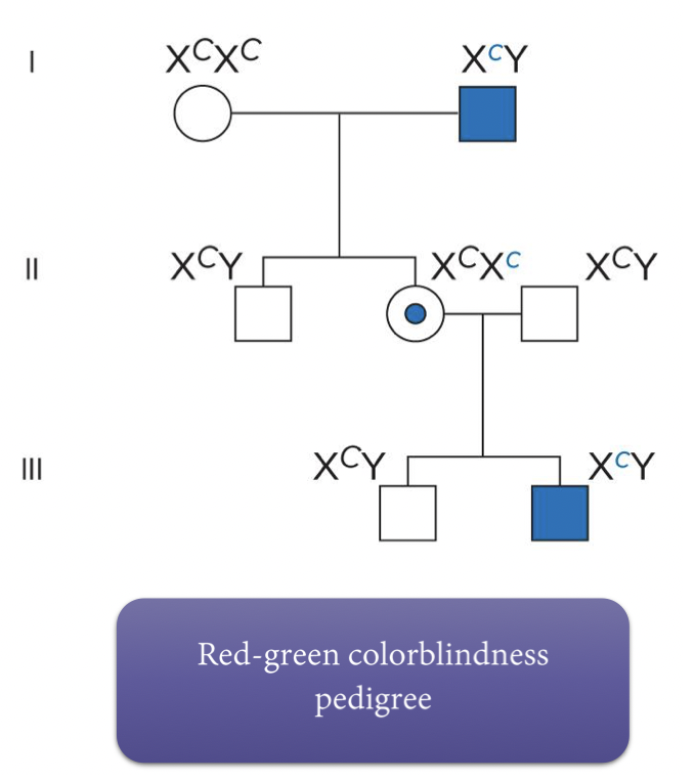
Pedigree patterns can indicate sex-linked inheritance. X-linked recessive mutation never passes from father to son, daughters of affected males are carriers. ½ of sons of carriers will inherit the trait. X-linked dominant trait seen in every generation, affected male will produce affected daughters and unaffected sons. Y-linked only in males.
Is a disease sex-linked or not?
If disease is successful, it never passes from father to son. Sons always receive X from their mother.
Daughters are normally carriers. Affected male will produce affected daughter. Mother carrier - 50% of sons will have disease.
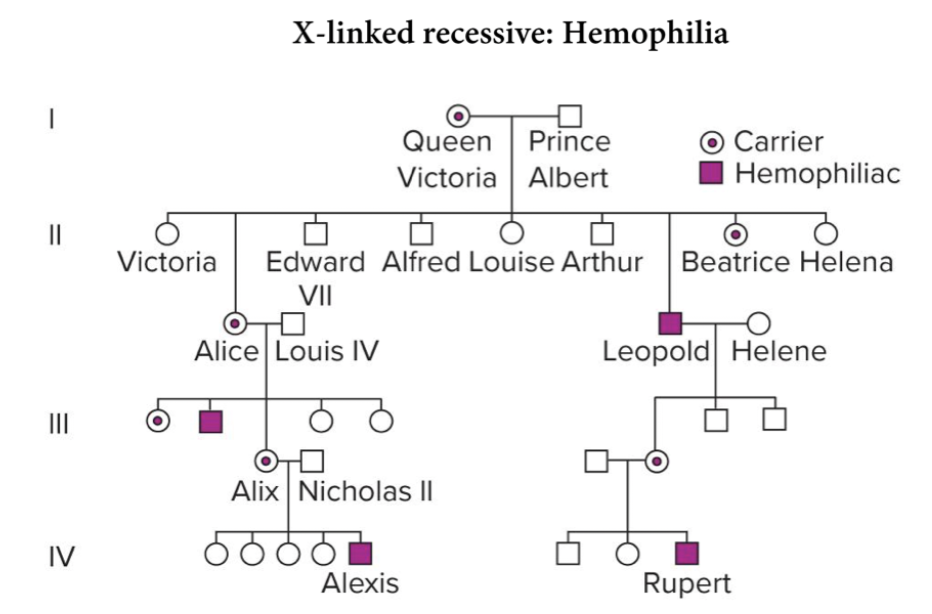
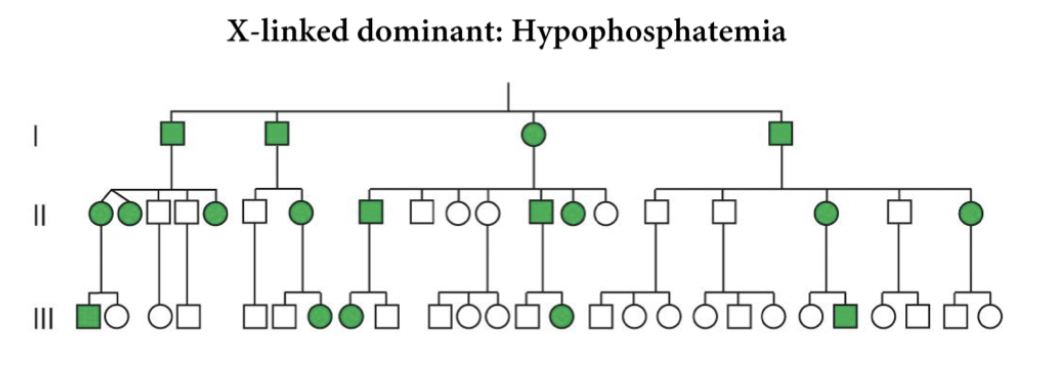
Human females have two X chromosomes and males have only one.
Dosage compensation: offsets what would be double the gene products in the homogametic versus heterogametic sex.
In female cells, on X is randomly inactivated. The inactive X chromosome is condensed into a Barr body.
Mechanisms that make X-link more viable. X been more resistant, dosage compensation. Majority of functions on body. Superactivation of chromatin - barr body, really tight.
In the early female embryo, each cell independently inactivates one X chromosome. Random.
Sex-limited traits: affect a structure or process found in only one sex. For example Drosophila stuck mutant males can’t separate from female after mating
Sex-influenced traits: Appear in both sexes but hormonal differences may cause difference. For example, male pattern baldness.
Two organelles with independent genomes: mitochondria and chloroplasts. haploid genomes. Almost exclusively inherited maternally. Mitochondrial disorders caused by mutations in the mitochondrial DNA are exclusively from mothers.
Very vital organelles. Exclusively inherited maternally because the cytoplasm comes from egg and not the sperm - usually only maternal because we get all are organelles from our mothers.
Maternal inheritance (mitochondrial): mitochondria contain their own DNA. All mitochondria are inherited from the mother. Mitochondria genomes are haploid. There is 100% chance that each child in the family will inherit a mitochondrial disease if the mother is affected. There is 0% chance that each child in the family will inherit a mitochondrial disease in the father is affected.
Linkage and Recombination
Linkage changes Mendelian ratios. Genes on the sex chromosomes are inherited differently than Mendelian traits. Still predictable once you know the mechanism of sex determination.
Other mechanisms also give rise to expectation from Mendelian patterns → Gene linkage.
Linkage is predictable, but may be less, just different type of predictable in terms of how they’re doing things.
Same chromosome exchange arms during meiosis. Not the case for all genes that are the same chromosomes.
Reason they do not do it independently - requires the closer two genes to cross over and occur. Not independent cuz they're the same. Crossing over requires physical distance.
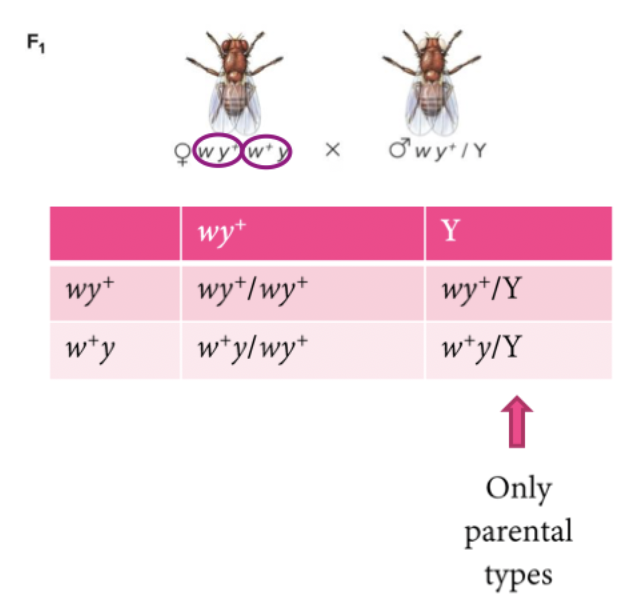
Gene linkage was discovered in X-linked Drosophila. Two X-linked genes in Drosophila. w+ (red eyes) and w (white eyes). y+ (brown body) and y (yellow body). F1 males get their only X chromosomes from their mothers, F1 females are dihybrids.
White gene, mutant phenotype, yellow gene. Cross female fly, mutant white and wild type yellow. All receive at least one yellow. Because female is white eye, all male are white eye because they get white eye from mothers.
If white and yellow genes assort independently, dihybrid females should produce four types of gametes. Regular type and the recombinant. If female in lower left corner get's their stuff normally, dihybrid female for both genes, the parental combinations she produces are what she has. They can recombinant to get what she got.
Female offspring have all dominant phenotypes for yellow. Can’t distinguish genotype. 100% wildtype yellow, 50% white eyes, 50% red eyes. We would expect all females dominant for yellow, but females have 2 X with 2 chromosomes. You can't tell which phenotype comes from which. Male only X, the phenotype will be exactly the genotype ratio. Allow them to make discovery of lineage.
Male offspring have phenotypes that reflect genotype. Males are hemizygous for X chromosomes, which means that recessive alleles are never hidden by dominant alleles. For females, 4 different genotypes, but 2 different phenotypes. Because males are hemizygous for X chromosome, they have 4 phenotypes.
The progeny from the F1 dihybrid cross did not yield the expected results. Remember, with crossing over and independent assortment we would expect equal 25% of each possible genotype in the progeny.
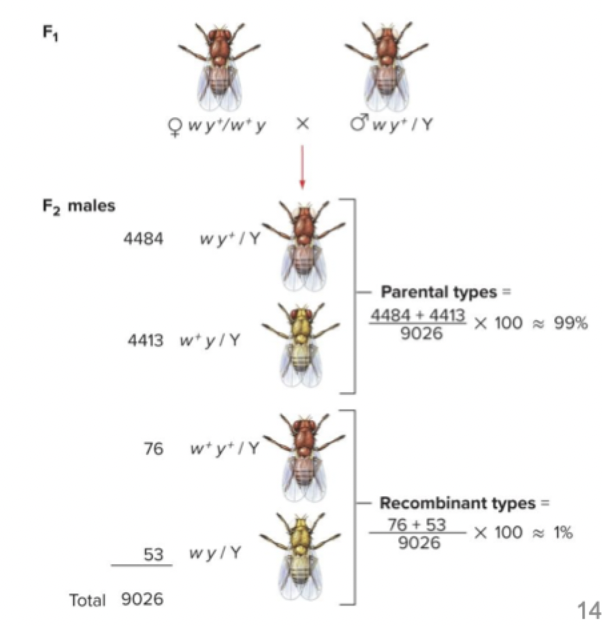
Linkage, 2 different genes something something
F1 dihybrid, if female made 4 different gametes, no gene linkage - 1:1:1:1 ratio, but they found they got 99% had parental, 1% had recombinant. The recombinant types are very rare. Two genes are linked. Can still happen sometimes, majority of the time they're too close together.
By looking at meals only, we get what we just talked about. By looking at the males only, we know that the genotypic rations are the phenotypes we see.
Actual progeny show 49.5% of each parental genotype and 0.5% of each recombinant genotype.
The w allele is bound to the y+ allele and w+ allele is bound to the y allele. Parental type gametes occur more frequently.
Independent assortment: Alleles of genes on different chromosomes always assort independently. Alleles of genes on the same chromosome need recombination assort independently. Have alleles on different chromosomes. Align randomly at plate.
Since yellow and white are on the same chromosome, without recombination we would expect to see only parental genotypes in the offspring (females would only make two types of gametes)
There is an even (with exceptions, but let’s assume so for now) probability of recombination occurring anywhere along the two homologous chromosomes during meiosis. Closer genes are physically, the likely is that recombination occurs between them.
Even probability the recombination will occur anywhere. 3 hypothetical genes, likelihood of genes, they're occur between B and C. Likelihood of it happening depends on closeness. If it doesn't occur, no assortment.
Parental type gametes occur more frequently when there is linkage. Parental more common > recombinant → probably linked.
Early evidence for recombination came from observing chromosomes with physical modifications.
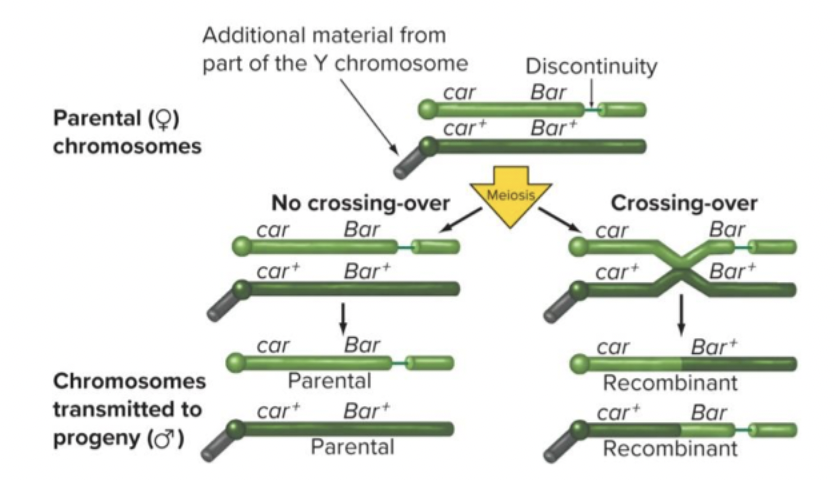
Crossing over (CO) will most likely happen somewhere along the two homologous chromosomes every meiosis, but it might not happen between two specific genes. Relative to those genes we call.
No crossing over: 4 parental gametes
Single crossing over: 2 parental and 2 recombinant gametes
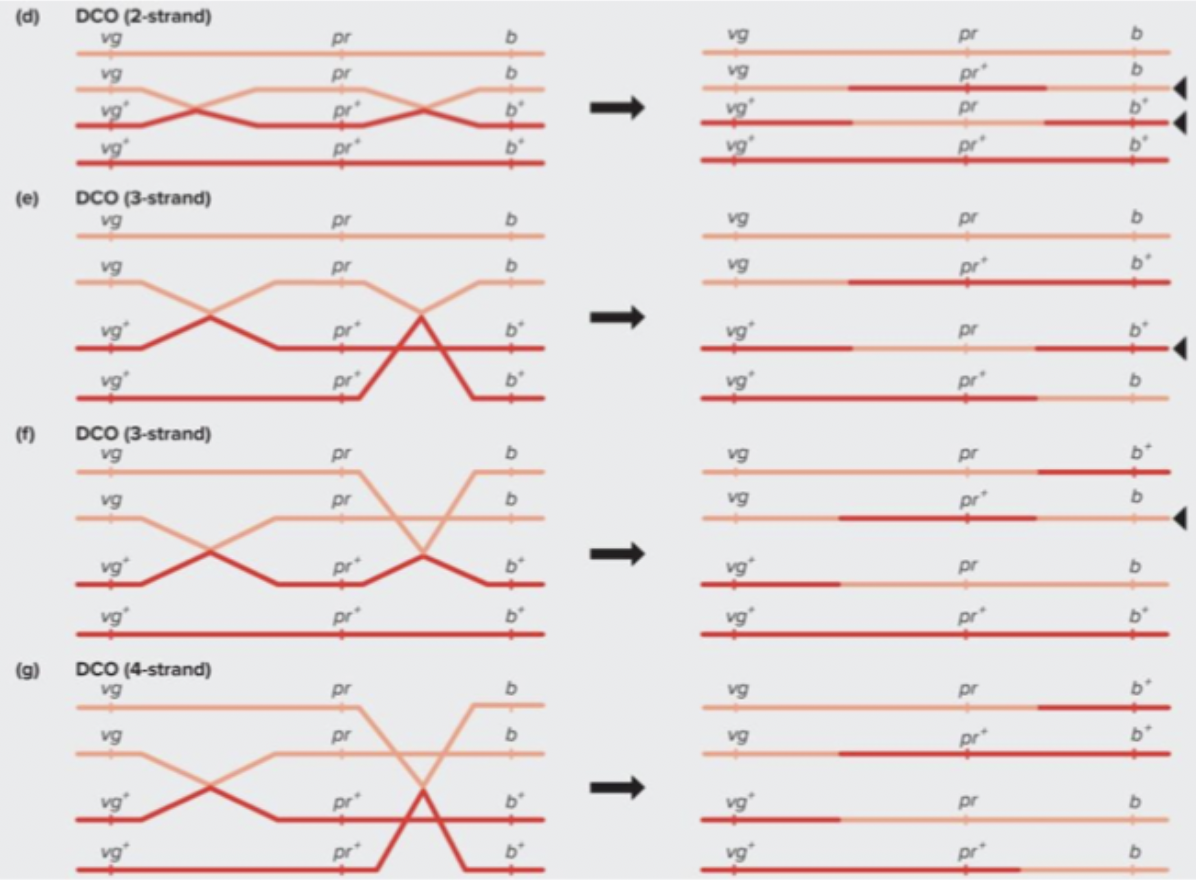
Double crossing over: varying proportions of parental and recombinant gametes.
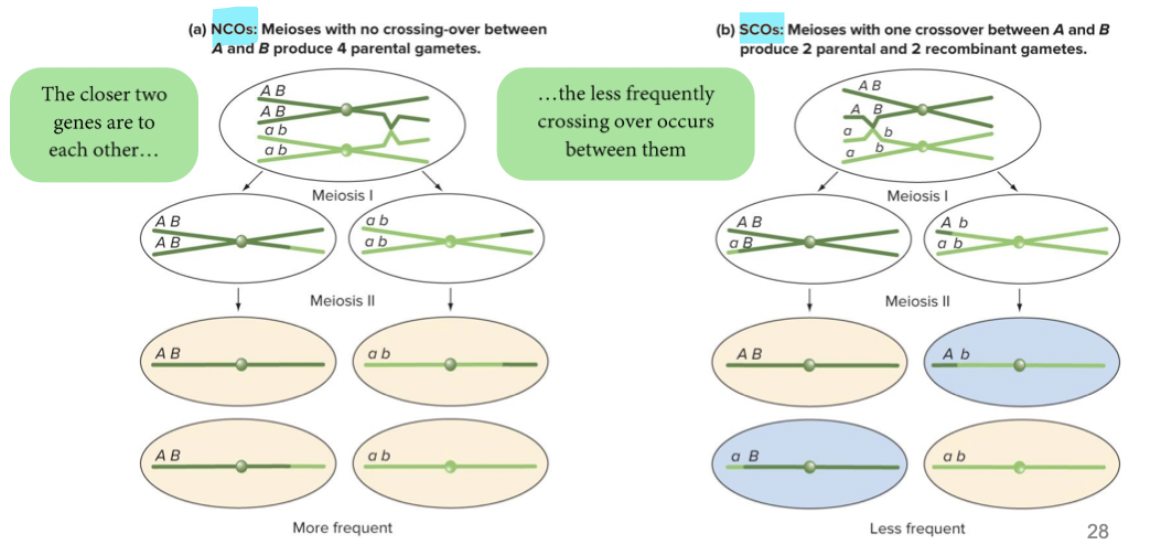
If genes are far enough apart, DCOs can occur. Possible double cross-over combinations still produce, on average, 50% recombinant gametes. Maximum frequency of recombinant gametes is 50%.
When you have double crossing over, they can happen on same strand. On sister chromatid with homologous chromosome. One you get on the end. Looks parental even though crossing over happened twice.
All 4 sister chromatids = 100% of recombinant gametes
If genes are sufficiently far apart on same chromosomes, one crossover likely gonna happen. They will then assort independently → unlinked. Parentals = Recombinants. If genes are sufficiently close together on the same chromosome and 0%<RF<50% → linked.
We assume how close they are based on recombinant frequency.
1 RF = 1 map unit (m.u.) = 1 centrimorgan (cM)
RF is a measure of physical distance between two genes. About 1.1% of recombinant frequency - 1.1 m.u.
To assess recombinant phenotypes for autosomal genes. Remember that in the yellow/white example, we had the advantage of males being hemizygous. Complicated with autosomal genes.
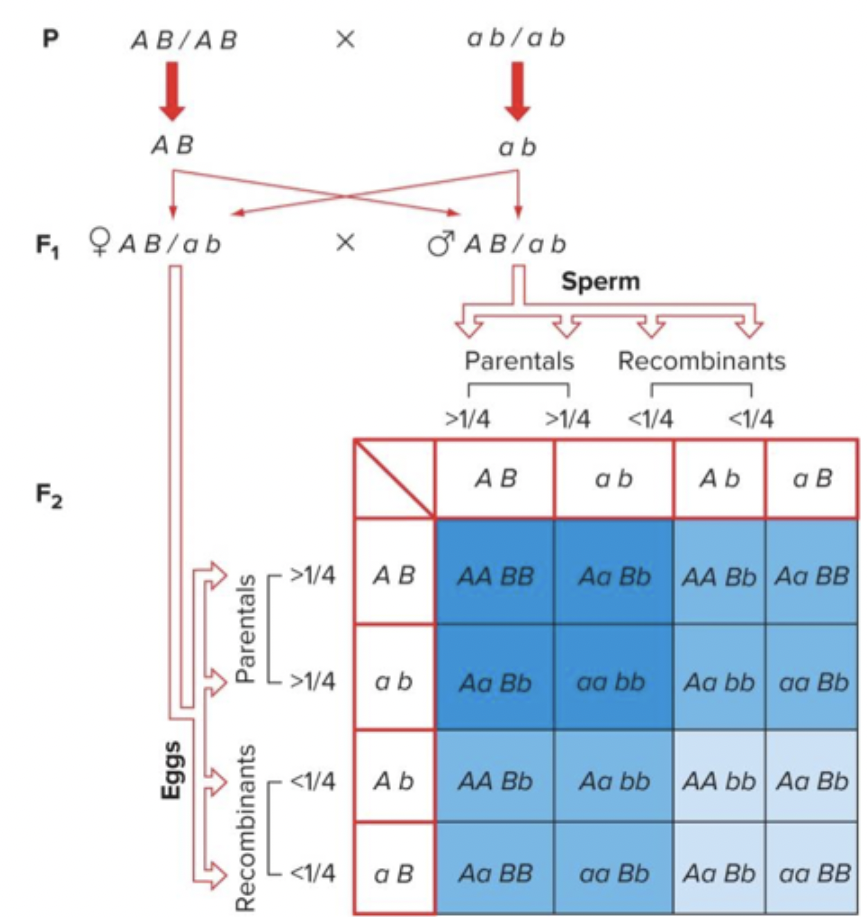
50% recombination (no linkage) in this scenario results in: individuals with two parental chromosomes (dark blue), one parental and one recombinant (medium blue), two recombinant (light blue).
Different ratio than 9:3:3:1. Dominant alleles hide where recombination occurred. Sometimes parental looks like RF.
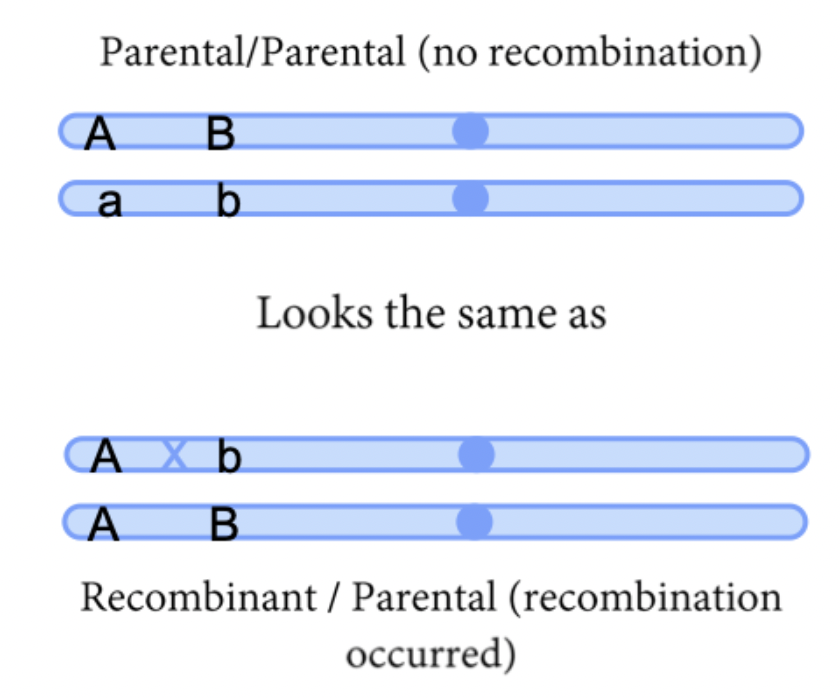
Test cross to see linkage - test F1 against an individual that is homozygous for both recessive genes. The phenotypes of progeny reveals the genotype. Then we can count the number of recombination events that occured in the females, because they are not obscured by dominant alleles coming from the male.
From male, progeny will only receive recessive combinaiton. Sex link can be any cross, and looking at male progeny only, while autosomal need a test cross. Cross double heterozygous to one that is double recessive. Recombination only occurs in females in drosophila.
Drosophila have no genetic methylation. Wildtype is brown body. Male is completely recessive/mutation. If we get individuals with black bodies and curved wing and brown body with straight wings. Recombinant is otherwise.
Gene mapping:
Start with known parental genotypes with mutations for two genes
Score the progeny for the number of recombinants
Divide the number of recombinants over total progeny to get recombination frequency (m.u.) between the two genes.
Repeat for another pair of genes
Eventually you will get a genetic map
Two point crosses: crossing two genes to calculate recombinant frequencies (m.u.)
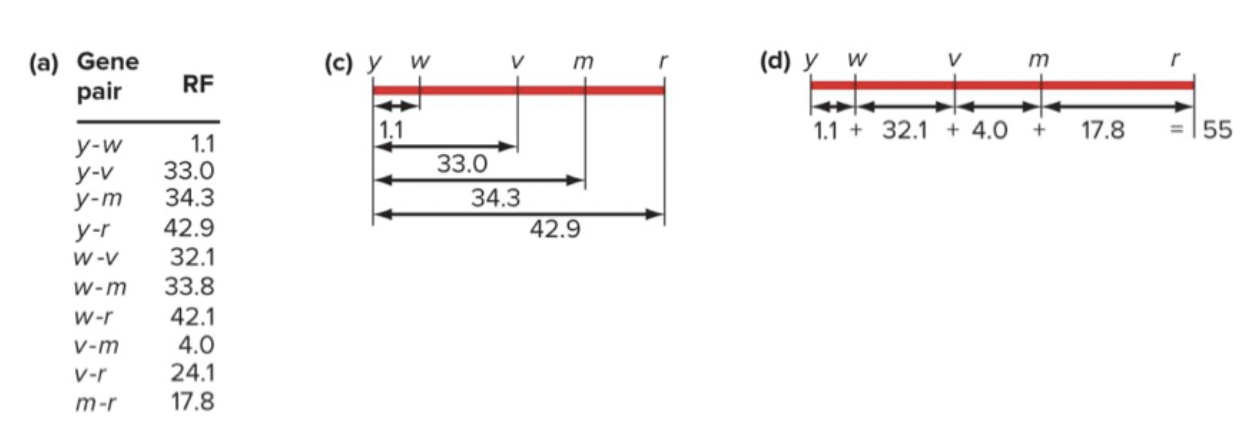
Sum up individual differences. Add up to 55 m.u., it should be 42.9. The actual distances between two genes do not always add up.
Limitations to two point crosses:
Pairwise crosses are time and labor consuming
Actual distances between genes do not always add up
If two genes are very close together, it is difficult to determine gene order.
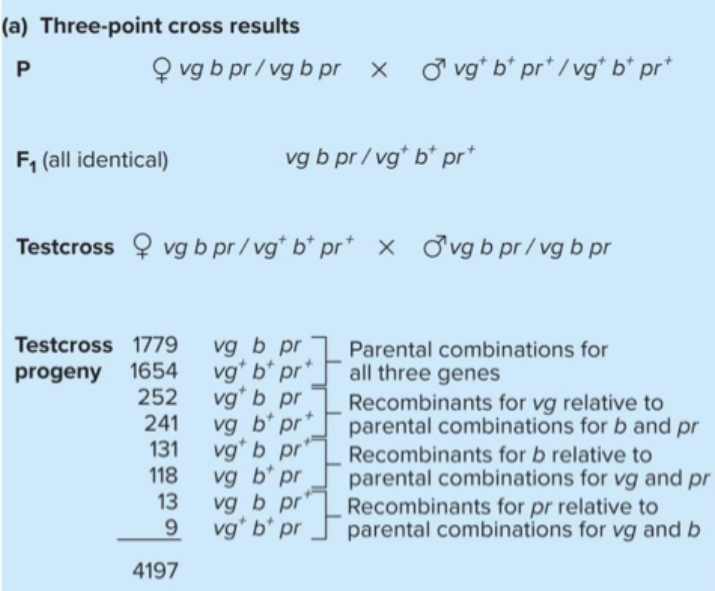
Three point crosses: cross individuals with mutations in three genes. Testcross progeny have four sets of reciprocal pairs of genotypes. Most frequent pair has parental configuration of alleles. Least frequent pair results from double crossovers. Examination of double crossover class reveals which gene is in the middle.
Fix this through doing 3 point crosses. Sum of individual differences doesn't add up. Four sets of reciprocal pairs of genotypes.Parental combinations - one gene is recombinant to the 2? Even if you don’t know parental combination, the most frequent classes are always parental. Least frequent is double crossover. If you examine double crossover, the gene that's different, always the one in the middle.
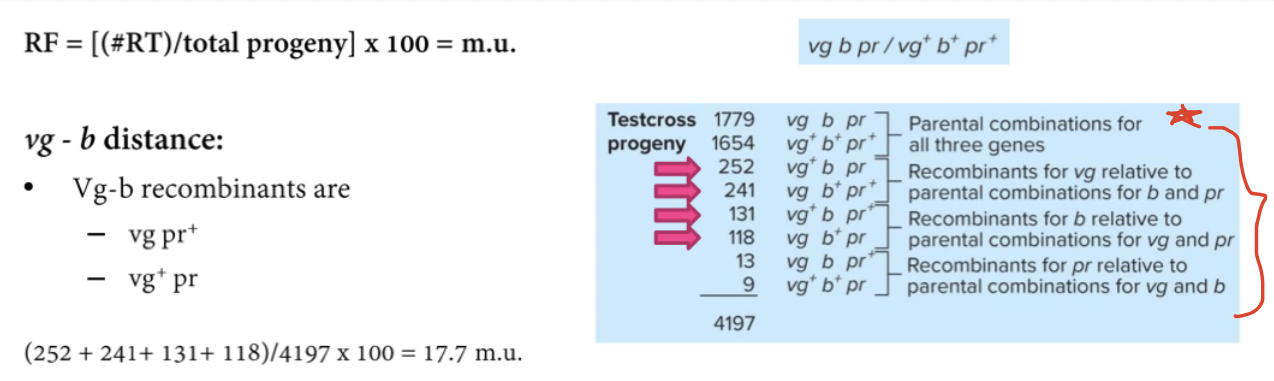
Key is to identify which alleles are recombinant and parental for two genes at a time. Take vg and pr first. Which ones are recombinant for vg and pr. All of the progeny divided by total number of progeny. We do the same thing other.
If RF for vg - pr and b-pr it’ll be 12.3 and 6.4 which is 18.7, but the cross says it should be 17.7.
When we do 3 point cross, sum of individual differences add up. When you do point crosses that are far apart, they’re the same. Back to schematic. Cross over still occur in individuals. Underestimating difference between some of those double crossovers.
Some double crossovers look like parental for the genes at the ends, even though crossover occurred (twice) → Recombination rate between end gene is underestimated.
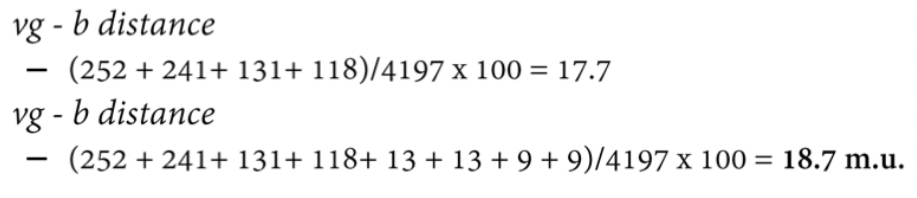
Add up the genes. Class that occurs the least often. If you add double crossover tab twice it add up, because it happens twice.
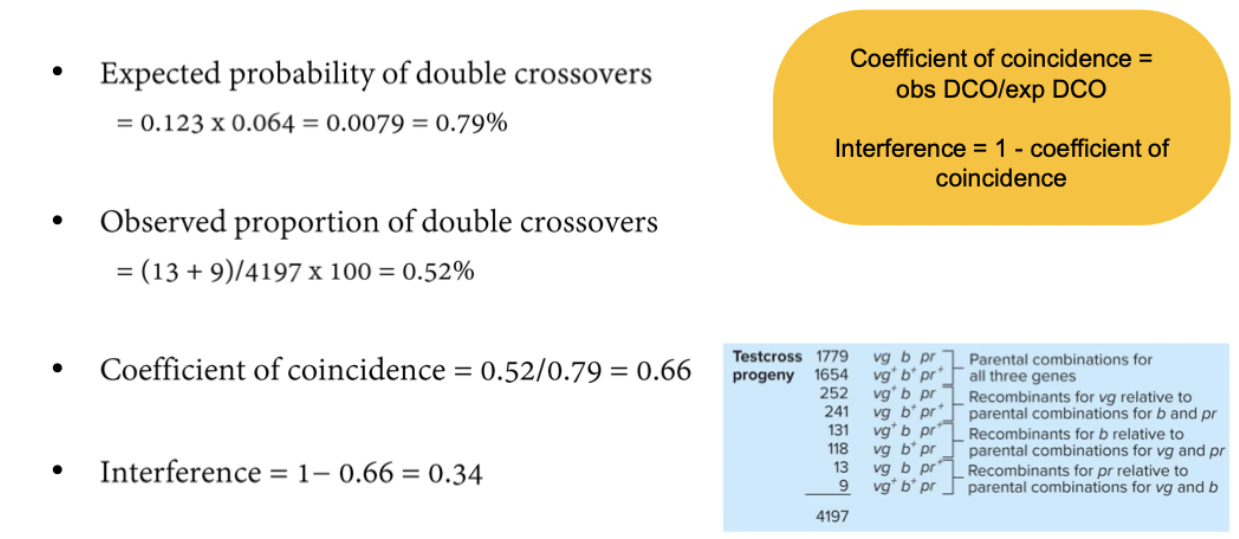
Chromosomal interference: occurence of crossover in one portion of a chromosome interferes with crossover in an adjacent part of the chromosome.
Compare observed and expected frequencies of double crossovers (DCO).
Coefficient of coincidence: observed DCO frequency/expected DCO frequency.
Interference = 1 - coefficient of coincidence
Expected probability of double crossovers is the product of the single crossover frequencies in each interval.
Genetic Maps vs. Physical
Order of genes revealed by genetic mapping corresponds to the actual order of genes along the chromosomes. Physical distance (amount of DNA) does not always show direct correspondence to genetic distance. Double, triple, and more crossovers. 50% limit on observable recombination frequency. Non-uniform recombination frequency across chromosomes. Recombination rates differ between species.
Very likely that crossing over occurs close together when they’re close together. Chromosomal map always respond correspondingly. Specific genes in the genomes with affects, how we produce so many antibodies.
Recombination hotspots
Genes A and B are separated by the same number of base pairs as genes B and C. A and B flank a recombination hotspot, they appear much more distant from each other than do genes B and C on a genetic map.
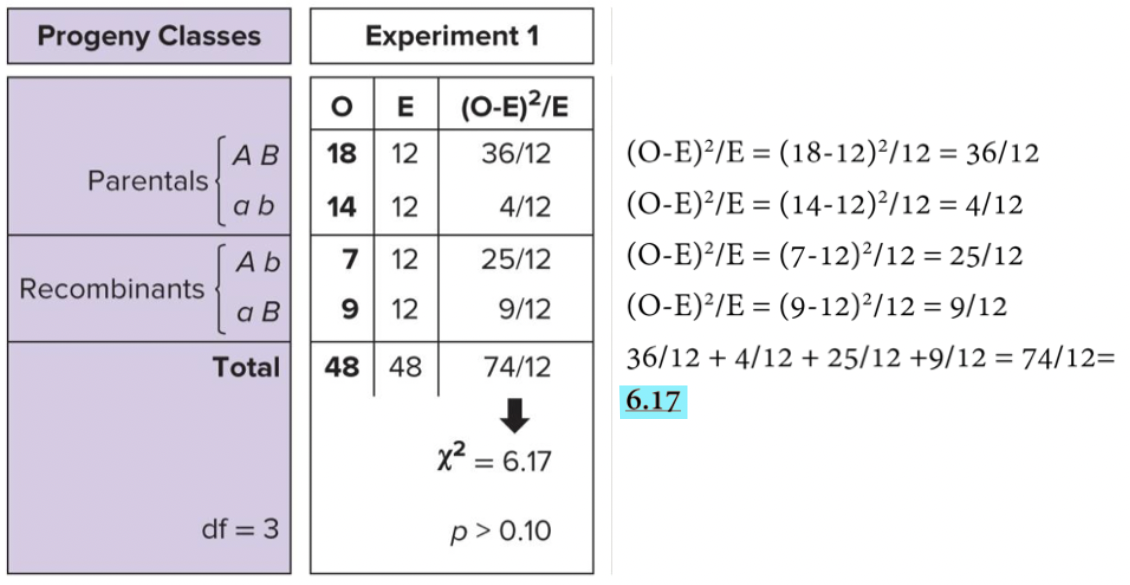
Chi-Square Test
Goodness of fit test. Significant = data differs from expected and you reject the null hypothesis (this means there is linkage)
Total number of progeny, how many classes of progeny, number of offspring observed in each class. Calculate number of offspring expected in each class if there is no linkage. 1:1:1:1 segregation.
sum of (O-E)² / E
Useful definitions
Syntenic genes: genes located on the same chromosome
Locus (pural loci): a specific location on a chromosome
Linkage groups: a group of genes that can all be linked to each other. As all genes on chromosome get mapped, the linkage group eventually converges to the whole chromosome. The genes at the ends of the chromosomes may not be linked, but they still are part of the same linkage group.
Lab Information
Flies
Thomas Hunt Morgan - 1933 Nobel Prize, physiology. Discovery of the white-eyed mutation in the fruit fly, Drosophila. Since then, (6 times to) 11 scientists have been awarded a Nobel Prize for work with fruit flies.
Drosophila benefits: Short generation time, many offspring, cost effective, largest number of transgenic tools available, genes and pathways are conserved.
Ways to anesthetize: Ice, CO2, Ether. Eggs hatch into larva, after third larva instar, they become pupa and turn into an adult fly. Coming out of the pupa can is called eclosing.
Drosophila characters:

Testing virginity: appear pale for less time than they remain virgins, may miss some virgins. Timing, female flies won’t mate several hours after they eclose (8 hours at 25oC, 16 hours at 18oC. hs-hid Y chromosome, heat shock pro-apoptotic gene on the Y.
Fly chromosome sex determination → Female (XX), Male wild-type (XY), Male infertile (XO), Female (XXY). Y, hs-hid virginizer stocks.
Drosophila have 4 pairs of chromosomes. Chromosome 1 is sex (XX, XY).
Wild-type: wt, +, OR not mentioned. Homologous chromosomes are written as a fraction when heterozygous.
Fourth chromosome usually ignored, not much impact, small.

RFLP
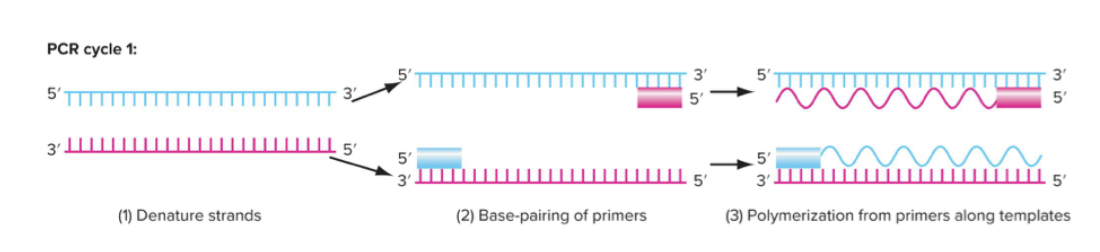
PCR - polymerase chain reaction, 1983 by Karry Millis. Study DNA sequence and genetic variation at a molecular level. Essential techniques in molecular biology and genetics, downstream techniques.
Method of making as many copies of a target region of DNA. Fast and extremely efficient - can amplify DNA from a single sperm or hair follicle.
Two oligonucleotide primers define region - complementary to one strand of DNA at one end, and the other complementary DNA at the other end.
Three steps to PCR:
Denature strands (raise temperature)
Base pairing of primers
Polymerization from primers along templates
35-40 cycles - amplifies exponentially
Fragment of interest isolated + enough DNA to visualize a fragment.
Results visualize with gel electrophoresis:
DNA migrates through Agrose gel. Migration distance depends on size (bigger, slower). May not amplify - smear.
DNA fragments are visualized with a fluorescent dye
Size of unknown fragments is determined by comparison to DNA markers with known size.
PCR products can be used to study DNA based on sequencing, size, and restriction enzyme.
Sequencing: Genotyping can identify carriers and homozygous individuals of sickle cell anemia.
Size: Determine number of trinucleotide repeats as a risk factor for Huntington’s disease. Careful primer design allows to detect chromosome rearrangements based on whether the PCR gives a product or no product. Cancer diagnostic. DNA fingerprinting - analyze 10-13 loci.
DNA Fingerprinting: Simple sequence repeat (SSR) loci that are highly polymorphic (many alleles with different number of repeats in the population). Individual carries only two alleles at a loci. 13 pairs of PCR primers are labeled with fluorescent dyes. 1 in 10 trillion chance (minus identical twins) of having same alleles at all 13 loci. product rule. CODIS database controlled by FBI. Crime scene.
Restriction enzymes: Fragments the genome at specific sites. Each restriction enzyme recognizes a specific sequence of bases anywhere within the genome. Cuts sugar-phosphate backbones of both strands, generated by digestion of DNA. Hundreds are available. Fragments are analyzed to see if a specific sequence was there or not
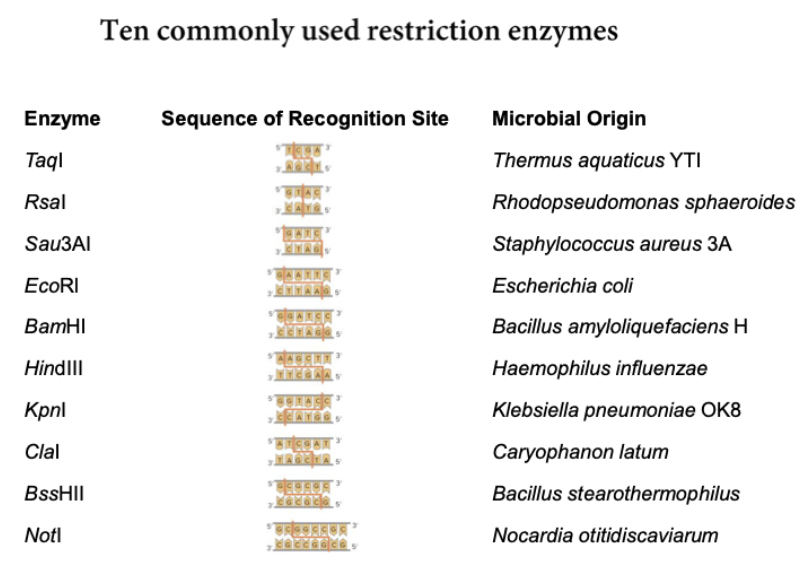
fragment length = 4n, n = number of bases in the recognition site.
Avg. restriction fragment size is 256 bp → 12 million fragments. 3 billion bp genome/256 = 12 million fragments.
Sticky or blunt ends can be cut. Sticky → they’re off, not cutting perfectly. Blunt → cut on same site of different strands of backbone
Uses of restriction enzymes: forensics, genotyping, paternity/maternity tests, study of animal and plant populations in ecology, cloning.